Abstract
Chromosome instability plays an important role in cancer by promoting the alterations in the genome required for tumor cell progression. The loss of telomeres that protect the ends of chromosomes and prevent chromosome fusion has been proposed as one mechanism for chromosome instability in cancer cells, however, there is little direct evidence to support this hypothesis. To investigate the relationship between spontaneous telomere loss and chromosome instability in human cancer cells, clones of the EJ-30 tumor cell line were isolated in which a herpes simplex virus thymidine kinase (HSV-tk) gene was integrated immediately adjacent to a telomere. Selection for HSV-tk-deficient cells with ganciclovir demonstrated a high rate of loss of the end these “marked” chromosomes (10-4 events/cell per generation). DNA sequence and cytogenetic analysis suggests that the loss of function of the HSV-tk gene most often involves telomere loss, sister chromatid fusion, and prolonged periods of chromosome instability. In some HSV-tk-deficient cells, telomeric repeat sequences were added on to the end of the truncated HSV-tk gene at a new location, whereas in others, no telomere was detected on the end of the marked chromosome. These results suggest that spontaneous telomere loss is a mechanism for chromosome instability in human cancer cells.
Keywords: chromosome fusion, breakage/fusion/bridge cycle, chromosome healing, telomere, chromosome instability
Introduction
Telomeres are DNA-protein complexes containing short DNA repeat sequences that are added on to the ends of chromosomes by the enzyme telomerase [1]. Telomeres serve multiple functions, including preventing chromosome fusion [2–4] and facilitating chromosome segregation [5,6]. In mammals, telomeres are maintained in the germ line but shorten with age in most somatic cells that do not express sufficient telomerase activity [3]. Telomere shortening has been proposed to be a signal for cell senescence [7], consistent with the extension in the life-span of primary human fibroblasts transfected with the gene for the catalytic subunit of telomerase [8]. Primary human cells that bypass senescence continue to show telomere shortening, and eventually enter “crisis,” which consists of increased chromosome fusion, aneuploidy, and death [2]. Thus, it has been proposed that for cells to become immortal they must not only avoid senescence but also regain the ability to maintain telomeres to avoid crisis [2,9]. In support of this hypothesis, it has been observed that the telomeres of immortal cell lines do not continue to shorten [2,10], and that cancer cells and immortal cell lines invariably maintain their telomeres, either by telomerase [2,9] or through an alternative mechanism [11–13].
Although telomere maintenance in cancer cells must be efficient enough to allow for continuous cell growth, in some instances telomeres may be lost completely or shortened to such an extent that they no longer protect the ends of chromosomes. Evidence that human cancer cells do not always properly maintain telomeres comes from the observation that many cancer cell lines have a high rate of “telomere associations” [3]. Telomere associations are chromosomes joined at or near their telomeres and have been proposed to be caused by telomere loss [4]. The complete loss of a telomere could occur through stochastic events due to high rates of double-strand breaks in the telomeric repeat sequences or subtelomeric DNA, or could occur gradually, due to altered expression or mutations in genes required for proper telomere maintenance. Consistent with the latter possibility, a large number of genes that have been found to be important in maintaining telomere length can also influence chromosome stability [14–18]. Telomere loss could play a role in the chromosome instability associated with tumor cell progression [19,20], because chromosomes involved in telomere associations have been found to have a high rate of rearrangement [21–24]. In fact, telomere loss was proposed more than 50 years ago to lead to chromosome instability through the initiation of breakage/fusion/bridge cycles [25]. Breakage/fusion/bridge cycles have been demonstrated to be involved in gene amplification in hamster cells [26–28], and have been demonstrated to be an important mechanism for chromosome instability in human cancer [29].
The extent of chromosome instability in cancer cells could also be influenced by their ability to perform chromosome healing, which involves the addition of telomeres to the ends of broken chromosomes. Chromosome healing has been proposed to play an important role in the cellular response to telomere loss through its ability to terminate breakage/fusion/bridge cycles [25]. Chromosome healing has been demonstrated to occur at sites with little or no complementarity to telomeric repeat sequences in a variety of different organisms [30,31], including yeast [30,31] and mouse embryonic stem (ES) cells [32]. Chromosome healing in yeast occurs primarily at sites distal to regions with more extensive homology to telomeric repeat sequences [33,34]; although chromosome healing can occur at virtually any location in strains with mutations in the Pif1p helicase [35]. Evidence for chromosome healing in humans comes from the analysis of terminal deletions associated with genetic disease [36,37]. In these studies, new telomeres were found to be added directly on to the ends of broken chromosomes at sites that usually contained 3 to 4 bp of complementarity to telomeric repeat sequences, although one site had no complementarity. Consistent with these studies, in vitro studies with extracts made from human cells demonstrated minimal sequence requirements for the de novo addition of telomeric repeat sequences to oligonucleotides with ends similar to one of the sites of chromosome healing on a human chromosome [38].
To investigate the rate and consequences of spontaneous telomere loss in human cancer cells, and to test the hypothesis that spontaneous telomere loss is a mechanism for chromosome instability in human cancer cells, we have established clones of a human tumor cell line that has a plasmid containing a selectable marker gene integrated immediately adjacent to a telomere. Subclones of these cell lines that had spontaneously lost the selectable marker gene were characterized by Southern blot analysis, nucleotide sequence analysis of the rescued plasmid sequences, and cytogenetic analysis using chromosome, plasmid, and telomere-specific probes to determine the nature of the events involved.
Materials and Methods
Cell Culture
The EJ-30 cell line (obtained from Dr. William Dewey, UCSF) was subcloned from the EJ bladder cell carcinoma cell line, which is also named MGH-U1 [39]. EJ-30 was grown in alpha-MEM (UCSF cell culture facility) supplemented with 5% fetal calf serum (Gibco), 5% newborn calf serum with iron (Gibco), 1 mM l-glutamine (Gibco), and gentimycin. Cells were propagated at 37°C in humidified incubators. The pNCT-tel plasmid was introduced into the cells by calcium phosphate-mediated transfection as previously described [40]. Clones containing the integrated pNCT-tel plasmid were selected with 400 µg/ml G418.
Plasmids
The plasmid used in our studies, pNCT-tel was derived from the pSXneo-1.6T2AG3 plasmid previously shown to form new telomeres on integration in human cells [41]. pNCT-tel contains an ampicillin-resistance gene for selection in bacteria, a neo gene for selection in mammalian cells using G418, a herpes simplex virus thymidine kinase (HSV-tk) gene, and 0.8 kb of telomeric repeat sequences. After linearization with the NotI restriction enzyme, the telomeric repeat sequences are oriented such that they can “seed” the formation of new telomeres on integration. The HSV-tk gene is immediately adjacent to the telomere and is under the control of the cytomegalovirus (CMV) transcriptional promoter derived from the pC1 expression vector (Promega). Selection for loss of the HSV-tk gene in medium containing ganciclovir [42,43] is used to identify cells that have lost the end of the chromosome. Selection for HSV-tk-deficient (HSV-tk-) cells was performed in medium containing 50 µM gancilovir, whereas selection for HSV-tk- cells that still contained the neo gene (neo+/HSV-tk-) was performed in medium containing both 50 µM ganciclovir and 400 µg/ml G418.
Fluctuation Analysis
The rate of formation of HSV-tk- cells was determined from the frequency of ganciclovir-resistant colonies in different subpopulations of the EJ-30 clones. The rate of formation of neo+/HSV-tk- cells was determined from the frequency of colonies resistant to both ganciclovir and G418 in these same subpopulations. Briefly, the cell lines were first plated at 50 cells per well in a 24-well tissue culture dish. After 7 days, the cells were resuspended and replated in six-well tissue culture dishes. On reaching near confluence, the cells were trypsinized, counted, and replated either at 2 or 3x105 cells/100 mm tissue culture dish (found to be below the cell density required for a bystander effect with ganciclovir) in duplicate in medium containing ganciclovir (50 µM) and in medium containing both ganciclovir (50 µM) and G418 (400 µg/ml). The cells were incubated, with changes every 5 days, for a period of 2 to 3 weeks. The cells were then fixed with methanol/acetic acid and the colonies were counted. The rate of appearance of HSV-tk- or neo+/HSV-tk- cells was calculated using the average number of resistant cells in 12 different subpopulations [44]. The equation used was r=aNt In (CaNt), where a is the rate of appearance of HSV-tk- or neo+/HSV-tk- cells, r represents the average number of colonies formed per subpopulation, Nt is the total number of cells in the population at the time of plating, and C is the number of subpopulations [44]. The average number of colonies formed per subpopulation was determined from the average number of colonies per plate corrected for the final number of cells (average number per platextotal number of cells/number of cells plated). The expected values for the ratio of the standard deviation over the average number of spontaneous events were calculated as previously described [44] using the equation: √C/ In (NtCa). The values (r, Nt) obtained for each subpopulation with ganciclovir alone were: A3 (231, 6.8x106), B3 (250, 7.7x106), B1–2 (3.7, 5.5x106), E1 (6.4, 9.5x106), and F1 (0.8, 6.9x106). The values obtained with ganciclovir and G418 together were A3 (0.6, 6.8x106), B3 (1.6, 1.65x106), B1–2 (0, 5.5x106), E1 (0, 9.5x106), and F1 (0.3, 6.9x106).
Southern Blot Analysis
Genomic DNA purified as previously described [45] was digested with restriction enzymes according to the manufacturer's instructions. For analysis of terminal fragments, the DNA was digested with BAL31 following the manufacturer's recommendations (Promega), extracted with phenol/chloroform, precipitated, resuspended, and digested with BamHI. Genomic DNA was fractionated by agarose gel electrophoresis using standard protocols, depurinated by treatment with 0.25 M HCl for 30 minutes and transferred in 0.5 M NaOH onto a charged nylon Hybond-N+ membrane (Amersham) using a vacuum transfer apparatus (Pharmacia). Prehybridization for 3 hours and hybridization overnight were performed at 65°C in 5xSSPE, 5xDenhardt's solution, 0.5% sodium dodecyl sulfate (SDS), and 0.25 mg/ml salmon sperm DNA. Probes were labeled with [α32P]dCTP (New England Nuclear) using a High Prime labeling kit (Boehringer-Mannheim). Filters were washed three times in 2xSSPE with 0.1% SDS at room temperature, twice in 1xSSPE with 0.1% SDS at 65°C, and twice in 0.1xSSC with 0.1% SDS at 65°C.
DNA Analysis
Plasmid rescue was performed by first digesting the DNA with the appropriate restriction enzyme. The DNA was then diluted to 1 µg/ml in ligase buffer (20 mM Tris 7.5, 10 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP) containing 20 U/ml ligase (Life Technologies) and incubated overnight at 16°C. The ligation reaction was then terminated by heating at 65°C for 10 minutes and the DNA concentrated by centrifugation using Ultrafree-MC 30,000 NMWL filtration units (Millipore). The DNA was then electroporated (2.5 kV, 200 Ω, 25 µf) using 0.2-cm cuvettes (BioRad) into electrocompetent bacteria: DH12s (Life Technologies) for DNA rescued from clone B3 and subclone G63, and SURE (Stratagene) for subclone G55.
Polymerase chain reaction (PCR) was performed by using one primer complementary to the HSV-tk gene, TACCTTATGGGCAGCATG for subclone G6 and AAATGCCCACGCTACTGC for subclone G8, and one primer complementary to the telomeric repeat sequences, AACCCTAACCCTAACCCT or CCTAACCCTAACCCTAAC. PCR was performed by an initial incubation of 2 minutes at 95°C, followed by 40 cycles of 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds, followed by one cycle of 95°C for 30 seconds, 62°C for 30 seconds, and 72°C for 2 minutes. PCR was performed with Taq polymerase (Perkin-Elmer) as previously described [46]. PCR products were excised from agarose gels and cloned into the pCRII cloning vector (Invitrogen) using protocols provided by the manufacturer. DNA sequence analysis of the cloned PCR fragments using universal primers in the pCRII vector was performed by the Biomolecular Resources Center, UCSF.
Fluorescence In Situ Hybridization
Preparation of metaphase chromosomes, staining with Giemsa for analysis of dicentric and ring chromosomes, and in situ hybridization of the integrated plasmid sequences were performed as previously described [47,48]. Telomere analysis was performed as previously described [49] using telomere-specific protein nucleic acid (PNA) probes labeled with Cy3 (Perseptive Biosystems).
Results
Establishment of Cell Clones with a Selectable Marker Gene Adjacent to a Telomere
Linearized plasmids containing telomeric repeat sequences on one end commonly form new telomeres when integrated on to the ends of truncated chromosomes in mammalian cells [41,50–52]. After introduction into the cell, these marked telomeres are elongated and maintained similarly to other telomeres [53]. This approach was used to establish clones of the EJ-30 human bladder cell carcinoma cell line that have selectable marker genes integrated immediately adjacent to a telomere. EJ-30 has a relatively normal karyotype (46 chromosomes), is deficient in p53 [54,55], and is telomerase positive (data not shown). The analysis of 50 metaphase spreads in EJ-30 demonstrated that 4% of the cells had chromosome fusions, i.e., dicentric or ring chromosomes (data not shown). Thus, EJ-30 has relatively few chromosome fusions for a human tumor cell line, since human tumor cell lines can have chromosome fusions in as many as 90% of the cells in the population [3].
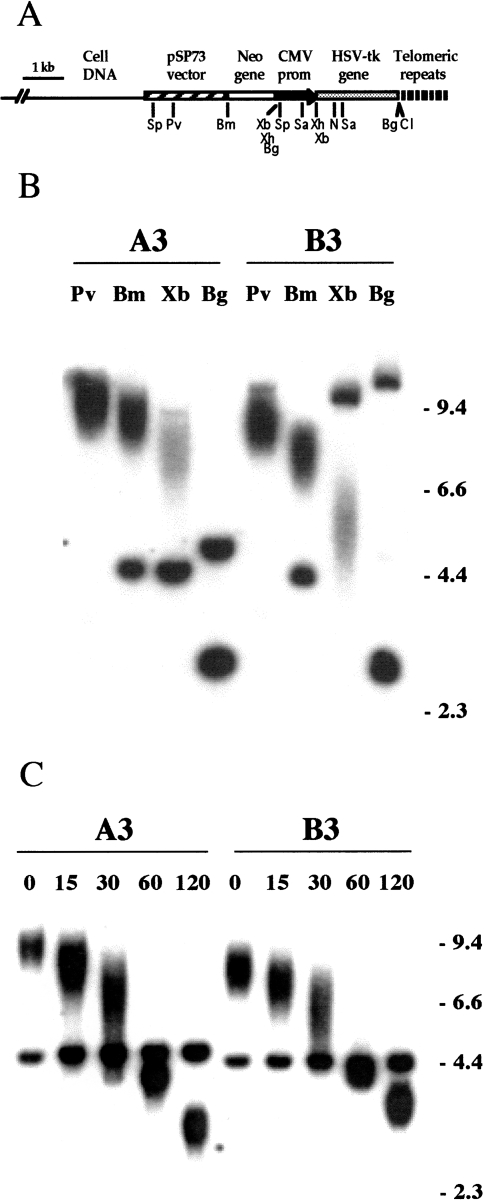
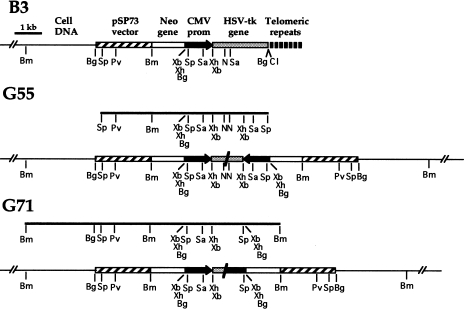
Transfection of EJ-30 with the linearized pNCT-tel plasmid (Figure 1A) resulted in the isolation of 72 G418-resistant subclones. Of these, 20 were found to be sensitive to ganciclovir and therefore contained the functional HSV-tk gene. Southern blot analysis, using the pNCT-Δ plasmid (the pNCT-tel plasmid without telomeric repeat sequences) as a probe identified five subclones with a single copy of the plasmid integrated at a telomere (data not shown). Cell clones with telomeric integration sites are evident from the presence of terminal restriction fragments that are heterogeneous in length because of variability in the length of the telomeric repeat sequences in different cells in the population [41,50,51,53]. The mapping of the integration site with various restriction enzymes in two of these clones, A3 and B3 (Figure 1B), indicated that the plasmid was integrated without rearrangement on the end of a chromosome. With PvuI, BamHI or XbaI, which cut once in the plasmid, there is one diffuse band and one discrete band that varies in length in the different clones. The length of the diffuse band is dependent on the length of the telomeric repeat sequences on the end of the chromosome, whereas the length of the discrete band is dependent on the location of the nearest restriction site within the genomic DNA. In both A3 and B3, the diffuse bands are progressively shorter as the plasmid sequences are cut closer to the telomere, so that the Pvul bands are longer than the BamHI bands, which are longer than the XbaI bands. Subtracting the length of the subtelomeric plasmid sequences (e.g., 3 kb with XbaI) from the mean length of the diffuse bands demonstrates that the mean length of the telomeric repeat sequences is approximately 5.0 kb in A3 and 3.0 kb in B3. Digestion with Bg/II, which cuts twice in the plasmid, once between the neo and HSV-tk genes and once between the HSV-tk gene and the telomeric repeat sequences, showed that both A3 and B3 contained the intact 3-kb fragment containing the HSV-tk gene.
Figure 1.
The characterization of pNCT-tel plasmid sequences integrated in the EJ-30 clones A3 and B3. (A) The structure of a telomere created by the integration of a single copy of the linearized pNCT-tel plasmid on the end of a chromosome. The location of the restriction sites BamHI (Bm), BgIII (Bg), ClaI (CI), NruI (N) PvuI (Pv), SacI (Sa), SspI (Sp), XbaI (Xb), and XhoI (Xh) in the plasmid are shown. (B) Southern blot analysis of genomic DNA from clones A3 and B3 digested with PvuI, BamHI, or XbaI, which cut once within the plasmid, or BgIII, which cuts at either end of the HSV-tk gene. Hybridization was performed with the pNCT-Δ plasmid probe that does not contain telomeric repeat sequences. (C) Digestion of genomic DNA from A3 and B3 with BAL31 exonuclease to identify terminal restriction fragments. After digestion of genomic DNA with BAL31, the DNA was digested with BamHI and Southern blot analysis was performed with the pNCT-Δ plasmid as a probe.
The telomeric integration sites in A3 and B3 were confirmed by digestion with BAL31 exonuclease. BAL31 is commonly used to identify telomeric sequences [50,56], because the ends of chromosomes are sensitive to digestion with exonuclease. Genomic DNAs from clones A3 and B3 were treated with BAL31 for various lengths of time before digestion with BamHI (Figure 1C). As expected, the diffuse bands that contain the telomeric repeat sequences are selectively reduced in size following treatment with BAL31 in both A3 and B3, and eventually reach sizes smaller than the 5.0- and 4.7-kb restriction fragments that represent the internal portion of the plasmid in A3 and B3, respectively. The accumulation of fragments of approximately 4 kb after 60 minutes indicates a slower rate of digestion of the subtelomeric plasmid sequences (4 kb with BamHI) relative to the telomeric repeat sequences, which has been previously reported [32,57]. This hypersensitivity of telomeric repeat sequences to BAL31 exonuclease is consistent with an altered secondary structure, as previously proposed from the sensitivity of telomeric repeat sequences to S1 endonuclease [58].
Analysis of HSV-tk- Subclones
Selection in medium containing 50 µM ganciclovir demonstrated that HSV-tk- cells were present at a frequency of approximately 10-3 in all five of the EJ-30 clones. Presumably, the loss of function of the HSV-tk gene in these cells results from spontaneous events and not selection in ganciclovir, because cells require several days to become resistant to ganciclovir following the loss of the HSV-tk gene [42,43]. Fluctuation analysis was performed to determine the rate of appearance of HSV-tk- cells and to confirm whether the loss of function of the HSV-tk gene occurred spontaneously or as a result of treatment with ganciclovir. In fluctuation analysis, subpopulations consisting of a small number of cells are passaged in culture and scored for the number of HSV-tk- cells after having grown to a large cell population. Fluctuation analysis demonstrated that the loss of function of the HSV-tk gene is a spontaneous event (Table 1). First, consistent with the loss of the HSV-tk gene in the different subpopulations at any time during passaging, the ratio of the standard deviation to the average number of HSV-tk- cells in the different subpopulations of A3 (0.45 and 0.37) and B3 (0.44 and 0.37) is much larger than the ratio observed in multiple platings of the same population (0.08 with 24 plates of clone B3). Second, it was observed that HSV-tk- cells accumulate in the population with increased passage (compare the frequency of HSV-tk- cells in experiments 1 and 2, Table 1), as expected for spontaneous events. Finally, the ratio of the standard deviation to the average number of HSV-tk- cells in the different subpopulations is close to that expected for spontaneous events (Table 1, Materials and Methods section).
Table 1.
Fluctuation Analysis of the Appearance of HSV-tk- Cells in the EJ-30 Clones A3 and B3.
| Clone | ||||
| B3 | B3 | A3 | A3 | |
| Experiment no. | 1 | 2 | 1 | 2 |
| No. cells/plate | 3x105 | 2x105 | 3x105 | 2x105 |
| No. colonies/plate | 110 | 177 | 217 | 353 |
| 198 | 166 | 230 | 129 | |
| 451 | 283 | 243 | 391 | |
| 136 | 220 | 174 | 131 | |
| 244 | 207 | 107 | 138 | |
| 144 | 260 | 198 | 286 | |
| 194 | 239 | 174 | 305 | |
| 141 | 236 | 174 | 134 | |
| 256 | 476 | 280 | 208 | |
| 333 | 208 | 187 | 281 | |
| 171 | 242 | 282 | 163 | |
| 433 | 286 | 230 | 235 | |
| Avg no. colonies | 234 | 250 | 208 | 231 |
| Avg no. cells | 2.0x106 | 7.7x106 | 1.7x106 | 6.8x106 |
| No. colonies/cult | 1568 | 9575 | 1040 | 7854 |
| Frequency | 7.8x10-4 | 1.2x10-3 | 6.1x10-4 | 1.2x10-3 |
| Rate | 1.0x10-4 | 1.3x10-4 | 1.0x10-4 | 1.3x10-4 |
| Observed SD/avg | 0.44 | 0.37 | 0.45 | 0.37 |
| Expected SD/avg | 0.49 | 0.32 | 0.24 | 0.40 |
Based on fluctuation analysis, both the A3 and B3 clones generated HSV-tk- cells at a rate of approximately 10-4 events/cell per generation (Tables 1 and 2). This rate is approximately 100-fold higher than in clones with interstitial integration sites (Table 2). Thus, the relatively high rate of generation of HSV-tk- cells in clones with telomeric integration sites is not due to a general instability of integrated plasmid sequences or a high rate of chromosome loss in EJ-30. Fluctuation analysis also demonstrated that most of the HSV-tk- cells had lost their resistance to G418. The rate of generation of neo+/HSV-tk- cells, as determined by selection with both G418 (400 µg/ml) and ganciclovir (50 µM), was approximately 100-fold lower than the rate of generation of HSV-tk- cells selected in ganciclovir alone (Table 2). This result was consistent with Southern blot analysis, which demonstrated that most of the HSV-tk- subclones selected in ganciclovir alone showed no hybridization with the pNCT-Δ plasmid probe and therefore had lost the entire plasmid (data not shown). In view of the much lower rate of generation of HSV-tk- cells in clones with interstitial integration sites (Table 2), the high rate of loss of the telomeric plasmid would appear to be due to deletions on the end of the marked chromosomes.
Table 2.
The Rate of Production of HSV-tk- Cells in EJ-30 Clones with Telomeric or Interstitial Integration Sites.
| Cell Clone | HSV-tk-* | neo+/HSV-tk-* |
| Telomeric | ||
| A3 | 1.3x10-4 | 1.6x10-6 |
| B3 | 1.3x10-4 | 6.9x10-7 |
| Interstitial† | ||
| B1–2 | 3.4x10-6 | <10-7 |
| E1 | 5.1x10-7 | <10-7 |
| F1 | 9.2x10-7 | 4.1x10-7 |
The rate of production of HSV-tk- or neo+/HSV-tk- cells was determined from the frequency of colonies resistant to ganciclovir or G418 and ganciclovir, respectively (see Materials and Methods section).
Clones used for analysis of interstitial integration sites lacked diffuse bands but had an intact HSV-tk gene, as demonstrated by Southern blot analysis and sensitivity to ganciclovir.
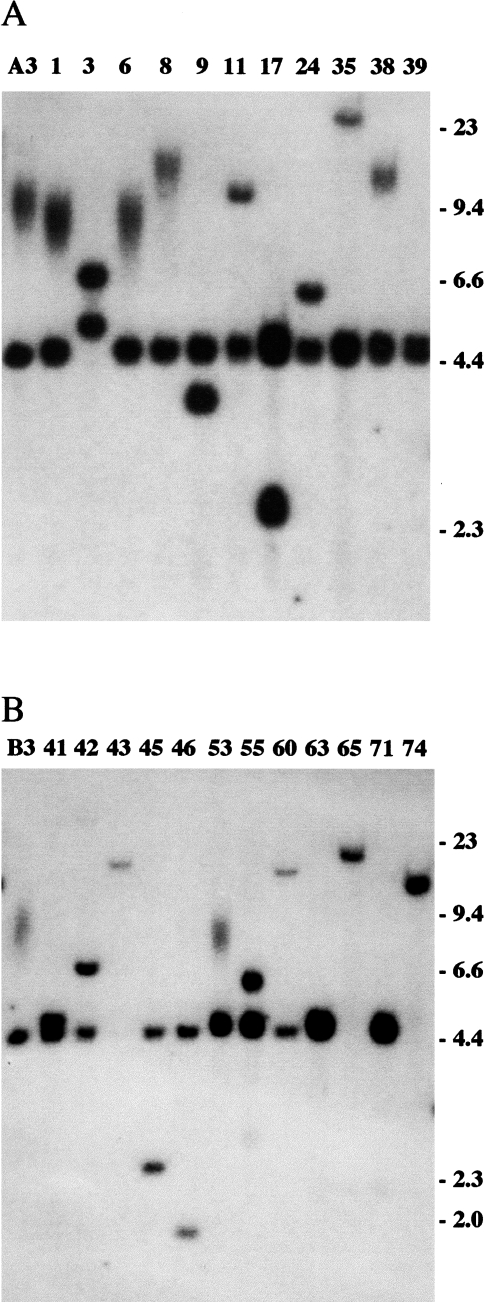
Further analysis of the types of events involved in the loss of function of the HSV-tk gene was confined to subclones that had retained some portion of the plasmid sequences to insure that a unique sequence tag remained on the end of the marked chromosome. Southern blot analysis of BamHI-digested genomic DNA from HSV-tk- subclones of A3 and B3 that had retained some portion of the plasmid sequences showed that most had lost the diffuse band and gained a discrete band that varied in size in the different subclones (Figure 2). Since most of these subclones retained the band generated from the internal portion of the plasmid, the most common event associated with the loss of function of the HSV-tk gene appeared to involve telomere loss and the joining of nontelomeric DNA on to the end of the truncated chromosome. However, some of these subclones showed no apparent change in the integrated plasmid sequences by BamHI digestion, including the A3 subclones G1, G6, G8, and G38 (Figure 2A), and the B3 subclone G53 (Figure 2B). The mean length of the diffuse band containing the HSV-tk gene often differs in these subclones. However, this variation does not necessarily arise from alterations in the HSV-tk gene, since subclones of human tumor cell lines often have telomeres that differ in length but return to the length found in the parental cell line with increasing time in culture [53]. Therefore, it is not possible to determine solely by Southern blot analysis after digestion with BamHI whether the loss of function of the HSV-tk gene in these subclones involves silencing, point mutations, internal deletions, or terminal deletions.
Figure 2.
Southern blot analysis of the integrated plasmid sequences in HSV-tk- subclones of A3 and B3 that retained some portion of the plasmid sequences. (A) Genomic DNA from A3 and 11 of its HSV-tk- subclones (G1–G39), and (B) genomic DNA from B3 and 12 of its HSV-tk- subclones (G41–G74) was digested with BamHI, which cuts once in the plasmid, and hybridization was performed with the pNCT-Δ probe. The absence of the diffuse band indicates the loss of the telomere, whereas the appearance of a new discrete band indicates the presence of nontelomeric DNA fused on to the end of the chromosome.
Analysis of neo+/HSV-tk- Subclones with No Apparent Change in the Plasmid Sequences
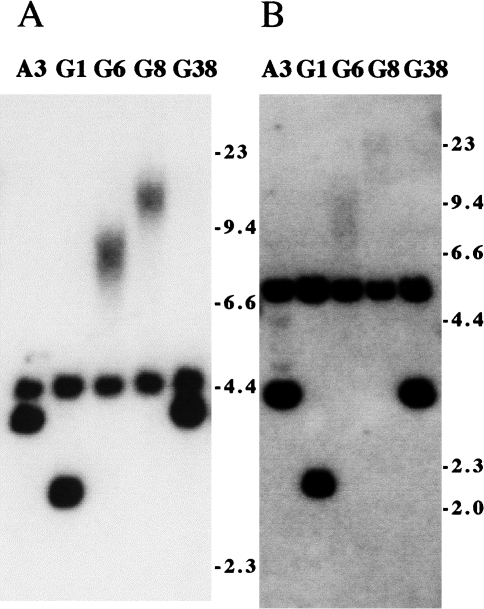
The status of the HSV-tk gene in the 10 HSV-tk- subclones with no apparent change in the integrated plasmid sequences by Southern blot analysis of BamHI-digested DNA was further investigated by digestion with the ClaI and Bg/II restriction enzymes (Figure 3). ClaI cuts the plasmid once immediately adjacent to the telomeric repeat sequences, and therefore together with BamHI should generate a 4.5-kb band if the integrated plasmid sequences are intact (see Figure 1A). Bg/II cuts twice in the plasmid, once between the HSV-tk and neo genes and once between the HSV-tk gene and the telomeric repeat sequences, and generates a 3.0-kb band containing the HSV-tk gene (Figure 1A). Of the 10 HSV-tk- subclones that showed no change with BamHI, five, including G6 and G8 derived from A3 (Figure 3), and T4, T12, and T21 derived from B3 (data not shown), showed diffuse bands in both the BamHI/ClaI and Bg/II digests and therefore had lost the ClaI and Bg/II sites between the HSV-tk gene and the telomeric repeat sequences (Table 3). These five subclones therefore have lost the end of the HSV-tk gene and have had telomeric repeat sequences added on to the end of the truncated HSV-tk gene at a new location. In two of the HSV-tk- subclones that showed no change with BamHI, including G1 derived from A3 (Figure 3) and T1 derived from B3 (data not shown), both the BamHI/ClaI and Bg/II bands were smaller than in the parental cell line (Table 3), demonstrating internal deletions within the HSV-tk gene. Three other HSV-tk- subclones that showed no change with BamHI, including G38 derived from A3 (Figure 3), and T14 and T26 derived from B3 (data not shown), were found to have no detectable change in the size of the fragment containing the HSV-tk gene using either BamHI/ ClaI or Bg/II (Table 3), and therefore appear to have undergone silencing or point mutations in the HSV-tk gene.
Figure 3.
Further analysis of HSV-tk- subclones that showed no detectable change in the plasmid sequences by Southern blot analysis of genomic DNA digested with BamHI. Genomic DNAs from A3 and four of its HSV-tk- subclones, G1, G6, G8, and G38, that showed no change with BamHI, were digested with (A) BamHI and ClaI or (B) BgIII, and hybridization was performed with the pNCT-Δ probe. The presence of diffuse bands indicates that although the plasmid is still telomeric, the BgIII and ClaI sites between the HSV-tk gene and the telomeric repeat sequences are no longer present.
Table 3.
The Number and Types of Events Seen in HSV-tk- Subclones of A3 and B3 that Retained Some Portion of the Plasmid Sequences.
| Type of Event | No. Subclones |
| Telomere loss/fusion* | 34 |
| Terminal deletions† | 5 |
| Internal deletions‡ | 2 |
| No apparent change§ | 3 |
Subclones with nontelomeric DNA joined to the end of the truncated HSV-tk gene.
Subclones with telomeric repeat sequences added on to the end of the truncated HSV-tk gene.
Subclones with deletions in the HSV-tk gene as detectable by Southern blot analysis.
Subclones with no apparent change in the HSV-tk gene by Southern blot analysis.
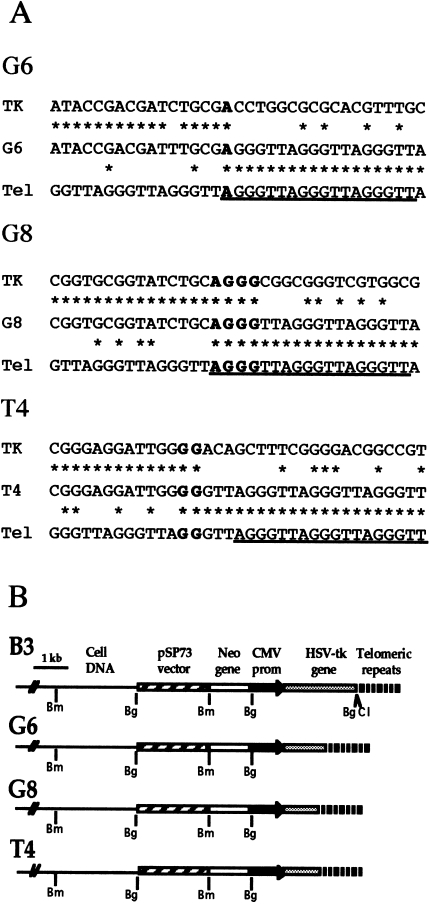
Three of the HSV-tk- subclones, G6 and G8 (Figure 3), and T4 (data not shown), that had telomeric repeat sequences added on to the end of the truncated HSV-tk gene were further analyzed by polymerase chain reaction (PCR) and nucleotide sequence analysis. The junctions between the plasmid and the telomeric repeat sequences were amplified using one primer complementary to various locations within the plasmid and another primer complementary to the telomeric repeat sequences (data not shown). Nucleotide sequence analysis of PCR products that were specific to these subclones demonstrated that the telomeric repeat sequences were added directly on to the end of the truncated chromosome (Figure 4A). In subclone G6, 1 bp of complementarity was found at the site of addition of the telomeric repeat sequences, whereas in subclones G8 and T4 there were 4 and 2 bp, respectively. In subclone G6, the terminal deletion resulted in the loss of 0.8 kb of DNA from the end of the HSV-tk gene, whereas in subclones G8 and T4, 1.2 kb was lost from the end of the HSV-tk gene (Figure 4B).
Figure 4.
Nucleotide sequence analysis of the sites of addition of telomeric repeat sequences following the loss of the end of the HSV-tk gene. (A) The nucleotide sequences at the sites of addition of the telomeric repeat sequences on to the end of the HSV-tk gene in subclones G6, G8, and T4 are compared with the sequence of the HSV-tk gene (TK) and telomeric repeat sequence (Tel). Regions of homology (asterisks) and short regions of complementarity between the plasmid and telomeric repeat sequences at the site of addition (bold) are indicated. The sequences of the telomere-specific primers used for PCR are also shown (underlined). (B) The structure of the integrated plasmid sequences in the parental clone B3 compared with the structure of the plasmid sequences in the G6, G8 and T4 subclones with terminal deletions involving the HSV-tk gene. The location of restriction sites for BamHI (Bm), BgIII (Bg) and ClaI (Cl) are shown.
Chromosome Fusion and Gene Amplification in Subclones with Spontaneous Telomere Loss
Plasmid rescue was performed to determine the origin of the nontelomeric DNA joined on to the end of the marked chromosome in two HSV-tk- subclones, G55 and G71. Consistent with Southern blot analysis (see Figure 1), the location of restriction sites in the DNA fragment rescued from the parental clone B3 indicated that a single copy of the plasmid was integrated at the telomere (Figure 5). In contrast, the restriction sites in the DNA fragment rescued from subclone G55 indicated that it contained an inverted repeat of plasmid sequences joined at almost identical sites in the HSV-tk gene (Figure 5). The head-to-head fusion of the plasmid in subclone G55 is consistent with Southern blot analysis of G55 genomic DNA digested with BamHI (see Figure 2B), which demonstrated the presence of a new band of the size expected (6 kb) for the novel junction fragment found in the rescued DNA. The restriction sites in the DNA fragment rescued from subclone G71 also indicated the presence of an inverted repeat of plasmid sequences (Figure 5). However, in subclone G71 the plasmid sequences appeared to be fused at sites approximately 1 kb apart, one site occurring in the HSV-tk gene and the other in the CMV promoter. As in subclone G55, the head-to-head fusion of the plasmid in subclone G71 is consistent with Southern blot analysis of G71 genomic DNA digested with BamHI (see Figure 2B). A single 4.5-kb band is observed, which would be expected since the novel junction fragment is the same size (4.5 kb) as the fragment containing the internal portion of the plasmid. The head-to-head orientation of the plasmid sequences in these subclones could have originated by fusion of sister chromatids following the loss of the telomere, as previously proposed by McClintock [25].
Figure 5.
Analysis of the DNA fused on to the end of the marked chromosome in the HSV-tk- subclones G55 and G71. The plasmid sequences rescued from subclone G55 after a partial Sspl digestion, or subclone G71 after partial BamHI digestion (bold lines), indicated the presence of an inverted repeat of the plasmid sequences in both subclones. The location of the restriction sites BamHI (Bm), BgIII (Bg), ClaI (Cl), NruI (N) PvuI (Pv), SacI (Sa), SspI (Sp), XbaI (Xb), and XhoI (Xh) are shown.
To confirm the presence of an inverted repeat within the plasmid DNA rescued from clones G55 and G71, nucleotide sequence analysis was performed on the fragments predicted to contain the junctions between the plasmid sequences. The 220-bp NruI fragment predicted to containing the junction between the inverted plasmid sequences in subclone G55 (see Figure 5) had the identical HSV-tk sequence at both ends in opposite orientations (data not shown). These results confirmed the presence of an inverted repeat resulting from a head-to-head fusion of the plasmid at nearly identical sites in the HSV-tk gene. However, the exact site of fusion could not be determined, because 70 to 80 bp in the center of the fragment could not be sequenced from either direction, apparently due to the presence of the inverted repeat. Nucleotide sequence analysis of the site of fusion of the plasmid sequences in subclone G71 also confirmed the head-to-head orientation of the plasmid sequences, with one plasmid fused in the HSV-tk gene and the other in the CMV promoter (data not shown). In addition, there was 175 bp of a repetitive human sequence located between the fused plasmid sequences (Genbank accession number AF187979). This short DNA fragment may have been inserted between the ends of the plasmid during nonhomologous end joining, because the insertion of short DNA fragments has previously been observed in mammalian cells by the analysis of mutations in the Aprt gene [59,60]. Alternatively, the presence of the short DNA fragment between the inverted plasmid sequences could mean that another type of recombination event preceded the fusion of the plasmid sequences.
Analysis of Chromosome Instability in Subclones with Spontaneous Telomere Loss
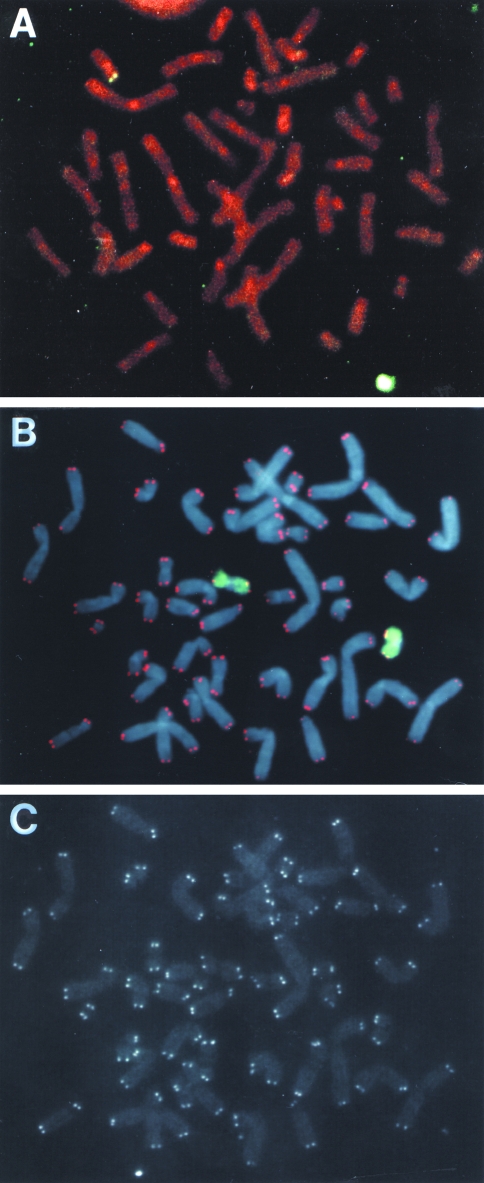
Fluorescence in situ hybridization was performed to further characterize the marked chromosome containing the integrated plasmid sequences in the parental B3 clone and three subclones with nontelomeric DNA joined on to the end of the marked chromosome. Hybridization with the pNCT-Δ plasmid probe showed that clone B3 had a single integration site near a telomere (Figure 6A), which was determined to be on the short arm of chromosome 16 by propidium iodide staining and hybridization with a chromosome-16-specific probe (data not shown). The analysis of 50 mitotic cells of clone B3 using the chromosome-16-specific probe identified only one cell that had a rearrangement of chromosome 16 (Table 4), demonstrating that the marked chromosome containing the integrated plasmid is relatively stable. Four different subclones of B3 that were selected at random also rarely contained detectable rearrangements in the marked chromosome (Table 4). Fluorescence in situ hybridization with both a telomere-specific PNA probe and a chromosome-16-specific probe was also used to analyze telomere status on the marked chromosome. PNA probes have been demonstrated to be highly sensitive and have previously been used to quantify telomere length in human cells [49,61]. The results demonstrated that in all of the cells observed, both homologs of chromosome 16 had telomeres on both ends (Figure 6B and C). Telomeres could also be seen on the ends of all the other chromosomes in virtually all of the cells.
Figure 6.
Analysis of the site of integration of the pNCT-tel plasmid in clone B3 using fluorescence in situ hybridization. (A) A single integration site of the pNCT-tel plasmid is observed in clone B3 at the end of the short arm of a chromosome. Hybridization was performed with the pNCT-Δ plasmid and chromosomes were counterstained with propidium iodide. The chromosome was identified by chromosome-specific probes to be chromosome 16 (data not shown). (B) Hybridization with both a telomere-specific PNA probe (red) and a chromosome-16-specific probe (green) demonstrated that telomeric repeat sequences are located on the ends of both homologs of chromosome 16 in clone B3. Chromosomes were counterstained with DAPI. (C) The hybridization signal observed with the telomere-specific probe alone showing detectable telomeres on both ends of all the chromosomes.
Table 4.
The Number of Mitoses With Aberrations in the Marked Chromosome Compared with the Number of Mitoses with Aberrations in Chromosome 1 or All Other Chromosomes in Clone B3 and Its Subclones.
| Chromosome 16* | |||
| Cell Clone | No Apparent Change | Rearranged | Dicentrics |
| B3 | 49 | 1 | 0 |
| B3-1 | 52 | 0 | 0 |
| B3-2 | 49 | 1 | 0 |
| B3-4 | 50 | 0 | 0 |
| B3-5 | 50 | 0 | 0 |
| G55 | 21 | 29 | 1 |
| G60 | 21 | 29 | 3 |
| G71 | 26 | 24 | 3 |
| Chromosome 1* | |||
| Cell Clone | No Apparent Change | Rearranged | Dicentrics |
| B3 | 50 | 0 | 0 |
| G55 | 50 | 0 | 0 |
| G60 | 6 | 0 | 0 |
| G71 | 49 | 1 | 0 |
| All chromosomes | |||
| Cell Clone | No Aberrations | Breaks† | Fusions‡ |
| B3 | 40 | 7 | 3 |
| G55 | 44 | 4 | 2 |
| G60 | 35 | 10 | 5 |
| G71 | 45 | 2 | 3 |
The analysis of rearrangements in specific chromosomes were performed by hybridization with a chromosome-specific libraries.
Breaks include both chromatid breaks and chromosome breaks.
Fusions include dicentric and ring chromosomes.
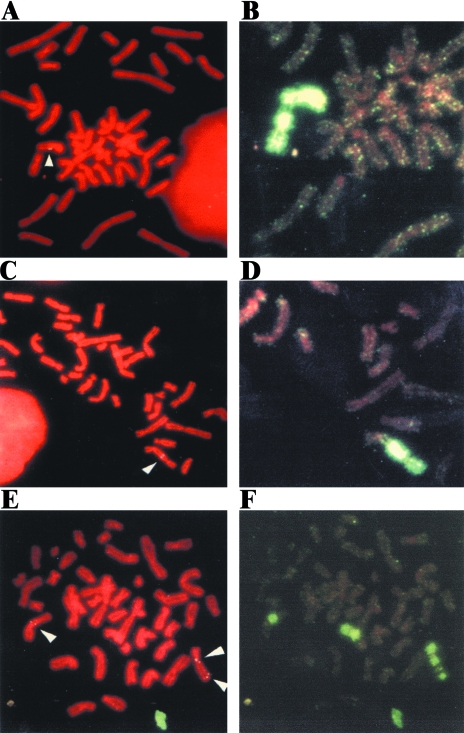
In contrast to the parental clone B3 or subclones selected at random, the marked chromosome was highly unstable in subclones G55, G60, and G71 (Table 4) in which nontelomeric DNA had been joined on to the end of the truncated HSV-tk gene (see Figures 2 and 5). Unlike the parental clone B3, in all three of these subclones the integrated plasmid sequences were located at interstitial sites in most cells in the population (Figure 7). Furthermore, chromosome painting showed a high degree of heterogeneity in the structure of the chromosome containing the integrated plasmid sequences (Figure 7, Table 4). The most common change involved the presence of fragments of chromosome 16 of varying lengths joined on to the end of the marked chromosome (Figure 7A and B). Combined with the demonstration of inverted repeats in the plasmid DNA rescued from subclones G55 and G71 (Figure 5), this observation strongly suggests that the DNA joined on to the end of the marked chromosome in these subclones resulted from sister chromatid fusion.
Figure 7.
Fluorescence in situ hybridization demonstrating the instability of the marked chromosome in subclones G60 and G71 that have nontelomeric DNA joined on to the end of the marked chromosome. Metaphase chromosomes from subclones G60 (A, B, E, F) and G71 (C, D) were hybridized with either the pNCT-Δ plasmid probe (A, C, E) or a chromosome-16-specific probe (B, D, F) and the chromosomes were counter stained with propidium iodide. Extensive heterogeneity was observed in the structure of the marked chromosome, including the presence of chromosome 16 fragments of different lengths joined on to the end (A, B), dicentric chromosomes involving the marked chromosome and other chromosomes (C, D), and amplification of the marked chromosome with chromosome-16-specific fragments interspersed with fragments of other chromosomes (E, F).
In some cells in the population, fragments of other chromosomes were also observed on the end of the marked chromosome (Figure 7C and D), often with fragments of chromosome 16 located between these fragments and the marked chromosome (see Figure 8E). In other cells, further amplification and/or fragmentation of the marked chromosome was observed (Figure 7E and F). Although the marked chromosome appeared to be normal in nearly 50% of the cells in all three subclones using the chromosome-16-specific painting probe (Table 4), further analysis showed that these “normal-looking” chromosomes had also undergone rearrangements: in situ hybridization with the plasmid probe commonly showed that the plasmid was no longer telomeric (data not shown) and hybridization with the telomere-specific probe showed that they often had no detectable telomere on one end (see below).
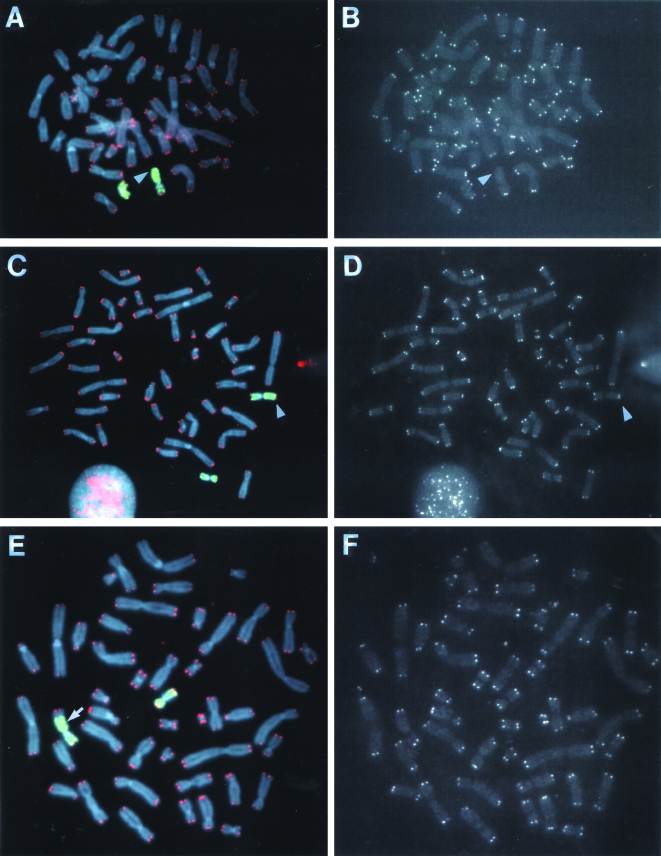
Figure 8.
Analysis of telomeres on the end of the unstable marked chromosome in subclone G71. Hybridization was performed with both a telomere-specific PNA probe (red) and a chromosome 16-specific probe (green). Chromosomes were counter stained with DAPI. Hybridization signals are shown for both probes together (A, C, and E) and with the telomere-specific PNA probe alone (B, D, and F). The results are shown for a cell (A, B) in which the DNA joined on to the end of the marked chromosome originated from chromosome 16 and had no detectable telomere on the end (arrow); a cell (C, D) in which the DNA joined on to the end of the marked chromosome originated from chromosome 16 and had a telomere on the end (arrow); and a cell (E, F) in which the DNA joined on to the end of the marked chromosome originated from both chromosome 16 and another chromosome and had a telomere on the end. The location of the interstitial plasmid sequences (as determined by hybridization with the pNCT-Δ probe, data not shown) at the junction between the marked chromosome and a fragment of chromosome 16 is shown (arrow, E).
The extensive heterogeneity in the structure of the marked chromosome in different cells in the population in subclones G55, G60, and G71 strongly suggests that the marked chromosome can be unstable for many generations following the loss of a telomere. A prolonged period of instability in the marked chromosome in these subclones is also indicated by: 1) the presence of dicentric chromosomes containing the marked chromosome in some cells in the population even after 20 generations (Table 4, Figure 7C and D), because dicentric chromosomes are generally unstable in mammalian cells [62]; 2) the amplification of the plasmid sequences in tandem arrays in some cells (Figure 7E); and 3) the presence of multiple fragments of chromosome 16 interspersed with fragments of other chromosomes (Figure 7F). In addition, many second-generation subclones isolated from subclone G71 continue to show a high degree of heterogeneity in the structure of the marked chromosome (data not shown), demonstrating that chromosome instability can continue for more than 20 generations.
In contrast to the marked chromosome, the other chromosomes in subclones G55, G60, and G71 were found to be relatively stable (Table 4). The analysis of all chromosomes by Giemsa staining showed no difference between the parental B3 clone and subclones G55, G60, and G71 in the frequency of chromatid or chromosome breaks, or the frequency of dicentric and ring chromosomes. Similarly, the use of a chromosome-specific probe for the analysis of chromosome 1 (Table 4) or chromosomes 12, 13, and 19 (data not shown), also showed that these chromosomes, unlike the marked chromosome, had few detectable rearrangements. Thus, the chromosome instability in the subclones in which the marked chromosome had undergone telomere loss and chromosome fusion is confined to the marked chromosome.
To determine whether telomeres are present on the marked chromosome in subclone G71, hybridization was performed with both a telomere-specific PNA probe and a chromosome-16-specific probe (Figure 8). In the parental B3 clone, telomeres are always detectable on both homologs of chromosome 16 (Figure 6B and C). In contrast, a telomere was not detectable on one end of a chromosome 16 in many cells (8 of 40) in subclone G71 (Figure 8A and B), despite the fact that telomeres were detectable on both ends of all the other chromosomes in virtually all of the cells observed. Telomeres were often observed on the end of the marked chromosome with fragments of chromosome 16 joined to the end (Figure 8C and D). Telomeres were also observed on the ends of fragments of other chromosomes joined on to the end of the marked chromosome (Figure 8E and F). Whether the acquisition of telomeres on the end of the marked chromosome in these cells resulted from de novo addition by telomerase or the capture of preexisting telomeres could not be determined. The absence of detectable telomeric repeat sequences at the junctions between the marked chromosome and the fragments of chromosome 16 (Figure 8C) or other chromosomes (Figure 8E) that contain telomeres on the end could mean that these fragments resulted either from translocation or from end-to-end fusion following telomere loss. Regardless of the mechanism involved, telomere restoration appears to be capable of stabilizing the marked chromosome in subclone G71, because the marked chromosome with telomeres on both ends is stable in some of the next generation of subclones isolated from subclone G71 (data not shown).
Discussion
The study of the loss of the telomeric plasmid sequences in the EJ-30 cell line suggests that this cell line undergoes spontaneous telomere loss and that telomere loss can result in prolonged periods of chromosome instability. The mechanism for the spontaneous telomere loss is unknown. The analysis of the length of the marked telomere in clone B3 during prolonged growth in culture demonstrated that it is maintained at a specific mean length (data not shown), as previously observed in two other telomerase-positive cancer cell lines [53,63]. However, despite this ability to maintain telomeres at a specific mean length, some telomeres can experience dramatic alterations in length and may become too short to protect the end of the chromosome [63]. The loss of the telomere would then result in extensive degradation of the subtelomeric DNA, followed by fusion between sister chromatids or other chromosomes. These changes in telomere length could result from mutations or altered expression of a large number of different genes that have been shown to influence telomere stability [14–18]. Although it cannot be completely ruled out that the presence of the plasmid sequences influences the rate of telomere loss, this does not appear to be the case. The presence of dicentric chromosomes in 4% of the cells in the parental EJ-30 cell line, and the fusion of the marked chromosome with other chromosomes (Figure 7), suggests that the telomere loss and chromosome instability seen in the marked chromosome also occurs on other chromosomes in this cell line. Consistent with these observations, the dynamics of the changes in length of marked telomeres generally reflects the dynamics of the endogenous telomeres and the frequency of telomere associations [13,53,63]. In addition, studies in yeast have demonstrated that normal telomere function requires only the presence of telomeric repeat sequences [64]. Telomere repeat sequences would also appear to be all that is necessary for normal telomere function in human cells, as demonstrated by the viability of humans with terminal deletions involving chromosome healing [36,37].
Chromosome Fusion and Chromosome Instability Associated with Telomere Loss
The most frequent event associated with the loss of function of the HSV-tk gene in the EJ-30 cell line is telomere loss and the joining of nontelomeric DNA on to the end of the truncated chromosome (Table 3). The DNA joined on to the end of the marked chromosome in two subclones, G55 and G71, consisted of inverted repeats of the end of the chromosome (Figure 5), indicating that sister chromatid fusion is often involved. Cytogenetic analysis also provides evidence for sister chromatid fusion, because in three different B3 subclones, G55, G60, and G71, the fragments joined on to the end of the marked chromosome 16 most often originated from chromosome 16.
The extensive heterogeneity in the structure of the marked chromosome in subclones G55, G60, and G71 (Table 4, Figures 7 and 8), demonstrates that the chromosomes with inverted repeats on their ends are often highly unstable. Decreased telomerase activity does not appear to be involved in this instability, since the level of telomerase activity in subclones G55, G60, and G71 is similar to that seen in the parental clone B3 (data not shown). Similarly, exposure of the cells to ganciclovir can also be ruled out, since the instability is confined to the marked chromosome (Table 4). A more likely explanation is that this chromosome instability is associated with telomere loss and sister chromatid fusion, consistent with previous studies that demonstrated a high rate of rearrangement of human chromosomes involved in telomere associations [21–24]. Thus, one possible mechanism for the chromosome instability in the marked chromosome is the breakage/fusion/bridge cycle. The breakage/fusion/bridge cycle was first described in plants by McClintock [25] and has subsequently been shown to be a mechanism of chromosome instability in rodent cells [26–28]. In this model, the loss of a telomere before or during DNA synthesis results in sister chromatids without telomeres. The sister chromatids then fuse at their ends, form a bridge during cell division, and break, leaving one daughter cell with an amplification and the other with a deletion of the end of the chromosome. This cycle then continues, leading to further chromosome rearrangements, until a new telomere is added, the chromosome is lost, or the cell dies. Several lines of evidence suggest that breakage/fusion/bridge cycles can occur after the loss of a telomere in EJ-30, including: 1) the analysis of the rescued plasmid DNA, which demonstrates the presence of inverted repeats (Figure 5); 2) cytogenetic analysis, which demonstrates fragments of chromosome 16 joined on to the end of the marked chromosome (Figures 7A and B and 8); and 3) the presence of tandem arrays of amplified plasmid sequences, as demonstrated by the presence of multiple spots on the same marked chromosome using hybridization with the pNCT-Δ plasmid (Figure 7E and F). The presence of dicentric chromosomes involving the marked chromosome and other chromosomes (Figure 7C and D), and the alternating chromosome-16-specific bands on some chromosomes (Figure 7F) also suggest that breakage/fusion/bridge cycles can occur between the marked chromosome and other chromosomes after the loss of a telomere.
Marked Chromosomes with No Telomeres or Telomeres Added at a New Location
An interesting observation arising from these studies is the presence of cells in the population in subclone G71 in which the marked chromosome lacks a detectable telomere on one end. Although it cannot be ruled out that telomeres are present but are too short to be detected, PNA probes are highly sensitive and have been shown to detect as little as 150 bp of telomeric repeat sequences [49]. Therefore, even if telomeres are present they may not be long enough to allow for the proper assembly of the proteins required for normal telomere function. One possible explanation for the absence of detectable telomeres on the ends of these marked chromosomes is that they contain fusions between the ends of the sister chromatids. Sister chromatid fusions would be expected in some marked chromosomes in subclones G55, G60, and G71 if they are continuing to undergo breakage/fusion/bridge cycles. Alternatively, the absence of detectable telomeres on the marked chromosome so many generations after the initial loss of a telomere could mean that chromosomes without telomeres can be maintained in the EJ-30 cell line without undergoing chromosome fusion. Chromosomes without telomeres have been previously demonstrated to be stably maintained through multiple cell divisions without fusion in Drosophila [65,66]. Chromosomes in yeast can also be maintained for up to 10 cell generations without a telomere as part of an adaptation process [33], which is limited to a single unrepairable double-strand break [67]. Extensive degradation resulting from the prolonged absence of a telomere, as previously reported in Drosophila [68], could explain the fact that the entire plasmid is commonly missing from the end of the chromosome in the HSV-tk- subclones selected in ganciclovir alone.
Approximately 1 in 10 of the HSV-tk- subclones that retained some portion of the plasmid had telomeric repeat sequences added directly on to the ends of the truncated chromosome (Figures 3 and 4, Table 3). It is not known at this time whether the addition of the telomere in these subclones occurred by de novo synthesis by telomerase or by capture of preexisting telomeres. Short regions of complementarity to telomeric repeat sequences were observed at the site of addition of the telomeric repeat sequences (Figure 4), as has been previously reported in chromosome healing in humans [36,37] and in mouse ES cells [32]. However, the presence of short regions of complementarity alone does not establish the mechanism involved. Although short regions of complementarity are commonly found at sites of de novo telomere addition [38], they are also commonly seen with nonhomologous end joining [59], a mechanism likely to be involved in telomere capture. Studies with human cell extracts have demonstrated that telomeric repeat sequences can be added on to the ends of oligonucleotides by de novo synthesis [38]. However, telomeric repeat sequences have also been found to be added on to the ends of transfected plasmid DNA [56] in a human cell line that was later shown to lack detectable telomerase activity [13]. Thus, additional studies are required to determine whether de novo addition of telomeres by telomerase plays a significant role in the mammalian cell response to telomere loss.
Factors Influencing Telomere Loss-Induced Chromosome Instability
The chromosome instability induced by telomere loss could be influenced by several factors. The ability of cells to regulate the cell cycle in response to chromosome damage could prevent the chromosome instability resulting from telomere loss, because the absence of a telomere or the presence of a dicentric chromosome would both be signals for a cell cycle arrest. Consistent with this possibility, a previous study found that most human cancer cell cultures that had breakage/fusion/bridge events were p53 negative [29]. Thus, the prolonged periods of chromosome instability in EJ-30 are likely to be dependent on the defect in p53 in this tumor cell line [54,55]. The ability of cells to adapt to the absence of a telomere on the end of a chromosome could also be important in determining the extent of the chromosome instability, since as mentioned earlier, chromosomes without telomeres can be maintained without fusions in both Drosophila [65,66] and yeast [33]. Finally, the ability of EJ-30 cells to restore lost telomeres could also affect chromosome instability, as originally proposed by McClintock [25]. Consistent with this hypothesis, chromosomes in EJ-30 that become unstable following the loss of a telomere can again become stable, as shown by the absence of heterogeneity in the marked chromosome with telomeres on both ends in subclones isolated from subclone G71 (data not shown). The importance of telomeres in chromosome instability in the EJ-30 cell line may therefore depend not only on factors that influence the rate of telomere loss, but also on the ability of these cells to respond to the presence of a lost telomere through cell cycle regulation, adaptation, and chromosome healing.
Acknowledgement
We thank Luis Martins for his helpful technical assistance.
Abbreviations
- ES
embryonic stem
- HSV-tk
herpes simplex virus thymidine kinase
- bp
base pair
- CMV
cytomegalovirus
- PNA
protein nucleic acid
Footnotes
The work in the J.P.M. laboratory was supported by grant number RO1 CA69044 from the National Cancer Institute, National Institutes of Health. The work in the L.S. laboratory was supported by contract number F14-CT950008 from the Commission of European Communities. B.F. was supported by training grant ES07106 from the National Institute of Environmental Health, National Institutes of Health, and G.P. was supported by Amplitech SA.
References
- 1.Blackburn EH, Greider CW, editors. Telomeres. Plainview, NY: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 2.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Lange T. Telomere dynamics and genome instability in human cancer. In: Blackburn EH , Greider CW, editors. Telomeres. Plainview, NY: Cold Spring Harbor Press; 1995. pp. 265–293. [Google Scholar]
- 4.Hastie ND, Allshire RC. Human telomeres: fusion and interstitial sites. Trends Genet. 1989;5:326–331. doi: 10.1016/0168-9525(89)90137-6. [DOI] [PubMed] [Google Scholar]
- 5.Conrad MN, Dominguez AM, Dresser ME. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 6.Kirk KE, Harmon BP, Reichardt IK, Sedat JW, Blackburn EH. Block in anaphase chromosome separation caused by a telomerase template mutation. Science. 1997;275:1478–1481. doi: 10.1126/science.275.5305.1478. [DOI] [PubMed] [Google Scholar]
- 7.Harley CB, Kim NW, Prowse KR, Weinrich SL, Hirsch KS, West MD, Becchetti S, Hirte HW, Counter CM, Greider CW, et al. Telomerase, cell immortality and cancer. Cold Spring Harbor Symp Quant Biol. 91:2900–2904. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 8.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 9.Harley CB. Telomeres and aging. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1995. pp. 247–263. [Google Scholar]
- 10.Klingelhutz AJ, Barber SA, Smith PP, Dyer K, McDougall JK. Restoration of telomeres in human papillomavirus-immortalized human anogenital epithelial cells. Mol Cell Biol. 1994;14:961–969. doi: 10.1128/mcb.14.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995;14:4240–4248. doi: 10.1002/j.1460-2075.1995.tb00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 13.Murnane JP, Sabatier L, Marder BA, Morgan WF. Telomere dynamics in an immortal human cell line. EMBO J. 1994;13:4953–4962. doi: 10.1002/j.1460-2075.1994.tb06822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 15.Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 17.Gravel S, Larrivée M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 18.Boulton SJ, Jackson SP. Components of the Ku-dependeant non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tlsty TD. Genomic instability and its role in neoplasia. Curr Top Microbiol Immunol. 1997;221:37–46. doi: 10.1007/978-3-642-60505-5_4. [DOI] [PubMed] [Google Scholar]
- 20.Cheng KC, Loeb LA. Genomic instability and tumor progression: mechanistic considerations. Adv Cancer Res. 1993;60:121–156. doi: 10.1016/s0065-230x(08)60824-6. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer JR, Goosen LS, Stine KC, Thomas JR. Telomere fusion as a mechanism for the progressive loss of the short arm of chromosome 11 in an anaplastic Wilms' tumor. Cancer. 1994;74:767–773. doi: 10.1002/1097-0142(19940715)74:2<767::aid-cncr2820740232>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 22.Sawyer JR, Roloson GJ, Bell JM, Thomas JR, Teo C, Chadduck WM. Telomeric associations in the progression of chromosome aberrations in pediatric solid tumors. Cancer Genet Cytogenet. 1996;90:1–13. doi: 10.1016/0165-4608(96)00058-1. [DOI] [PubMed] [Google Scholar]
- 23.Sawyer JR, Swanson CM, Lukacs JL, Hassed SJ, Curtis MA, North PE, Kozlowski KJ, Pihoker C. Telomeric fusion and chromosome instability in multiple tissues of a patient with mosaic Ullrich-Turner syndrome. Am J Med Genet. 1997;69:383–387. [PubMed] [Google Scholar]
- 24.Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B. Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood. 1998;91:1732–1741. [PubMed] [Google Scholar]
- 25.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;41:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 27.Smith KA, Stark MB, Gorman PA, Stark GR. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toledo F, Roscouet D, Buttin G, Debatisse M. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melek M, Shippen DE. Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. BioEssays. 1996;18:301–308. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- 31.Murray AW, Claus TE, Szostak JW. Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4642–4650. doi: 10.1128/mcb.8.11.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 34.Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 35.Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 36.Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, Galanello R, Cao A, Wood WG, Ayyub H, Higgs DR. Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet. 1994;55:505–512. [PMC free article] [PubMed] [Google Scholar]
- 37.Wong AC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE. Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am J Hum Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- 38.Morin GB. Recognition of a chromosome truncation site associated with α-thalassaimia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 39.O'Toole CM, Povey S, Hepburn P, Franks LM. Identity of some human bladder cancer cell lines. Nature. 1983;301:429–430. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- 40.Wigler M, Pellicer A, Silverstein S, Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as a donor. Cell. 1978;14:725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- 41.Hanish JP, Yanowitz JL, De Lange T. Stringent sequence requirements for the formation of human telomeres. Proc Natl Acad Sci USA. 1994;91:8861–8865. doi: 10.1073/pnas.91.19.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murata S, Matsuzaki T, Takai S, Yaoita H, Noda M. A new retroviral vector for detecting mutations and chromosomal instability in mammalian cells. Mutat Res. 1995;334:375–383. doi: 10.1016/0165-1161(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 43.Brisebois JJ, DuBow MS. Selection for spontaneous null mutations in a chromosomally integrated HSV-1 thymidine kinase gene yields deletions and a mutation caused by intragenic illegitimate recombination. Mutat Res. 1993;287:191–205. doi: 10.1016/0027-5107(93)90012-5. [DOI] [PubMed] [Google Scholar]
- 44.Capizzi RL, Jameson JW. A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat Res. 1972;17:147–148. doi: 10.1016/0027-5107(73)90265-0. [DOI] [PubMed] [Google Scholar]
- 45.Murnane JP, Fuller LF, Painter RB. Establishment and characterization of a permanent pSVori--transformed ataxia-telangiectasia cell line. Exp Cell Res. 1985;158:119–126. doi: 10.1016/0014-4827(85)90437-9. [DOI] [PubMed] [Google Scholar]
- 46.Richard CW, III, Withers DA, Meeker TC, Maurer S, Evans GA, Myers RM, Cox DR. A radiation hybrid map of the proximal long arm of human chromosome 11 containing the multiple endocrine neoplasia type 1 (MEN-1) and bcl-1 disease loci. Am J Hum Genet. 1991;49:1189–1196. [PMC free article] [PubMed] [Google Scholar]
- 47.Dutrillaux B, Couturier J. La pratique de l'analysee chromosomique. Paris: Masson; 1981. [Google Scholar]
- 48.Lemieux N, Dutrillaux B, Viegas-Pequignot E. A simple method for simultaneous R- or G-banding and fluorescence in situ hybridization of small single-copy genes. Cytogenet Cell Genet. 1992;59:311–312. doi: 10.1159/000133277. [DOI] [PubMed] [Google Scholar]
- 49.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little M-T, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 50.Barnett MA, Buckle J, Evans EP, Porter ACG, Rout D, Smith AG, Brown WRA. Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farr C, Fantes J, Goodfellow P, Cooke H. Functional reintroduction of human telomeres into mammalian cells. Proc Natl Acad Sci USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprung CN, Bryan TM, Reddel RR, Murnane JP. Normal telomere maintenance in immortal ataxia telangiectasia cell line. Mutat Res. 1997;379:177–184. doi: 10.1016/s0027-5107(97)00119-x. [DOI] [PubMed] [Google Scholar]
- 53.Sprung CN, Sabatier L, Murnane JP. Telomere dynamics in human cancer cell line. Exp Cell Res. 1999;247:29–37. doi: 10.1006/excr.1998.4293. [DOI] [PubMed] [Google Scholar]
- 54.McIlwrath AJ, Vasey PA, Ross GM, Brown R. Cell cycle arrests and radiosensitivity of human tumor cell lines: dependence on wild-type p53 for radiosensitivity. Cancer Res. 1994;54:3718–3722. [PubMed] [Google Scholar]
- 55.Cooper MJ, Haluschak JJ, Johnson D, Schwartz S, Morrison LJ, Lippa M, Hatzivassiliou G, Tan J. p53 mutations in bladder carcinoma cell lines. Oncol Res. 1994;6:569–579. [PubMed] [Google Scholar]
- 56.Murnane JP, Yu L-C. Acquisition of telomere repeat sequences by transfected DNA integrated at the site of a chromosome break. Mol Cell Biol. 1993;13:977–983. doi: 10.1128/mcb.13.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown WRA, MacKinnon PJ, Villasante A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 58.Budarf M, Blackburn E. S1 nuclease sensitivity of a double-stranded telomeric DNA sequence. Nucleic Acids Res. 1987;15:6273–6292. doi: 10.1093/nar/15.15.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meuth M. Illegitimate recombination in mammalian cells. In: Berg DE, Howe MM, editors. Mobile DNA. Washington, DC: American Society for Microbiology; 1989. pp. 833–860. [Google Scholar]
- 60.Phillips JW, Morgan WF. Illegitimate recombination induced by DNA double-strand breaks in a mammalian chromosome. Mol Cell Biol. 1994;14:5794–5803. doi: 10.1128/mcb.14.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martens UM, Zijlmans JMJM, Poon SSS, Dragowska W, Yui J, Chavez EA, Ward RK, Lansdorp PM. Short telomeres on human chromosome 17p. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- 62.Spruill MD, Ramsey MJ, Swiger RR, Nath J, Tucker JD. The persistence of aberrrations in mice induced by gamma radiation as measured by chromosome painting. Mutat Res. 1996;356:135–145. doi: 10.1016/0027-5107(95)00218-9. [DOI] [PubMed] [Google Scholar]
- 63.Sprung CN, Afshar G, Chavez EA, Lansdorp P, Sabatier L, Murnane JP. Telomere instability in a human cancer cell line. Mutat Res. 1999;429:209–223. doi: 10.1016/s0027-5107(99)00115-3. [DOI] [PubMed] [Google Scholar]
- 64.Murray AW, Szostak JW. Construction of artificial chromosomes in yeast. Nature. 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- 65.Levis RW. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58:791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 66.Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci USA. 1990;87:1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 68.Biessmann H, Mason JM. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 1988;7:1081–1086. doi: 10.1002/j.1460-2075.1988.tb02916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]