Abstract
Expression of the human telomerase RNA component gene, hTERC is essential for telomerase activity. The hTERC gene is expressed during embryogenesis and then downregulated during normal development, leaving most adult somatic cells devoid of hTERC expression. During oncogenesis, however, hTERC is re-expressed consequently contributing to the unrestricted proliferative capacity of many human cancers. Thus the identification of the molecular basis for the regulation of the telomerase RNA component gene in normal cells and its deregulation in cancer cells is of immediate interest. We have previously cloned the hTERC promoter and in this study have identified several transcription factors that modulate the expression of hTERC. We demonstrate that NF-Y binding to the CCAAT region of the hTERC promoter is essential for promoter activity. Sp1 and the retinoblastoma protein (pRb) are activators of the hTERC promoter and Sp3 is a potent repressor. These factors appear to act in a species-specific manner. Whereas Sp1 and Sp3 act on the human, bovine, and mouse TERC promoters, pRb activates only the human and bovine promoter, and NF-Y is only essential for the human TERC gene.
Keywords: telomerase, NF-Y, hTERC, retinoblastoma, hTR
Introduction
Telomeres are nucleoprotein complexes at the end of linear chromosomes and are considered essential for maintaining chromosomal integrity [1]. In the event of critical telomere-sequence loss during cell division, a cell-cycle arrest known as replicative senescence is induced [1,2]. Telomerase is a ribonucleoprotein reverse transcriptase that has the ability to synthesize telomeric sequence and thus counterbalance telomere attrition. Human telomerase activity in vitro can be reconstituted by the RNA subunit, hTERC, and the catalytic protein component encoded by the hTERT gene, although it is likely these core subunits will be augmented by additional factors to function optimally [1,2]. hTERC gene expression is regulated during normal human development, with the high levels of expression found during embryogenesis decreasing as tissue differentiation occurs. By the 10th postnatal week, the adult pattern of hTERC expression is evident with only primary spermatocytes retaining high levels of expression [3]. Although hTERC expression is absent from most adult somatic cells, low hTERC expression can be detected in some cells such as the regenerative regions of epithelium and activated lymphocytes [3–5]. However, during carcinogenesis the repression of hTERC in the cancer cell is lost or deregulated, thus contributing to the almost universal reactivation of telomerase in cancer cells [2,6]. Recent studies have indicated that transcriptional regulation may have a role to play in the control of hTERC gene expression [7,8]. Thus the identification of the molecular basis for hTERC gene regulation in normal cells and its deregulation in cancer cells is of immediate interest. It is also an emerging theme in transcriptional control that regulation results from the functional interactions between groups of transcription factors. Thus, the aim of the present study was to identify multiple transcription factors that regulate the hTERC gene promoter.
Materials and Methods
Site-Directed Mutagenesis and Constructs
The human promoter luciferase constructs, hProm876, hProm176, hProm120, and mouse TERC promoter construct mProm628 were made by inserting PCR products into pGL-3 Basic (Promega Southampton, U.K.). Details of the primers used can be obtained from the authors. Orientation and sequence were confirmed by sequencing. The (hProm176h10m1) construct was generated using the site-directed mutagenesis kit (QuikChange, Stratagene Cambridge, UK) to introduce a four-bp mutation at -58/-55. The primer used for construction of this site-replaced mutant is shown in Figure 1b (h10m1). It was subcloned into the pGL3 luciferase reporter vector and sequenced to confirm the mutation.
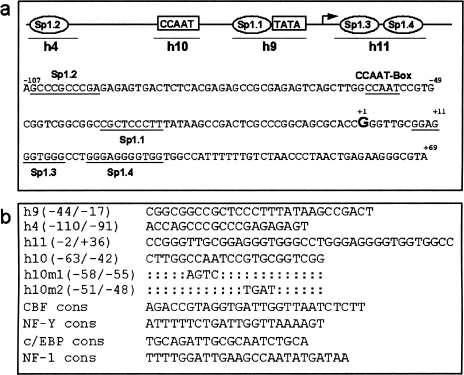
Figure 1.
(a) hTERC promoter sequence and potential transcription factor recognition sites. A total of 176 bp of the human promoter sequence is shown and potential regulatory motifs, identified by computer analysis, are underlined on the sequence. The transcriptional start site is marked as “+1.” Four potential Sp1 sites are shown and termed Sp1.1 to Sp1.4. Sp1.4 contains two overlapping Sp1 consensus sites. The names of the oligonucleotides used in electrophoresis mobility shift assays (EMSA) are indicated under their respective sites. (b) Oligonucleotides used in EMSA and mutagenesis studies of the hTERC promoter. The sequence of the wild-type oligonucleotides covering potential transcription factor recognition sites are shown. Oligonucleotide h9 covers Sp1.1 and the adjacent TATA-box, h10 covers the CCAAT box, h4 covers Sp1.2 and h11 covers the closely opposed Sp1.3 and Sp1.4 sites. Mutations, h10m1 and h10m2, were introduced into the wild-type oligonucleotide h10 and used in EMSA and in the construction of the mutant reporter construct (hProm176h10m1).
Preparation of Nuclear Protein Extracts
HeLa cells were lysed in TMS (5 mM Tris-HCl pH 7.5, 2.5 mM MgCl2, 125 mM sucrose plus 0.25% Triton X-100), and nuclei were harvested by centrifugation. Nuclei were resuspended in 5 ml of TMS and 0.1 volumes of 4 m NaCl. After centrifugation, ammonium sulfate was added and the precipitate pelleted and redissolved in E50 buffer (50 mM ammonium sulphate, 20 mM KOH-Hepes, pH 7.9, 5 mM MgCl2, 0.1 mM EDTA, 0.1% (v/v) Brij 35, 20% (v/v) glycerol, 1 mM DTT).
Electrophoresis Mobility Shift Assay
Electrophoresis mobility shift assays (EMSA) were performed by using the EMSA kit (Promega). HeLa nuclear proteins were incubated in 15 µl of reaction containing 4% glycerol, 1 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.5 mM EDTA, 50 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 2.0 µg poly(dl-dC) with or without 100-fold molar excess of unlabeled DNA competitors on ice for 10 min, followed by addition of the radiolabeled probe. For supershift assays, antibodies against Sp1, Sp3, Ap-2, Ets2, c/EBPα, NF-1, NF-YC (Santa Cruz Biotechnology Santa Cruz, CA), NF-YA, and NF-YB, were added to the reaction mixture 25 minutes before the addition of the probe. All DNA-protein complexes were resolved by electrophoresis on 5% native polyacrylamide. Thee/EBP, CBF, and NF-1 sequences used in EMSA are from Santa Cruz Biotechnology: sc-2525, sc-2591, and sc-2553. The NF-Y sequence is from Geneka Biotechnology Inc. (Montreal, Canada).
Transfection Assays
Transfections were carried out using Superfect transfection reagent (Qiagen West Sussex, UK) as previously described, [8]. To standardize the amount of hTERC plasmid DNA in transfection assays, purified plasmid DNA was first quantified by UV spectrophotometry. Then 1.0 µg of plasmid DNA was digested with XhoI/HindIII and fragments resolved on a 6% PAGE gel to detect the 176-bp promoter fragment. The amount of each DNA sample was analyzed by PhosphorImager with ImageQuant software analysis (Molecular Dynamics Bio-Rad, Hemel Hempstead, UK). For cotransfections, 3 µg of hProm176 or 1 µg of hProm876 reporter plasmids was cotransfected with 0.01 to 3 µg of pNF-YAm29, pNF-Y13, pCMV-Sp1, pCMV-Sp3, pSV40-Rb expression vectors. The hProm176 construct was used throughout this study but the hProm867 construct was also used to extend the analysis of the minimal human promoter construct and test whether the maximal promoter responded to modifier genes in a similar way to the minimal promoter. Both human constructs responded in cotransfection experiments in identical ways.
The Great EscAPe SEAP System (Clontech Basingstoke, UK) was used to control for transfection efficiency in experiments with the mutant promoter constructs, the pSV40-Rb vector, the pNF-Y13 and pNF-YAm29 expression vectors. In keeping with others we found that the Sp1 and Sp3 expression vectors modulated the activity of the SV40 promoter used in the pSEAP2-Control vector [9,10]. For these experiments semiquantitative PCR of the transfected plasmid was carried out using luciferase-specific primers and products adjusted relative to input genomic DNA.
The means and standard deviations of duplicate samples from representative transfections are shown and all experiments were carried out at least three times in duplicate. The experiments shown in Figure 4b used retinoblastoma protein (pRb) variants developed by Sellers et al. [16].
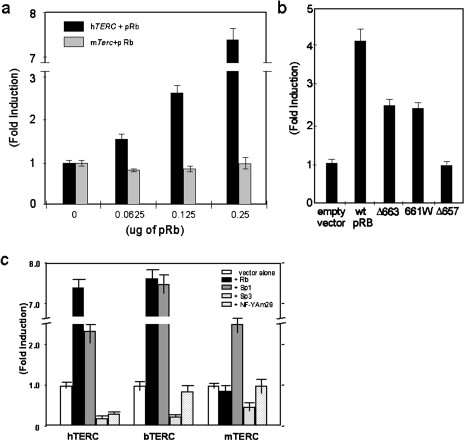
Figure 4.
(a) Activation of hTERC promoter activity by pRb. The pRb/p53 negative cell line 5637 was transfected with a fixed amount of the hTERC-luciferase plasmid and increasing concentrations of pRb expression vector. In addition, the mouse telomerase (mTerc) promoter-luciferase construct mProm628 was also analyzed for its response to pRb [8]. The data is expressed as fold induction of luciferase activity relative to the promoter alone. The pSEAP2-Control vector was used as an internal control for transfection efficiency. (b) Differential activation of hTERC promoter activity by wild-type pRb and three variants, Δ663, 661W, and Δ657. The pRb/p53 negative cell line 5637 was transfected with 3 fig of the hTERC-luciferase plasmid (phProm176) and 0.5 µg of each pRb expression vector. The data is expressed as fold induction of luciferase activity relative to the expression vector alone (empty vector). The pSEAP2-Control vector was used as an internal control for transfection efficiency. (c) Functional divergence in the transcriptional regulation of TERC genes between species. Promoter-luciferase constructs for the human (hProm867), mouse (mProm628) and bovine TERC genes were compared for their regulation by Sp1, Sp3, NF-YAm29, and pRb. The activity of the promoters in response to cotransfection with the transcriptional regulators is shown as fold induction relative to the promoters alone. The pRb/p53 negative cell line 5637 was used in all experiments and the mean luciferase activity normalized for protein and transfection efficiency is shown with the standard deviation of duplicate samples.
Cloning of bTERC
A PCR product encompassing the 320 bp of the bTERC promoter was amplified using the primers bTRf1 GCGCTCGAGCCGTACCTGGCTTTTAAGAG and bTRr1 CGAAGCTTAGATGAGAAATGGCTGCCAC and bovine genomic DNA as a template (Clontech). Primers were designed from the bTERC gene sequence [11]. This was then cloned into luciferase reporter construct pGL3 Basic (Promega). Orientation and sequence of the insert was checked by sequencing. The bTERC promoter sequence has been submitted to GenBank, accession number AF176663.
Results
We have previously identified the proximal region of the hTERC promoter necessary to direct hTERC transcription [8]. DNase I footprinting identified a 176-bp region within this which contained four protected regions (data not shown). Computer analysis revealed that these corresponded to four putative Sp1 binding sites and a CCAAT box (Figure 1: h4, h9, h11, h10). In the present study a combination of gel shift assays, mutation analysis, and transfection assays have been used to identify key regulators of hTERC gene expression.
The NF-Y Transcriptional Complex Interacts with the hTERC CCAAT Box and This Interaction Is Central to hTERC Transcriptional Efficiency
EMSA was used to identify whether cellular proteins interact with the CCAAT box that is found upstream of the transcriptional start site. Protein extract from the HeLa cell line was used with a radiolabeled oligonucleotide corresponding to the wild-type hTERC CCAAT box sequence (h10). A protein-DNA complex was identified (Figure 2a, lane 1) and specificity of binding confirmed by competition studies. Competition by the wild-type oligonucleotide itself and by an oligonucleotide with a mutation outside the CCAAT box (h10m2) abolished the complex (Figure 2a, lanes 2 and 4). In contrast the oligonucleotide with a mutation within the CCAAT box (h10m1) was unable to compete for protein binding (Figure 2a, lane 3).
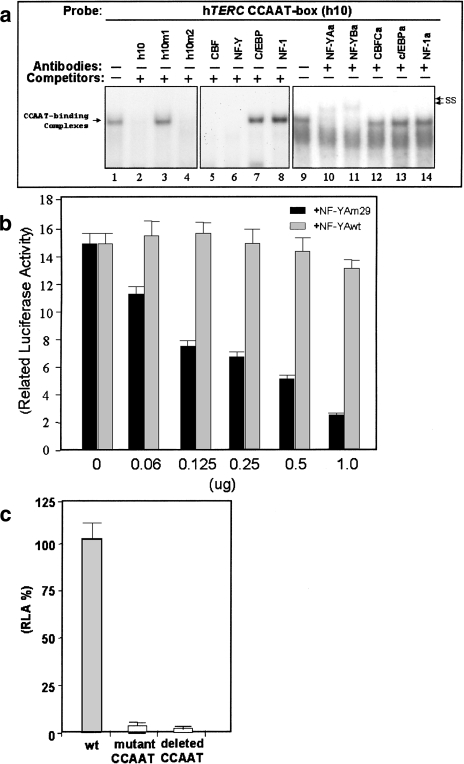
Figure 2.
(a) Identification of nuclear proteins binding to the CCAAT box. HeLa nuclear extract was mixed with radiolabeled oligonucleotide probe and analyzed by EMSA. Specific DNA-protein complexes are indicated by the arrow on the left. Complexes supershifted by preincubation with antibodies are shown on the right by arrows marked “ss.” The oligonucleotide h10 was used as a probe and the oligonucleotides used as competitors are indicated at the top of lanes 2 to 8. The antibodies used in supershifts are indicated above lanes 9 to 14. (b) Inhibition of hTERC promoter activity by a dominant-negative mutant of NF-YA termed NF-YAm29. 5637 cells were transfected with fixed amount of the hTERC-luciferase plasmid hProm176 and increasing concentrations of the NF-YAm29 dominant-negative vector (black columns) or wild-type NF-YA vector (light grey columns). For each transfection the mean and standard deviation for duplicate samples is shown. (c) Functional analysis of the CCAAT box. Promoter activities of a mutant CCAAT box construct (hProm176h10m1) and shorter promoter construct with deletion of the CCAAT box (hProm120) were assayed by transfection into the bladder carcinoma cell line 5637 and compared to the wild-type promoter activity (hProm176). Promoter activities of the constructs are shown as percentage luciferase activity of the wild-type promoter alone. The pSEAP2-Control vector was used as an internal control for transfection efficiency.
To ascertain the likely components of the hTERC CCAAT-binding complex, three candidate CCAAT-binding complexes were investigated; NF-Y (also known as CBF and CP1), c/EBP, and NF-1. Two consensus oligonucleotides for NF-Y (termed NF-Y and CBF) were able to compete for hTERC CCAAT binding (Figure 2a, lanes 5 and 6), whereas consensus oligonucleotides for c/EBP and NF-1 were not (lanes 7 and 8). Antibody supershift analysis revealed that the hTERC CCAAT-binding complex contained two subunits of the NF-Y protein complex, NF-YA and NF-YB. Nuclear extracts were preincubated with antibodies specific for the three components of the NF-Y complex, NF-YA, NF-YB and NF-YC, and with antibodies specific to other CCAAT-binding complexes c/EBP and NF-1. Disruption of the CCAAT complex and supershift was only seen with antibodies to the NF-Y complex components NF-YA and NF-YB (lanes 10 and 11). Antibodies to the NF-YC subunit of NF-Y did not disrupt the complex (lane 12). This has been observed by others and is possibly due to inaccessibility of the C subunit to the antibody on complex formation [12]. Antibodies to NF-1 and c/EBP had no effect on complex formation (lanes 13 and 14).
To evaluate the functional significance of NF-Y complex formation on the hTERC promoter, we used a dominant negative mutant of NF-YA (NF-YAm29) in transfection experiments [13]. As shown in Figure 2b, cotransfection of NF-YAm29 specifically represses hTERC promoter activity five- to seven-fold in a dose-dependent manner, whereas cotransfection of the wild-type NF-YA subunit had no effect on promoter activity. These results confirm that NF-Y is the major factor that transactivates the hTERC promoter through the CCAAT box. The functional significance of the CCAAT box and its interaction with NF-Y was also confirmed by introducing a mutation into the basal promoter region. This was cloned into a luciferase reporter construct (hProm176-h10m1). Similarly a promoter construct with 56-bp deletion of the 5′ end, including the CCAAT box region, was generated (hProm120). Promoter activities of the mutant and deleted CCAAT box constructs were assayed by transfection into the bladder carcinoma cell line 5637 and compared to the wild-type promoter activity. Mutation or deletion of the CCAAT box sequence element completely abolishes hTERC promoter activity (Figure 2c), suggesting that this element is the site of action for an activator of hTERC promoter activity and central to hTERC transcriptional efficiency.
The Sp1 and Sp3 Transcription Factors Interact with Multiple Sites within the hTERC Promoter and Can Modulate Promoter Activity
To identify cellular proteins interacting with the four putative Sp1 sites in the hTERC promoter, EMSA were performed with three oligonucleotides (Figure 1b: h9, h4, and h11) spanning the four sites (Figure 1a and b). Protein complexes characteristic of binding of Sp1 and Sp3 [14] were seen with all three oligonucleotides (Figure 3a, lanes 1, 5, and 10). To confirm the identity of the proteins binding the Sp1 sites, nuclear extracts were preincubated with anti-bodies to the transcriptional regulators Sp1 and Sp3 and with control antibodies specific for Ets2 and Ap2. Specific disruption of complex formation was only seen when preincubation occurred with antibodies against Sp1 and Sp3 (Figure 3a, lanes 2, 3, 6, 7, 11, 12, and 13). This suggests that the Sp family of transcription factors is involved in the regulation of hTERC gene expression. To test this hypothesis, expression vectors for Sp1 and Sp3 were cotransfected into 5637 cells with the hTERC-luciferase construct. Figure 3b demonstrates dose-dependent activation of the promoter by Sp1 and dose-dependent repression by Sp3.
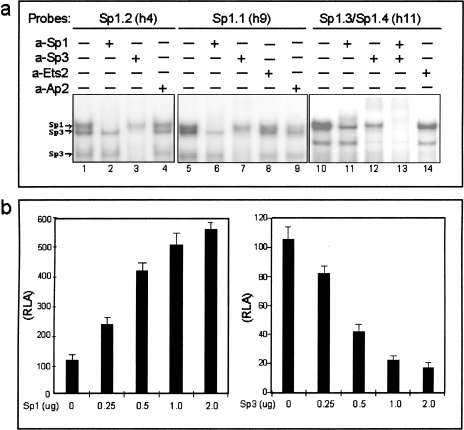
Figure 3.
(a) Identification of nuclear proteins binding to the Sp1 sites. HeLa nuclear extract was mixed with radiolabeled oligonucleotide probe and analyzed by EMSA. The proteins bound to the probe were identified by preincubation of nuclear extracts with antibodies specific for Sp1, Sp3, or control antibodies specific for Ets2 and Ap2. Complexes specific to Sp1 and Sp3 are indicated to the left of the panel. The antibodies used are indicated at the top of the figure. (b) The Sp1 transcription factor upregulates and Sp3 transcription factor downregulates the hTERC promoter. The wild-type hTERC-luciferase plasmid was cotransfected with increasing amounts of either an Sp1 or Sp3 expression vector (0.25 µg to 2.0 µg) into the 5637 cell line. The mean luciferase activity normalized for protein and transfection efficiency is shown with the standard deviation of duplicate samples.
The Retinoblastoma Gene Product Activates the hTERC Promoter
The pRb is considered to have important roles in normal transcriptional regulation and in the progression of a normal mortal cell to an immortal cancer cell [2,15]. The ability of pRb to modulate the hTERC promoter was therefore assessed. The pRb/p53 null bladder cell line, 5637, was cotransfected with a fixed amount of the hTERC-luciferase plasmid and increasing amounts of a pRb expression vector. Figure 4a shows that expression of the pRb is a dose-dependent activator of the hTERC promoter. To investigate the transcriptional activation of hTERC by pRb in more detail, a series of pRb variants developed by Sellers et al. [16] was used. Variants Δ663 and 661W are unable to bind E2F and therefore do not cause cell-cycle arrest, yet they retain the ability to activate gene transcription. Variant Δ657 is a null mutant of pRb, it is unable to bind E2F and cannot activate gene transcription [16]. As can be seen from Figure 4b, transcriptional activation of the hTERC promoter by pRb is not dependent on E2F binding as it is observed with variants that retain transcriptional activation capabilities but which are unable to bind E2F (Δ663 and 661W). The null mutant of pRb (Δ657), which is unable to bind E2F or to activate gene transcription in the assays used by Sellers et al., is similarly unable to activate the hTERC promoter. The specificity of pRb to activate the hTERC promoter was tested by comparing the human and mouse telomerase RNA gene promoters, which, at the sequence level, show no significant homology [8]. As can be seen from Figure 4a, the mouse promoter does not respond to the pRb thus demonstrating a functional difference in the regulation of the mouse and human telomerase RNA genes. In addition, the specificity of hTERC activation by pRb was confirmed by cotransfection of the hTERC promoter construct containing the mutation in the CCAAT box (hProm120) with pRb. As shown in Figure 2C, mutation of the CCAAT box abolishes promoter activity. The expression of pRb is unable to activate hTERC promoter activity in the presence of the mutated CCAAT box, thus confirming the functional specificity of the pRb activation on the wild-type promoter (data not shown).
Functional Divergence in the Transcriptional Regulation of TERC Genes between Species
Despite conservation in telomerase enzyme function across species, there may be fundamental differences in telomerase regulation between mouse and human [8,17]. To gain a clearer insight into the regulation of telomerase RNA genes, the response of the human, mouse, and bovine telomerase RNA gene promoters to the transcriptional regulators pRb, NF-Y, Sp1, and Sp3 were compared in transient transfection experiments. The bovine telomerase RNA gene promoter was included in this study as it is more closely related to the human gene in terms of sequence and may therefore be of value in dissecting promoter function [11]. The human wild-type promoter construct shown in Figure 4 is the full-length 867-bp plasmid, hProm867 [8] to allow comparison of the full-length promoter of each species. The mouse telomerase (mTerc) promoter luciferase construct used was mProm628 [8], and a luciferase reporter construct containing bovine telomerase RNA gene promoter sequences (bTERC) was constructed as detailed in the Materials and Methods section.
Significant functional divergence was observed between species; the human and bovine promoters are activated by pRb whereas the mouse promoter is not (Figure 4c). Sp1 activates all three promoters and Sp3 represses all three promoters. Interestingly, NF-Y regulation appears to be specific to the human promoter.
Discussion
Reactivation of telomerase is an almost universal event in carcinogenesis. Though other proteins are involved in optimal function of the enzyme, the RNA subunit and the catalytic protein components are sufficient to reconstitute enzyme activity. Consistent with this, both components are expressed at high levels in the majority of cancers. In this study, several factors capable of modulating hTERC promoter activity in cell culture have been identified thus providing an insight into the possible molecular mechanisms for hTERC gene regulation in normal cells and its deregulation in cancer cells. The mechanisms regulating normal expression and repression may, however, differ from those involved in loss of repression and activation of expression in oncogenesis.
The present study shows that the CCAAT box is essential for basal promoter function. We have demonstrated that this can be recognized by the NF-Y complex and inhibition of NF-Y activity abrogates promoter activity. NF-Y is a heteromeric protein composed of three subunits, NF-YA, NF-YB and NF-YC [18]. NF-Y activity is found to change during senescence, and in response to inducing agents [18,19]. It binds and controls a number of cell-cycle regulated promoters and may be directly involved in oncogenesis [18,20,21]. NF-Y may exert its effect through alterations in promoter architecture. Chromatin structure can restrict the access of transcription-associated proteins to promoters and histone acetylation is a major mechanism for antagonism of chromatin-mediated repression. NF-Y is able to associate with nucleosomes, and to inhibit nucleosome formation in in vitro models [22]. It can also interact with the histone acetyltransferases, p300, GCN5, and P/CAF, [12,23,24] and may provide a means of targeting these to specific promoters. NF-Y has also been shown to increase the affinity of structurally unrelated activators, including Sp1, to their binding sites [25–27]. It would be of interest to examine whether the regulation of telomerase genes in development and in mortal and immortal cells is related to chromatin structure and if so, whether this is mediated by NF-Y.
Sp1 and Sp3 represent just two members of a growing family of mammalian Kruppel-like transcription factors that bind GC-rich sequence elements, and are involved in the regulation of tissue specific and housekeeping genes. Sp proteins have been implicated in cell-cycle regulation, chromatin modeling, and maintenance of methylation-free islands [14]. In addition, Sp1 knockouts show that Sp1 is required for early mouse development [14]. Transcriptional regulation by the Sp family of proteins is complex, but in general Sp1 activates gene expression, whereas Sp3 represses it [14,28]. We have identified Sp protein-binding sites within the hTERC promoter. We have demonstrated that Sp1 and Sp3 can bind to these sites and that Sp1 activates and Sp3 represses gene expression. Thus fluctuations in Sp1/Sp3 ratios or alteration in their relative binding affinities could modulate expression in vivo. This is of particular significance because Sp1 has recently been demonstrated to activate the human telomerase protein component promoter (hTERT) [29] and we have therefore identified an overlapping, though not necessarily co-coordinated, control mechanism.
Our finding that pRb is an activator of the hTERC promoter was somewhat surprising given its well-known function as a tumor suppresser. Protein Rb negatively regulates transcription of a number of genes that play a role in cell-cycle progression by forming complexes with members of the E2F transcription family. However, pRb also has a vital role in normal development, a time when hTERC is highly expressed, and the ability of pRb to coactivate a number of genes including TGF-beta2, the retinoblastoma gene itself, cyclin D1, and the Werner helicase gene is well described [15,16,30–34]. How pRb activates transcription is not fully understood but in keeping with the results of Sellers et al., the ability of pRb to activate the hTERC promoter is independent of its ability to bind E2F. Protein Rb has not been shown to bind directly to the promoters that it activates [35] though certain sequence elements have been identified as being involved in pRb modulation in other promoters [30,36]. pRb may act by sequestering an inhibitor of other transcription factors [37]. The identification of hTERC as another gene that is activated by pRb may contribute to the investigation of this aspect of pRb function.
The functional significance of the ability of pRb to activate the hTERC promoter in oncogenesis is not clear given that dysfunction of some part of the pRb signal transduction pathway is very common in cancer. However, the complete loss of pRb itself is relatively rare and even certain germ line mutations in RB-1 can give rise to a protein defective in E2F-binding yet retaining the ability to activate transcription [16]. Furthermore, we have demonstrated that pRb is a modulator of promoter activity but it is not essential for transcription. One might predict that when pRb-transactivating function is lost from the cancer cell, compensation for the reduced hTERC promoter activity might occur. Indeed, the hTERC gene can be amplified in human cancers [38]. It could be that this is selected for to increase hTERC expression in the absence of the positive regulatory influences (such as pRb) or to titrate out negative regulatory factors (such as Sp3).
Telomerase enzyme function is conserved across species but there are significant differences in gene expression between species with high levels of expression in the mouse compared to the human [39,40]. We have demonstrated a marked divergence in the regulation of the human and the mouse TERC gene. This is perhaps not that surprising because the two promoters show no significant sequence homology [8] and, in particular, a section of the human and bovine promoters lying between the transcription start site and the template region is absent in the mouse promoter. Phylogenic analysis of the TERC sequences of several species described by Chen et al. [41] reveals that this area is also absent in other rodents. Whether this section of the promoter is responsible for the species variation remains to be investigated. The divergence in the regulation of telomerase RNA genes between species needs to be taken into account when studying telomerase in animal models of development and disease states [17].
Conclusion
Our results indicate that multiple signals including Sp1, Sp3, pRb, and NF-Y are able to contribute to the regulation of hTERC gene expression. Thus, alterations in the relative contributions of Sp1, Sp3, pRb, and NF-Y in a cell, or signal-transduction-specific manner, may be relevant to hTERC gene expression. This information provides a focus for studies designed to uncover how cancer cells reactivate or upregulate hTERC gene expression. In addition, an understanding of the molecular basis for hTERC gene regulation may be of value in the manipulation of telomerase expression, for example in age-related disease and for developing rational therapies to treat cancer through the exploitation of hTERC gene transcription [6,8].
Acknowledgements
We thank Dr. William Kaelin, Jr. and Dr. Robert White for the pRb expression vectors, Dr. Roberto Mantovani for NF-Y expression vectors and antibodies, Dr. Guntram Suske for the Sp1 and Sp3 expression vectors, and Dr. L. Bryce for critical reading of the manuscript.
Abbreviations
- EMSA
electrophoretic mobility shift assay
Footnotes
This work was supported by the Cancer Research Campaign (UK) and Glasgow University.
References
- 1.O'Reilly M, Teichmann SA, Rhodes D. Telomerases. Curr Opin Struct Biol. 1999;9:56–65. doi: 10.1016/s0959-440x(99)80008-6. [DOI] [PubMed] [Google Scholar]
- 2.Holt SE, Shay JW. Role of telomerase in cellular proliferation and cancer. J Cell Physiol. 1999;180:10–18. doi: 10.1002/(SICI)1097-4652(199907)180:1<10::AID-JCP2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 3.Yashima K, Maitra A, Rogers BB, Timmons CF, Rathi A, Pinar H, Wright WE, Shay JW, Gazdar AF. Expression of the RNA component of telomerase during human development and differentiation. Cell Growth Differ. 1998;9:805–813. [PubMed] [Google Scholar]
- 4.Weng N, Levine BL, June CH, Hodes RJ. Regulation of telomerase RNA template expression in human T lymphocyte development and activation. J Immunol. 1997;158:3215–3220. [PubMed] [Google Scholar]
- 5.Ogoshi M, Le T, Shay JW, Taylor RS. In situ hybridization analysis of the expression of human telomerase RNA in normal and pathologic conditions of the skin. J Invest Dermatol. 1998;110:818–823. doi: 10.1046/j.1523-1747.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- 6.Sarvesvaran J, Going JJ, Milroy R, Kaye SB, Keith WN. Is small cell lung cancer the perfect target for anti-telomerase treatment? Carcinogenesis. 1999;20:1649–1652. doi: 10.1093/carcin/20.8.1649. [DOI] [PubMed] [Google Scholar]
- 7.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol. 1999;19:3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao JQ, Hoare SF, McFarlane R, Muir S, Parkinson EK, Black DM, Keith WN. Cloning and characterization of human and mouse telomerase RNA gene promoter sequences. Oncogene. 1998;16:1345–1350. doi: 10.1038/sj.onc.1201892. [DOI] [PubMed] [Google Scholar]
- 9.Chen SJ, Artlett CM, Jimenez SA, Varga J. Modulation of human alpha 1(I) procollagen gene activity by inter-action with Sp1 and Sp3 transcription factors in vitro. Gene. 1998;215:101–110. doi: 10.1016/s0378-1119(98)00268-6. [DOI] [PubMed] [Google Scholar]
- 10.Conn KJ, Rich CB, Jensen DE, Fontanilla MR, Bashir MM, Rosenbloom J, Foster JA. Insulin-like growth factor-I regulates transcription of the elastin gene through a putative retinoblastoma control element — a role for Sp3 acting as a repressor of elastin gene transcription. J Biol Chem. 1996;271:28853–28860. doi: 10.1074/jbc.271.46.28853. [DOI] [PubMed] [Google Scholar]
- 11.Tsao DA, Wu CW, Lin YS. Molecular cloning of bovine telomerase RNA. Gene. 1998;221:51–58. doi: 10.1016/s0378-1119(98)00432-6. [DOI] [PubMed] [Google Scholar]
- 12.Faniello MC, Bevilacqua MA, Condorelli G, de Crombrugghe B, Maity SN, Awedimento VE, Cimino F, Costanzo F. The B subunit of the CAAT-binding factor NFY binds the central segment of the Co-activator p300. J Biol Chem. 1999;274:7623–7626. doi: 10.1074/jbc.274.12.7623. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 14.Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 15.Sellers WR, Kaelin WG. RB [corrected] as a modulator of transcription [published erratum appears in Biochim Biophys Acta 1996 Dec 9;1288(3):E-1] Biochim Biophys Acta. 1996;1288:M1–M5. doi: 10.1016/0304-419x(96)00014-5. [DOI] [PubMed] [Google Scholar]
- 16.Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WGJ. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA [see comments] Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 18.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 19.Good LF, Chen KY. Cell cycle- and age-dependent transcriptional regulation of human thymidine kinase gene: the role of NF-Y in the CBP/tk binding complex. Biol Signals. 1996;5:163–169. doi: 10.1159/000109214. [DOI] [PubMed] [Google Scholar]
- 20.Gu Z, Kuntz-Simon G, Rommelaere J, Cornells J. Oncogenic transformation-dependent expression of a transcription factor NF-Y subunit. Mol Carcinog. 1999;24:294–299. doi: 10.1002/(sici)1098-2744(199904)24:4<294::aid-mc7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 21.Pang JH, Good LF, Chen KY. The age-dependent binding of CBP/tk, a CCAAT binding protein, is deregulated in transformed and immortalized mammalian cells but absent in premature aging cells. Exp Gerontol. 1996;31:97–109. doi: 10.1016/0531-5565(95)02019-5. [DOI] [PubMed] [Google Scholar]
- 22.Motta MC, Caretti G, Badaracco GF, Mantovani R. Interactions of the CCAAT-binding trimer NF-Y with nucleosomes. J Biol Chem. 1999;274:1326–1333. doi: 10.1074/jbc.274.3.1326. [DOI] [PubMed] [Google Scholar]
- 23.Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 25.Reith W, Siegrist CA, Durand B, Barras E, Mach B. Function of major histocompatibility complex class-ii promoters requires cooperative binding between factors Rfx and Nf-Y. Proc Natl Acad Sci USA. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright KL, Moore TL, Vilen BJ, Brown AM, Ting JPY. Major histocompatibility complex class ii-associated invariant chain gene-expression is up-regulated by cooperative interactions of Sp1 and Nf-Y. J Biol Chem. 1995;270:20978–20986. doi: 10.1074/jbc.270.36.20978. [DOI] [PubMed] [Google Scholar]
- 27.Dooley KA, Millinder S, Osborne TF. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor nuclear factor Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 28.De Luca P, Majello B, Lania L. Sp3 represses transcription when tethered to promoter DNA or targeted to promoter proximal RNA. J Biol Chem. 1996;271:8533–8536. doi: 10.1074/jbc.271.15.8533. [DOI] [PubMed] [Google Scholar]
- 29.Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, Ariga H, Inoue M. Spl cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SJ, Wagner S, Liu F, O'Reilly MA, Robbins PD, Green MR. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature. 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 31.Müller H, Lukas J, Schneider A, Warthoe P, Bartek J, Eilers M, Strauss M. Cyclin D1 expression is regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1994;91:2945–2949. doi: 10.1073/pnas.91.8.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamabe Y, Shimamoto A, Goto M, Yokota J, Sugawara M, Furuichi Y. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol Cell Biol. 1998;18:6191–6200. doi: 10.1128/mcb.18.11.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 34.Park K, Choe J, Osifchin NE, Templeton DJ, Robbins PD, Kim SJ. The human retinoblastoma susceptibility gene promoter is positively autoregulated by its own product. J Biol Chem. 1994;269:6083–6088. [PubMed] [Google Scholar]
- 35.Kaelin WG. Functions of the retinoblastoma protein. Bioessays. 1999;21:50–958. doi: 10.1002/(SICI)1521-1878(199911)21:11<950::AID-BIES7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 36.Udvadia AJ, Rogers KT, Horowitz JM. A common set of nuclear factors bind to promoter elements regulated by the retinoblastoma protein. Cell Growth Differ. 1992;3:597–608. [PubMed] [Google Scholar]
- 37.Chen L, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y, Grunwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soder AI, Hoare SF, Muir S, Going JJ, Parkinson EK, Keith WN. Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene. 1997;14:1013–1021. doi: 10.1038/sj.onc.1201066. [DOI] [PubMed] [Google Scholar]
- 39.Blasco MA, Funk W, Villeponteau B, Greider CW. Functional-characterization and developmental regulation of mouse telomerase Rna. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- 40.Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]