Abstract
We have developed a “comparative growth assay” that complements current assays of drug effects based on cytotoxicity. A co-culture of two cell lines, one of which is fluorescently labeled, is exposed to a cytotoxic agent and the proportion of fluorescent cells is compared with that of a baseline unexposed co-culture. For demonstration purposes, two HCT116 cell lines (an hMLH1 homozygous and an hMLH1 heterozygous mutant), altered by insertion of vector alone or the same vector carrying an insert for the expression of enhanced green fluorescent protein (EGFP), were exposed to numerous “anti-cancer” agents. The assay was further validated in a system of two cell lines differing only in the expression of the breast cancer resistance protein (BRCP). The assay allowed the estimation of the duration of action of a particular agent. Assessment of the agent's differential activity over a given time in culture could be expressed as a selection rate, which we chose to describe on an “average selection per day” basis. We conclude that this assay: 1) provides insight into the differential dynamic effects of chemotherapeutic agents or radiation; and 2) allows, through the use of matched cell lines, the investigation of critical physiologic features that govern cell sensitivity.
Keywords: growth assay, mismatch repair, BCRP, multiagent resistance, chemosensitivity
Introduction
Assaying effects of putative anticancer agents on the in vitro growth and survival of appropriate cell populations represents a critical tool in their pre-clinical evaluation [1,2]. Exposing multiple cell populations simultaneously to a particular agent (in co-culture) provides data complementary to unitary in vitro growth assays. Co-culture experiments have become more logistically straightforward with the recent development of vectors expressing autofluorescent proteins [3], which allow permanent labeling of living cells and their progeny [4]. Thus, it has become practical to grow two or more cell populations side by side in co-culture, expose them simultaneously to anticancer agents, and track their differential growth rate over time. Fink has used co-cultures to model the differential effect of mismatch repair (MMR) proficiency on in vivo [5] and in vitro [6] cytotoxicity of cisplatin. We developed the co-culture technique into a comparative growth assay and report here its ability to quantify the ability of anticancer agents to select relatively resistant cell populations.
The comparative growth assay was evaluated extensively in a model system of two matched cell lines differing only in a single gene alteration, namely in the presence of the an intact mismatch repair (MMR) hMLH1 gene [7]. The assay was validated in a system of two cell lines differing only in the expression of the breast cancer resistance protein (BRCP) [8]. The hMLH1 gene belongs to the DNA MMR system [9,10] consisting of hMSH2, hMLH1, hPMS1, hPMS2, and GTBP (hMSH6) proteins which correct, on average, 99.9% of all mismatches occurring in the newly synthesized strand [11]. A functional deficiency of the MMR system, found in 5% to 40% of primary and metastatic human cancers [12–15] and in most cancers of patients with the hereditary nonpolyposis colon cancer (HNPCC) syndrome, induces a resistance, not only toward alkylating agents as originally proposed [7,16], but to a wide variety of other common anticancer therapies as well [17–22]. Comparing the effect of standard agents used for the treatment of solid tumors on two colorectal cell lines that differ in nucleotide MMR competence thus offers a distinct test of the comparative growth assay. The BRCP is a recently described adenosine triphosphate-binding cassette halftransporter associated with resistance to mitoxantrone and anthracyclines [8,23]. Comparing the effect of mitoxantrone on two breast cancer cell lines differing in the expression of BCRP offers an independent validation of this assay.
Materials and Methods
Cell Lines
Two sublines of HCT 116 (ATCC no. CCL-247), a poorly differentiated human colon cancer cell line which is MMR-deficient due to a homozygous mutation of the hMLH1 gene located on chromosome 3 [7], carrying intact p53 and p21 genes [24], a mutated ras oncogene [25], and a mutated TGF-βRII gene [26], producing CEA [27], TGF-α [28], TGF-β1, and TGF-β2 [29], were used throughout all experiments. One of the sublines, HCT 116+ch3 (“N”), has its MMR deficiency corrected by introducing a single human fibroblast chromosome 3 tagged with a neomycin resistance gene in pSV2 and containing the hMLH1 gene (rendering it hMLH1±). Consequently, the TGF-βRII gene residing on chromosome 3 is in this subline, also ±. The other subline was left uncorrected by transferring a similarly neoresistant chromosome 2 (HCT 116+ch2=“O”) [7]. The cells were grown in McCoy's 5A modified media (Gibco BRL, Gaithersburg) with 10% fetal bovine serum (Gibco BRL) (=“complete media”) and 200 µg/ml of the neomycin equivalent Geneticin (Gibco BRL). Geneticin assured the continuous presence of the transferred chromosomes 2 and 3, respectively. The cells were split 1:20 once a week. After every fourth weekly passage, a new frozen vial was thawed to avoid selection of particular sublines.
Vectors
The vector pcDNA3.1/Zeo(+) (Invitrogen Corporation, Carlsbad, CA), equipped with a CMV promoter/enhancer and a bleomycin family (Zeocin) resistance gene encoding a 13-kDa protein which binds to Zeocin, thus preventing it from binding to DNA, was used for all constructs. The source of the enhanced green fluorescent protein (EGFP) gene [30] was the pEGFP-C1 vector (Clontech, Palo Alto, CA). Constructs containing a (CA)12 repeat preceding the EGFP gene (pcDNA3.1-(CA)12-EGFP) yielded stable fluorescent EGFP transfectants with the highest frequency (data not shown).
Construction of Control and Fluorescent Cell Lines
To obtain Zeocin-resistant cell lines, transfections were done with pcDNA3.1. To obtain fluorescent and Zeocin-resistant cell lines, cells were transfected with pcDNA3.1-(CA)12-EGFP. In this way, Zeocin-resistant fluorescent (O+) and nonfluorescent (O) HCT116+chromosome 2, and fluorescent (N+) and nonfluorescent (N) HCT116+chromosome 3 cell lines were generated. For transfections, plasmids pcDNA3.1 orpcDNA3.1-(CA)12-EGFP were linearized in the unique Pvul site of the Amp resistance gene, mixed with the LipofectAmin reagent (Gibco BRL) and transfected according to manufacturer's instructions. A Southern blot analysis using a random-primed EGFP probe has shown the presence of single EGFP integrations with one or very few copies per integration in both fluorescent lines (not shown).
BCRP Cell Lines
To evaluate the effect of the BCRP on mitoxantrone resistance, double-transfected MCF-7 cell lines were used. Cell lines supposed to overexpress the BCRP were transfected with a pcDNA3 vector containing the BCRP-clone 8 [8] and a Geneticin resistance gene (B), whereas sensitive cells were transfected with the plasmid lacking BCRP (V). Each cell line was then transfected either with a plasmid expressing the EGFP in pcDNA3.1/Zeo (+) or with the plasmid not containing EGFP. Thus, four different cell lines were created (B+, V+, B, V).
The Assay
The comparative growth assay (Figure 1) was designed to enable the evaluation of the relative sensitivity of different cell lines to growth-retarding agents. As a model, the relative sensitivity to antitumor agents of two sublines (O and N) of the HCT116 colon cancer cell line differing mainly in one primary genotypic variable, the expression of a functioning hMLH1 gene, was evaluated. To assure an absolute identity of conditions for both of the tested sublines, the sublines to be compared were grown in co-culture during the experiment. To be able to distinguish cells belonging to one cell line from cells belonging to the other one, cells of one subline were fluorescently marked (+) by the expressed product of a stably integrated EGFP gene. The proportion of cells belonging to the marked cell line could then be evaluated by flow cytometry and expressed as odds marked/unmarked. To focus on the effect of the drug or radiation, odds marked/unmarked in co-culture experiments influenced by the studied agent were compared to the same odds of baseline co-culture experiments in which the cells were exposed to media only — thus yielding an odds ratio. To exclude the possibility of an interaction between the EGFP and genes differentially expressed in the compared cell lines, each experiment was additionally performed in an inverted schema in which the originally unlabelled cell line was labeled and vice versa. Co-cultures of the same sublines differing only in the presence of the EGFP (O+/O and N+/N) served as controls for “clonal variation” and EGFP influence on cell line growth. Thus, up to four different kinds of co-cultures were evaluated in each experiment and compared to appropriate unexposed baseline co-cultures, but in every case evaluating the effect of an agent, at least the O+/N and N+/O combinations were tested. Similarly, B+/B and V+/V co-cultures served as controls for the BCRP experiment and V+/B, B+/V co-cultures were used to indicate the differential effect of mitoxantrone.
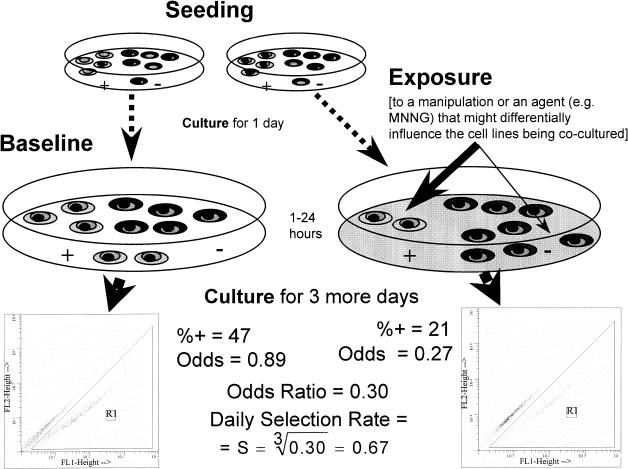
Figure 1.
A schematic depiction of a typical comparative growth assay. The comparative growth assay was designed to measure the differential growth inhibition, or the selective pressure cytotoxic agents exert on two distinct cell populations. Co-cultures of a nonfluorescent cell line (-) mixed with a cell line tagged with a fluorescent marker (+) are seeded. One day later, one of two identical co-cultures is exposed to a study agent and the other one (baseline) is not. After a period of exposure (e.g., 1 hour), the agent is washed away and the co-cultures are grown for 3 more days. The ratio of fluorescent-to-nonfluorescent cells is determined by flow cytometry (insets). If the studied agent is more growth-inhibitory against one cell type than against the other, then the ratio of fluorescent-to-nonfluorescent cells becomes different in the exposed co-culture compared to the ratio in the undisturbed baseline co-culture. The selection rate expressed as an average selection per day is calculated for the given example. The calculation assumes a 3-day observation period (see Appendix A for derivation of this equation).
Cells of each of the four sublines (N, N+, O, and O+) to be examined were grown to subconfluency then washed twice with Ca2+, Mg2+ free PBS, removed from culture dishes by Trypsin-EDTA (Gibco BRL) and strained through a sterile 40 µm cell strainer (Becton Dickinson, Franklin Lakes, NJ) to achieve a single cell suspension. Cell numbers were adjusted and the abovementioned cell line combinations mixed at the intended seeding ratio. One hundred thousand cells of the co-culture in 2 ml of complete media with Zeocin and Geneticin were seeded per well of a six-well plate, or 4x104 cells in 1 ml of complete media with antibiotics were seeded into one well of a 12-well plate (Becton Dickinson). The cells were incubated at 37°C in a 5% CO2 air mixture. After 24 hours, the media were replaced by complete media containing the antibiotics and the drug to be tested and incubated for a specific exposure time. After the exposure time had elapsed, individual wells were washed once with the complete media not containing the tested drug, and grown in the same media for an additional 1 to 4 days. Baseline co-cultures had their control media changed in the same intervals. After the total 2- to 5-day incubation period, cells of both the exposed and baseline co-cultures were washed with PBS and detached by Trypsin-EDTA (Gibco BRL) not containing phenol red. The cells were then strained through a 35-µm strainer cap mounted on top of a 12x75 mm flow tube (Becton Dickinson) and subjected to flow cytometry. Three replicas were used for each co-culture.
Flow Cytometry
Flow cytometry was performed on a FACSort (Becton Dickinson Immunocytometry Systems, San Jose, CA) equipped with a 20-mW Argon laser tuned to 488 nm. Fluorescence amplifiers were in logarithmic mode. The trigger threshold was set on forward scatter. No compensation was used. Five to ten thousand cells of each co-culture were analyzed for every data point. The HP Reader 2.04 program (Verity Software House, Inc., Topsham, ME) facilitated the transfer of “listmode” files to an IBM-compatible computer. Listmode data were analyzed by WinList 3.0 (Verity Software House) under Windows 95. The population of fluorescent cells was distinguished from that of nonfluorescent cells in a correlation diagram with pulse height of green fluorescence plotted against pulse height of red fluorescence. Nonfluorescently transfected healthy as well as dying cells (which were yellow autofluorescent) projected into the diagonal of the scatter diagram distinct from the population of fluorescent EGFP-transfected cells which were more green and less red, thus projecting off the diagonal (Figure 1, insets). In such an arrangement, one EGFP-expressing cell can easily be detected among 10,000 cells not expressing the EGFP (unpublished observation). The proportion of fluorescent cells was recorded as a percentage. These percentages were automatically stored in an Excel (Microsoft Corporation, Redmond, WA) file by Verity Winlist software.
Evaluation
The percentage of fluorescent cells in each co-culture was expressed as the odds of fluorescent versus nonfluorescent cells using the formula odds(f/nf)=%f/(100-%f). The odds value for the co-culture exposed to a drug was divided by the average odds values of unexposed baseline co-cultures yielding an odds ratio. An odds ratio of 1 states that the growth rates relative to each other, of two cell lines exposed to a given drug, do not differ from the relative growth rates obtained when the cell lines were exposed only to the diluent of the given drug (for derivation, see Appendix). Using odds ratios makes the results independent of the seeding ratio (of the two compared cell lines) as long as the same seeding ratio is used in both the exposed and unexposed co-cultures and is in the optimal range. Using odds ratios also allows determination of the duration of the selective effect. In a plot depicting the log of the odds ratios against time, a plateau suggests loss of differential drug effect (Figure 2), whereas a constant slope suggests a continuing differential effect at the evaluated time points (for rationale, see Appendix).
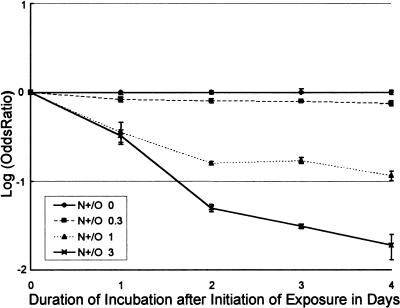
Figure 2.
Log odds ratio plotted against time depicts the duration of the selective effect (N+/O co-culture). To establish the duration of agents' selective effects on two cell lines differing in their MMR proficiency, the logarithm of the ratio of odds of finding a green cell in a N+/O co-culture exposed to MNNG at different concentrations and compared to an unexposed N+/O co-culture was plotted against the MNNG concentration. All co-cultures were exposed to MNNG for 2 hours and further incubated for 48 to 120 hours. The selective effect, evident as a downward slope of the line (see Appendix) lasts for at least 120 hours for the MNNG concentration of 3 µM (triangles) and at least 72 hours for the MNNG concentration of 1 µM (triangles).
For time intervals of sustained continuous differential drug effect, the daily selection rate (S) can be calculated as D root of the odds ratio, where D is the number of days following initial exposure to the tested agent. (S) is a measure of selection expressed as an average selection per day. In cases where a specific agent exerts a constant effect, this measure will be a precise reflection of the selection process. In these cases, computation of this “daily” selection rate allows comparison of the most prolonged experiment with other experiments of shorter durations to a reasonable approximation (Figure 3). In cases where the selection is “discontinuous” (e.g., all the differential effects occur in the first or last day of co-culture), this measure will only represent an average value of the effect over the entire length of the observation period. Using selection rates also creates a framework for comparison of the effect of different agents on the same cell line combinations.
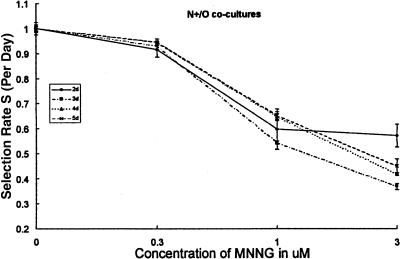
Figure 3.
The comparative growth assay allows for estimates of the differential growth inhibitory effect of an agent that can be independent of the interval since initiation of exposure. Co-cultures of two cell lines, “N” (MMR-competent) and “O” (MMR-defective), differing in the status of their hMLH1 gene were exposed for 1 hour to varying concentrations of MNNG or media containing only diluent. They were subsequently grown in the absence of the cytotoxic agent. N was carrying a vector-expressing EGFP rendering the cell line fluorescent green (N+, see Materials and Methods section). The odds of finding a green cell in the exposed culture was compared to the odds of finding a green cell in the control culture and expressed as an odds ratio. The co-cultures were assayed 2, 3, 4, and 5 days after initiating exposure and the odds of fluorescent/nonfluorescent established. The D (days elapsed since exposure) root of the odds ratio is the selection rate per day, S. The data show selection by MNNG against the MMR-competent fluorescent cell line (N+) (the comparative chance of finding a green cell in the exposed co-culture decreases as the concentration of MNNG increases). S depends on the concentration of MNNG and not on the time of evaluation (at least over the course of this 5-day experiment) with the possible exception of 3 µM concentration of MNNG analyzed at 48 hours following exposure. Using selection rate (S) expressed as an average on a per day basis allows comparisons of experiments done with different follow-up times and reflects the time span of the drug's selective activity.
A selection rate S(O+/N) of 1.06 (=1+0.06) states that the fluorescent O cell line (HCT116+ch2=MMR-deficient) tagged with EGFP (+) grew, on average, 6% per day faster than the nonfluorescent MMR-proficient N cell line. An S(O+/N) of 0.94 (=1-0.06) shows that the tagged proficient cell line grew slower by an average of 6% per day of the experiment. When the dose-effect relationship was examined, the daily selection rates (S) were plotted for each co-culture combination against drug concentrations.
Statistics
It was reassuring to see dose-effect relationships obtained for O+/N co-cultures to be roughly mirror images (or at least mirror “trends”) of those for N+/O co-cultures. The same was true for B+/V and V+/B. Simple t-tests (assuming equal variances) were used to rule out the null hypothesis stating that the ratio of the labeled and unlabeled cells in co-cultures under a given drug concentration has not changed from that of baseline (no drug added).
Drugs Used
Drugs were diluted in either the supplied diluent or in dimethylsulfoxide (Sigma, St. Louis, MO). N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) (1-methyl-3-nitro-1-nitrosoguanidine; Aldrich, Milwaukee, WI) is a carcinogenic DNA alkylator capable of forming O6-methylated purines. Gemcitabine (Eli Lilly, Indianapolis, IN) is a dCyd antagonist that incorporates into DNA, interferes with DNA chain elongation, and inhibits ribonucleotide reductase. 6-Thioguanine [20], a purine antimetabolite which is prone to methylation after incorporation into DNA [20], was purchased from Glaxo Wellcome Co. (Research Triangle Park, NC). The antimetabolite, fludarabine phosphate, was obtained from Berlex Laboratories (Wayne, NJ); 5-fluorouracil from Roche Pharmaceuticals (Nutley, NJ). Doxorubicin hydrochloride, an intercalating topoisomerase II, inhibitor was obtained from Pharmacia, Inc. (Columbus, OH). Etoposide, a nonintercalating topoisomerase II inhibitor, was obtained from Bristol-Myers Squibb (Princetown, NJ). Paclitaxel (Bristol-Myers Squibb) is a microtubule-stabilizing agent. Cisplatin is a bifunctional platinum complex forming intrastrand or interstrand cross-links or adducts (Bristol-Myers Squibb). Irinotecan, a prodrug with a low genuine activity of a topoisomerase I inhibitor, was obtained from Pharmacia and Upjohn (Kalamazoo, MI). Mitoxantrone (Immunex Corp., Seattle, WA), a DNA intercalator, was used in the BCRP experiments.
Radiation Used
The cell lines were irradiated by a Co60 source to the stated dose of 2.5, 5, 7.5, 10, and 12.5 Gy at a dose intensity of 20 Gy/hour.
Results
Establishing the Optimal Seeding Ratio
To optimize the comparative growth assay, the effect of the actual seeding ratio of both cell populations participating in the co-culture was assessed in terms of the ability to follow the growth of each line accurately. The relationship between the reproducibility of measurements and the actual odds (fluorescent versus nonfluorescent) at the time of evaluation was assessed in preliminary experiments with MNNG exposure of a co-culture of fluorescent “O” cells and nonfluorescent “N” cells (O+/N). Variation coefficients of odds of triple replicas were plotted against the average odds (Figure 4). The scatter diagram shows that the lowest variation (1% to 6%) is achieved if the odds fluorescent/nonfluorescent at the time of evaluation by flow cytometry is above 0.18 (demarcated by a dashed line) and less than 0.82. Seeding co-cultures at a ratio of fluorescent cells to nonfluorescent cells of 1:1 turned out to be satisfactory in most cases, as odds at evaluation usually fell within this range. Experiments with long follow-up times sometimes necessitated changing the seeding ratio by initially under-representing the cell population that is more resistant to the studied agent to assure that the odds at evaluation of both the exposed and baseline co-cultures would fall into the range of minimal variation (i.e., there would still be enough measurable cells of the more sensitive cell line).
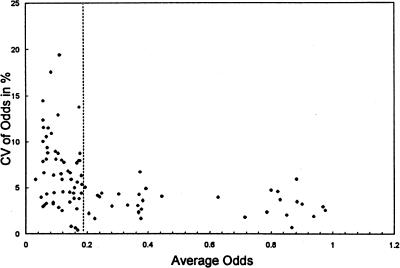
Figure 4.
Establishing the optimal ratio of fluorescent/nonfluorescent cells in co-cultures for reproducible flow cytometry analyses. The comparative growth assay, which is based on co-culture of fluorescent and nonfluorescent cell lines, uses their ratio (expressed as odds) for evaluation. The odds at evaluation are obviously dependent not only on the differential growth rate of the observed cell lines but on the initial (seeding) ratio as well. To establish a range of odds at the time of FACS analyses associated with a low variation, data from preliminary experiments with a 1 hour MNNG exposure of an O+/N co-culture done in triplicates were utilized. The variation coefficient (CV=100x standard deviation÷mean) of triplicate odds was plotted against its average odds. The scatter diagram shows that the lowest variation (1% to 6%) is achieved when the odds fluorescent/nonfluorescent at the time of evaluation by flow cytometry are above 0.18. There was more variation within the measurement if the ratio of the fluorescent cells to the nonfluorescent ones at the time of flow cytometry was below 0.18. As the same variation can be expected in the inverse situation of scarce nonfluorescent cells, the same restriction applies on the upper end of the scale. To generate results with low variation, the experiment has to be set up (by adjusting the seeding ratio) to keep the odds ratio at evaluation in the range of 0.18 to (1-0.18).
Effect of MNNG
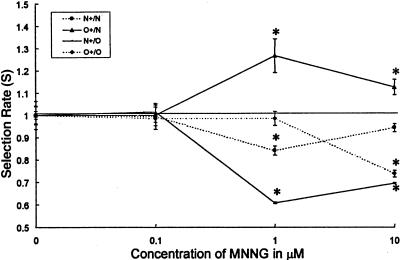
The assay was first tested using MNNG, reported to be less toxic to MMR-deficient than MMR-proficient cells [31]. A 1-hour exposure of MNNG inhibited the growth of MMR-proficient HCT-116+ch3 (N) cells more than that of the MMR-deficient HCT-116+ch2 (O) cells. The dose-effect is shown in a graph in which the estimated average selection rate per day S is plotted against the drug concentration (Figure 5). The lowest MNNG concentration, the effect of which can be documented, is 0.3 µM (data not shown) added to the co-culture for 1 hour and evaluated after 48 to 96 hours. Compared to the baseline unexposed co-culture of O+/N cells, O+/N co-cultures, exposed to MNNG at the concentration of 1 µM for 1 hour and assayed after a 3-day interval incubation, demonstrated a relatively increased survival of O+ cells (by 27% per day) compared to N cells (selection rate S(O+/N) was 1.27). The selection rate decreased with higher concentrations of MNNG; S(O+/N) was 1.12 at 10 µM of MNNG. That the effect is not due to the presence of EGFP is suggested by the inverse (N+/O) co-culture. Under these circumstances, MNNG at 1 µM decreased the relative presence of N+ cells by 39% per day and at 10 µM by 31% per day. However, a possible negative selective interaction of EGFP expression and MNNG is suggested at 1 µM for the N+/N combination and at 10 µM for the O+/O combination. MNNG has been used as quality control in assessing the differential effect of anticancer agents on “N” and “O” cell lines throughout further experiments.
Figure 5.
Dose-effect relationship for MNNG. The differential growth inhibitory effect of MNNG was studied by utilizing the comparative growth assay in which the growth of fluorescently labeled cells (+) is compared with unlabelled cells in the presence or absence (baseline) of the tested agent. Four different co-cultures, two experimental, O+/N, N+/O, and two control, N+/N and O+/O, were exposed for 1 hour to different concentrations of MNNG and compared after 3 days to the same co-cultures which were left untreated. Means of the calculated selection factor averaged per day (S) with 95% confidence limits are plotted against concentrations of MNNG. MNNG favors the growth or survival of MMR-deficient O cells, so that fluorescently tagged O cells become more prevalent in a co-culture (O+/N) (solid line with triangles) as compared to the baseline unexposed co-culture. Inversely fluorescently tagged N cells (N+) decrease in prevalence in a N+/O co-culture (solid line with squares). The prevalence of fluorescent cells in a mix of fluorescent and nonfluorescent MMR-deficient cells (O+/O) (dashed line with triangles) changed orders of magnitude less significantly. The same was true for the prevalence of fluorescent cells in a mix of fluorescent and nonfluorescent cells MMR-competent cells (N+/N) (dashed line with rectangles). MNNG is ineffective at O.1 µM and effective between 1 and 10 µM. At high concentration (10 µM), the differential sensitivity is diminished, as the cytotoxic effect of the agent for each individual cell line is more pronounced. *p<.0001. A possible negative selective interaction of EGFP and MNNG is suggested by the N+/N combination at 1 µM MNNG and the O+/O combination at 10 µM MNNG.
Duration of the Differential Effect
The comparative growth assay can be used to estimate the duration for which a single exposure to a studied drug exerts its differential growth effect on the co-culture. A compound that exerts a constant selective pressure on the co-culture yields a straight line in a log (odds ratio) versus time plot. If the compound favors the unlabeled cell line, the plot has a downward slope. The time point in which the selective pressure has ceased is evident in such a plot as the point at which the slope becomes parallel with the x-axis (for rationale, see Appendix). Using this feature of the comparative growth assay, we could show, e.g., that the selective effect of a 2-hour exposure to 1 µM MNNG lasts at least 72 hours but for a 3-µM concentration, it appears to last for at least 120 hours (Figure 2).
6-Thioguanine and Cisplatin
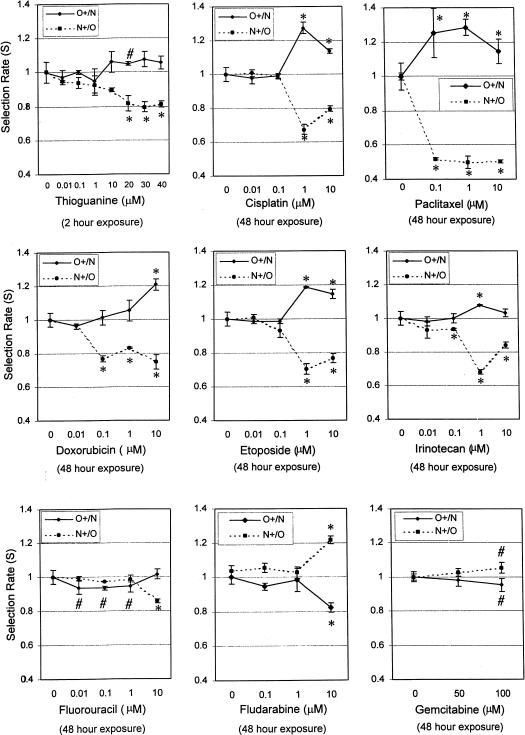
6-Thioguanine present in the co-culture for 2 hours was more toxic for the MMR-proficient cell line (N) than for the MMR-deficient cell line (O). At 20 µM, O cells were selected for by approximately 5% per day (S(O+/N) was 1.05). S(O+/N) was 1.07 at 30 µM, and 1.06 at 40 µM of 6-thioguanine. N cells were selected against by 19% a day (S(N+/O) was 0.81) at 20 µM of 6-thioguanine. S(N+/O) was 0.80 at 30 µM and 0.81 at 40 µM. The lack of apparent mirror image effect of 6-thioguanine at 20, 30, and 40 µM on S suggests an influence of the co-culture type (O+/N versus N+/O), e.g., an effect of clonal variation or EGFP/6-TG contributory interaction. A 1 hour exposure to cisplatin at a concentration of 10 µM but not 1 µM selected for MMR-deficient cells in the O+/N co-culture by 4% a day and analogously in the N+/O co-culture, it decreased the number of labeled N+ cells by 6% a day (not shown). A 48-hour exposure to cisplatin at a 1 µM concentration selected for O cells by 27% a day in the O+/N combination and by 33% a day in the N+/O co-culture. At a dose of 10 µM, the effect was less prominent with 14% per day selection in the O+/N co-culture and 20% per day in the N+/O co-culture (Figure 6). Thus, our results obtained with the comparative growth assay support the notion that alkylators and cisplatin select for hMLH1-deficient cells.
Figure 6.
Drugs selecting in vitro for MMR-deficient cells and drugs selecting for MMR-proficient cells. The differential growth inhibitory effects against MMR-deficient and proficient cells of cytotoxic agents that are components of common anticancer regimens were investigated by the comparative growth assay. Co-cultures of fluorescently labeled MMR-deficient O cells and unlabelled MMR-proficient N cells (O+/N, continuous line) and vice versa (N+/O, dotted line) were exposed to 6-thioguanine for 2 hours at the indicated concentrations and to all other drugs for 48 hours. Assay times varied between 48 and 72 hours. The selection factor averaged per day (S) with 95% confidence limits was plotted against the drug concentration. Drugs selecting for MMR-deficient cells: MNNG and 6-thioguanine were evaluated after a 1-hour incubation and cisplatin, doxorubicin, paclitaxel, etoposide, irinotecan, 5-fluorouracil evaluated after a 48-hour exposure. Drugs selecting for MMR-deficient cells: Fludarabine (rectangles) at a concentration of 10 µM and gemcitabine (triangles) at concentrations of 100 µM. *p<.005; #p<.05.
Other Agents Selecting for Mismatch-Repair-Deficient Cells
Representative nonalkylating cytotoxic drugs were examined in the comparative growth assay for differential effect on the growth of MMR-deficient (O) and proficient (N) cells. A 1-hour exposure to etoposide at a concentration of 10 µM evaluated after 72 hours demonstrated a selection for MMR deficient cells, S(O+/N)=1.05 and S(N+/O)=0.94 (p<.002) (not shown). Doxorubicin, etoposide, paclitaxel, and irinotecan, each individually exposed for 48 hours at concentrations of 1 and 10 µM, increased the prevalence of MMR-deficient cells evaluated after 72 hours in the O+/N co-cultures and decreased the prevalence of mismatch proficient cells in N+/O co-cultures (Figure 6). The selective effect for doxorubicin was most pronounced at 10 µM (S(O+/N)=1.21; S(N+/O)=0.75), for etoposide at 1 µM (S(O+/N)=1.19; S(N+/O)=0.70) for paclitaxel at 1 µM (S(O+/N)=1.29; S(N+/O)=0.50). and for irinotecan at 1 µM (S(O+/N)=1.08; S(N+/O)=0.68). Fluorouracil, at evaluated concentrations and a 48-hour exposure, was to a lesser extent (see 10 µM concentration) toxic to MMR-proficient cells. The comparative growth assay showed no mirror image trend, crossing of the dose-effect curves obtained with incremental drug concentrations for the two co-cultures, and a highly significant difference from baseline only for the N+/O combination at the highest concentration tested (Figure 6).
Drugs Selecting Against MMR-Deficient Cells
Fludarabine and gemcitabine had a marginal effect in preferentially slowing down the growth of MMR-deficient cells. Fludarabine, at a concentration of 10 µM and a 48-hour exposure, selected against the MMR-deficient cells by an estimated 18% a day in the O+/N co-culture and for MMR-proficient cell by 21% a day in the N+/O co-culture (p<.001). Gemcitabine, at 100 µM for a 48-hour exposure, induced a growth advantage for MMR-proficient cells of 5% a day, S(N+/O)=1.05, S(O+/N)=0.95 (p<.05) (Figure 6).
Effect of Ionizing Radiation
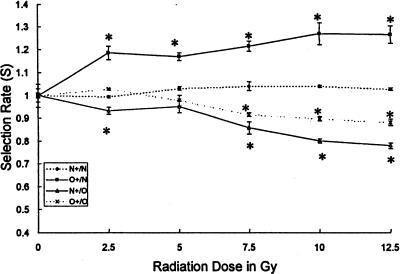
Gamma irradiation slowed the growth of the population of MMR-proficient cells significantly more than that of MMR-deficient cells. The selective advantage of MMR-deficient cells estimated on an average per day basis was, at the lowest radiation dose used (2.5 Gy), 18% per day in the O+/N combination and 7% per day in the N+/O combination. It reached 27% per day and 20% per day at the dose of 10 Gy. The differential dose-related effect thus increased with the dose used (Figure 7). The differential effect exerted by radiation at increasing doses was highly statistically significant (p<.001).
Figure 7.
Differential effect of radiation on co-cultures of MMR-deficient (O) and MMR-proficient (N) cell lines. The differential effect of radiation on a MMR-deficient and a MMR-proficient cell line was determined by the comparative growth assay. The cell lines were cultured for 72 hours after the radiation exposure. The selection rate is shown plotted against the radiation dose. Control co-cultures (dashed lines), O+/N co-culture (rectangles), N+/O co-culture (triangles). A selection for MMR-deficient cells is observed with irradiation. A dose-effect relationship is seen. *p<.0001.
Effect of BCRP on Mitoxantrone Resistance
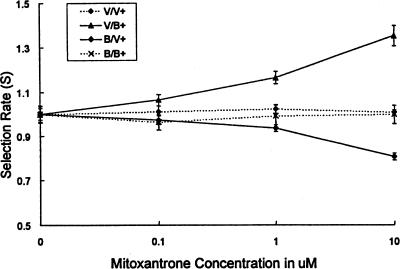
To verify the validity of the comparative growth assay in a different system, MCF-7 breast cancer cell lines made to overexpress BCRP (B) were compared to vector control lines (V) in the comparative growth assay. Mitoxantrone at concentrations 0.1, 1, and 10 nM was present for 48 hours of co-culture (Figure 8). Mitoxantrone at concentrations of 1 (p<.01) and 10 (p<.001) mM selected for BCRP-transfected fluorescent cells and against vector-transfected fluorescent cells, thus supporting the notion that BCRP is involved in mitoxantrone resistance.
Figure 8.
Relative resistance to mitoxantrone provided by BCRP overexpression. Using a quadruplet of cell lines either overexpressing the breast cancer resistance protein (B) or transfected with a control vector only (V), made either to express the EGFP (+) or no fluorescent proteins, differential toxicity to mitoxantrone was studied. Whereas control combinations not differing in the expression of BCRP show no selection for green cells by mitoxantrone, green cells overexpressing BCRP (B+) are selected for, and green cells transfected with the vector only are selected against (V+).
Discussion
Overview
The comparative growth assay was developed to enable the examination of the differential effects cytotoxic agents exert on cell populations or cell lines differing in a clearly defined trait often driven by the expression or mutation of a single gene. Advantage was taken of the ability to stably label one of the cell lines with the green fluorescent protein and thus recognize its progeny in co-culture. The assay is complementary and supplementary to clonogenic, cytotoxic dye exclusion, and metabolic assays. The sensitivity of the assay is due to uniformity of experimental conditions: both cell types are exposed together in the same cultures with one unlabeled cell line contrasted with a fluorescently labeled cell line. The resolution between the fluorescent and nonfluorescent cell lines is provided by the very high intensity of fluorescence of EGFP-marked cells and the wavelength of the emitted fluorescent light (green) which is easily distinguishable by flow cytometry from the autofluorescence of sick or dying cells (yellow) (unpublished observation).
Using odds instead of percentages streamlined all calculations. Using an odds ratio (the ratio of odds of finding a green cell in the exposed co-culture to the same odds in the unexposed co-culture) offers the additional advantage of making the assay relatively independent of the seeding ratio and allowing for an estimate of the duration of the differential effect in an odds ratio time plot. For example, in our model of matched cell lines differing predominantly in expression of the hMLH1 gene, the differential effect of a 1-hour exposure to MNNG lasts from 3 to 5 days, depending on the concentration of MNNG.
The direct relationship between the odds ratio and the estimated average selection rate (S) (for experiments with sustained differential effect for the whole duration of the experiment) facilitated: 1) the comparison of experiments with different incubation times (Figure 3), and 2) presentation of the results in a potentially clinically meaningful way. In comparison to other published assays, greater sensitivity was achieved. For example, differential effects of MNNG and cisplatin could be easily documented at one order of magnitude lower concentrations than in published reports [31].
Overexpression of EGFP itself might change the growth rate of the fluorescently marked sublines. This would not change the results of the assay as odds ratios with baseline co-cultures are evaluated. To evaluate the possibility that the expression of the EGFP protein specifically changes drug sensitivity of tested cell lines in the context of hMLH1 presence or absence, the assay was also done in mirror image in which the originally labeled cell line is unlabeled and vice versa. The lack of a precise mirror image between the results obtained for O+/N and N+/O co-cultures exposed to some drugs might be due to random variation, evaluation at less then optimal conditions (odds not in the interval of 0.18–0.82), and additional mutations that occurred during the selection of GFP expressing sublines. Performing the assay with identical subline co-culture combinations O+/O and N+/N also provides information relevant to interpretation of EGFP effects and clonal variation.
The validation of the comparative growth assay in mitoxantrone-treated co-cultures containing BCRP-overexpressing cells points toward the general utility of the assay.
MMR and Sensitivity to Chemotherapeutic Agents and Radiation
As an initial test for the ability of this assay to distinguish between the responses of different cell types placed in co-culture, we investigated the response to some current standard chemotherapeutic agents and ionizing radiation of two colorectal cancer cell lines that differ primarily in a single gene function (hMLH1) that defines one pathway of malignant transformation. Thus, the interplay of the action of different cytotoxic drugs with MMR deficiency could be studied in this model and compared to data generated by other laboratories often using the same cell lines but different assays. About 15% of primary sporadic colorectal tumors [13,32–40] and most tumors arising in patients with HNPCC demonstrate MMR deficiency [12,41]. In this assay, most cytotoxic drugs believed to cause DNA damage and induce G2-arrest and apoptosis through the MMR system and radiation were more toxic to cells expressing a functional hMLH1 (MMR competent). Two agents, gemcitabine and fludarabine, elicited a modest preferential slowing of the growth of MMR-deficient cells.
Small volume co-cultures of MMR-proficient and -deficient cells exposed to ionizing radiation demonstrate differential radiation sensitivity in a dose-related manner. These features of the comparative growth assay could possibly be used to develop systems for biologic dosimetry.
Possible Implications of the Selection for MMR Deficiency
Our data, as well as data from many other laboratories [5,7,19,42], suggest that cytotoxic agents used in the treatment of malignancies can be divided into three groups: 1) agents that are preferentially cytotoxic for MMR-proficient cells and thus might select for MMR-deficient cells (6-thioguanine [6,20,43], carboplatin, cisplatin [14,22], busulfan, procarbazine, and MNNG [44], temozolomide [44], etoposide [45,46], doxorubicin [18,22,46], irinotecan, paclitaxel, fluorouracil, and ionizing radiation [47]); 2) agents that are neutral (oxaliplatin [19] and BCNU [44]); and 3) agents that select for MMR-proficient cells (fludarabine and gemcitabine). Whereas our data were derived from the comparative growth assay, other studies used clonogenic assays [19], established the MMR status of cell lines selected for resistance to different agents [44], or used animal models [5].
While we did not attempt, in this initial study, to duplicate particular chemotherapeutic regimens in terms of dose intensity or duration, if selection for MMR-defective cells were to occur at the clinical level, it would have clear implications for the design of drug combinations for front line therapy, adjuvant therapy, and salvage therapy. So far, few clinical findings support the prediction that MMR deficiency contributes to chemotherapy and radiotherapy resistance and that recurrent tumors following such therapy are more often MMR-deficient than primary tumors. The replication error (RER) status [48] of recurrent tumors following radiotherapy or chemotherapy with a selective agent compared with the RER status of the primary tumor has not been extensively reported even though such a study is in progress for a specific malignancy [49]. A three-fold, though statistically insignificant, increase in the proportion of hMLH1-defective ovarian tumors was observed in samples taken at second look laparotomy after chemotherapy compared to samples taken before chemotherapy [14]. Decreased expression of MMR genes was found by reverse transcription polymerase chain reaction in the majority of gliomas [50]. While in this study we have looked at the effect of a number of standard agents, an equal utility of this assay might be found in the screening for “lead” compounds that demonstrate a profound differential effect on the growth of MMR-defective versus MMR-competent cells (or on any other cell combinations that differ in a definable and clinically relevant way).
Furthermore, while we have demonstrated the utility of this assay on cell lines differing in their ability to recognize and repair nucleotide mismatches and in cell lines differing in the expression of a drug transporter protein, we believe that this assay is well-suited for assaying different responsiveness to cytotoxic agents of any two cell lines differing in a clearly defined way.
Acknowledgements
We thank Bert Vogelstein for the HCT116+ch2 and HCT116+ch3 cell lines that were prepared by Koi in Boland's laboratory [7]. We thank L. Austin Doyle and Douglas D. Ross for the gift of BCRP-expressing and control cell lines. We thank Allen Chen for irradiating co-cultures of the experiment.
Abbreviations
- MMR
mismatch repair
- BCRP
breast cancer resistance protein
Appendix
Let us assume that two populations of cells, one white (nonfluorescent) with white progeny and the other fluorescently marked (green with green progeny), grown under identical conditions in co-culture. The number of green cells, NGg, after Gg generations of green cell division can be written as:
and the number of white cells, NGw, after Gw generations of white cell division, as:
where N0 is the total number of cells plated, Pg and Pw are the proportions of green and white cells, respectively, at seeding, and fg and fw are the fractions of green and white cells, respectively, that are undergoing cell replication. The odds of finding a green cell in a co-culture are then:
The odds of finding a green cell in an unexposed (baseline) co-culture can be written as:
where fgb is the growth fraction of green cells and fwb is the growth fraction of white cells under baseline (b) conditions. The odds of finding a green cell in the coculture exposed (e) to the agent of interest is similarly given by:
assuming (for computational simplicity) that the fraction of cycling cells rather than the generation time is changing under the influence of the drug.
The odds ratio (OR) exposure/baseline, i.e., the ratio of the odds of finding a green cell in the co-culture exposed to the studied agent compared to the odds of finding a green cell in the uninfluenced baseline co-culture, is:
The odds ratio is obviously independent of the seeding ratio (Pg/Pw) if both the baseline and exposed co-cultures are seeded at the same seeding ratio (with the same cell mix).
One can assume, for computational convenience, that:
as any deviation from this assumption is accommodated for in the f factors. It follows then that:
where f is the ratio of the drug effect exerted on green cells (fge/fgb) to that exerted on white cells (fwe/fwb), i.e., the drug-induced selection factor per generation.
As the number of generations is a linear function of time:
where t is time, it can bewritten:
where S is the selection rate, and for time given in days (t=D), the selection rate's unit is selection per day:
If a given drug exerts its effect evenly over the whole observation time, an assumption that can only be validated by direct observation and plotting as described below, then selection averaged over a period of time t(St) is independent of the number of days of observation.
In the absence of any differential effect of the tested agent against green and white cells:
and therefore:
Depiction of the duration of the selective effect is achieved by plotting log(OR) against time. As:
log(S) is the slope of the plotted line. It becomes horizontal (log(1)=0) when the selection effect ceases to exists.
The generation time might differ for different cell populations. This does not create a fundamental obstacle as the growth fraction of the individual cell populations can be altered in (f)G to reflect changes in G in order to keep the calculation limited to a single variable (f).
Estimation of the selection rate (S) (expressed as selection per day) from experimental data is straightforward. First, odds of drug exposed and baseline co-cultures are calculated from percentages of fluorescent cells according to the formula:
then the odds ratio is calculated as the ratio of the odds of the exposed co-culture and the odds of the baseline co-culture. The daily selection rate (S) is calculated as the “D” root of the odds ratio, where D is the time after initiation of exposure in days.
References
- 1.Bosanquet AG, Burlton AR, Bell PB, Harris AL. Ex vivo cytotoxic drug evaluation by DiSC assay to expedite identification of clinical targets: results with 8-chloro-cAMP. Br J Cancer. 1997;76:511–518. doi: 10.1038/bjc.1997.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinstein JN, Myers TG, PM OC, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, Buolamwini JK, van Osdol WW, Monks AP, Scudiero DA, Sausville EA, Zaharevitz DW, Bunow B, Viswanadhan VN, Johnson GS, Wittes RE, Paull KD. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Prasher DC. Using GFP to see the light. Trends Genet. 1995;11:320–323. doi: 10.1016/s0168-9525(00)89090-3. [DOI] [PubMed] [Google Scholar]
- 5.Fink D, Zheng H, Nebel S, Norris PS, Aebi S, Lin TP, Nehme A, Christen RD, Haas M, MacLeod CL, Howell SB. In vitro and in vivo resistance to cisplatin in cells that have lost DNA mismatch repair. Cancer Res. 1997;57:1841–1845. [PubMed] [Google Scholar]
- 6.Fink D, Nebel S, Norris PS, Aebi S, Kim HK, Haas M, Howell SB. The effect of different chemotherapeutic agents on the enrichment of DNA mismatch repair-deficient tumour cells. Br J Cancer. 1998;77:703–708. doi: 10.1038/bjc.1998.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 8.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 10.Modrich P. Mismatch repair, genetic stability and tumour avoidance. Philos Trans R Soc London B Biol Sci. 1995;347:89–95. doi: 10.1098/rstb.1995.0014. [DOI] [PubMed] [Google Scholar]
- 11.Fishel R, Kolodner RD. Identification of mismatch repair genes and their role in the development of cancer. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 12.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 13.Liu B, Nicolaides NC, Markowitz S, Willson JK, Parsons RE, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton SR, et al. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 14.Brown R, Hirst GL, Gallagher WM, McIlwrath AJ, Margison GP, van der Zee AG, Anthoney DA. hMLH1 expression and cellular responses of ovarian tumour cells to treatment with cytotoxic anticancer agents. Oncogene. 1997;15:45–52. doi: 10.1038/sj.onc.1201167. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaides NC, Littman SJ, Modrich P, Kinzler KW, Vogelstein B. A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol Cell Biol. 1998;18:1635–1641. doi: 10.1128/mcb.18.3.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci USA. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, Christen RD, Boland CR, Koi M, Fishel R, Howell SB. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 18.Drummond JT, Anthoney A, Brown R, Modrich P. Cisplatin and adriamycin resistance are associated with MutLalpha and mismatch repair deficiency in an ovarian tumor cell line. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 19.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 20.Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng Q, Edwards M, Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 21.Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem Biol. 1996;3:579–589. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 22.Anthoney DA, McIlwrath AJ, Gallagher WM, Edlin AR, Brown R. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res. 1996;56:1374–1381. [PubMed] [Google Scholar]
- 23.Ross DD, Yang W, Abruzzo LV, Dalton WS, Schneider E, Lage H, Dietel M, Greenberger L, Cole SP, Doyle LA. Atypical multidrug resistance: breast cancer resistance protein messenger RNA expression in mitoxantrone-selected cell lines. J Natl Cancer Inst. 1999;91:429–433. doi: 10.1093/jnci/91.5.429. [DOI] [PubMed] [Google Scholar]
- 24.Take Y, Kumano M, Teraoka H, Nishimura S, Okuyama A. DNA-dependent protein kinase inhibitor (OK-1035) suppresses p21 expression in HCT116 cells containing wild-type p53 induced by adriamycin. Biochem Biophys Res Commun. 1996;221:207–212. doi: 10.1006/bbrc.1996.0575. [DOI] [PubMed] [Google Scholar]
- 25.Nagasu T, Yoshimatsu K, Rowell C, Lewis MD, Garcia AM. Inhibition of human tumor xenograft growth by treatment with the farnesyl transferase inhibitor B956. Cancer Res. 1995;55:5310–5314. [PubMed] [Google Scholar]
- 26.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 27.Chakrabarty S, Chen JW, Chen XY, Trujillo JM, Lin PF. Modulation of differentiation-related responses in human colon carcinoma cells by protein kinase inhibitor H-7. Anticancer Res. 1992;12:97–104. [PubMed] [Google Scholar]
- 28.Zorbas MA, Yeoman LC. Growth control in a human colon carcinoma cell line mediated by cell- associated transforming growth factor-alpha (TGF alpha) Exp Cell Res. 1993;206:49–57. doi: 10.1006/excr.1993.1119. [DOI] [PubMed] [Google Scholar]
- 29.Sun L, Wu S, Coleman K, Fields KC, Humphrey LE, Brattain MG. Autocrine transforming growth factor-beta 1 and beta 2 expression is increased by cell crowding and quiescence in colon carcinoma cells. Exp Cell Res. 1994;214:215–224. doi: 10.1006/excr.1994.1251. [DOI] [PubMed] [Google Scholar]
- 30.Zhang G, Gurtu V, Kain SR. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun. 1996;227:707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- 31.Carethers JM, Hawn MT, Chauhan DP, Luce MC, Marra G, Koi M, Boland CR. Competency in mismatch repair prohibits clonal expansion of cancer cells treated with N-methyl-N′-nitro-N-nitrosoguanidine. J Clin Invest. 1996;98:199–206. doi: 10.1172/JCI118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merlo A, Mabry M, Gabrielson E, Vollmer R, Baylin SB, Sidransky D. Frequent microsatellite instability in primary small cell lung cancer. Cancer Res. 1994;54:2098–2101. [PubMed] [Google Scholar]
- 33.Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 34.Ishwad CS, Ferrell RE, Rossie KM, Appel BN, Johnson JT, Myers EN, Law JC, Srivastava S, Gollin SM. Microsatellite instability in oral cancer. Int J Cancer. 1995;64:332–335. doi: 10.1002/ijc.2910640509. [DOI] [PubMed] [Google Scholar]
- 35.Huddart RA, Wooster R, Horwich A, Cooper CS. Microsatellite instability in human testicular germ cell tumours. Br J Cancer. 1995;72:642–645. doi: 10.1038/bjc.1995.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Farrington SM, Petersen GM, Hamilton SR, Parsons R, Papadopoulos N, Fujiwara T, Jen J, Kinzler KW, Wyllie AH, et al. Genetic instability occurs in the majority of young patients with colorectal cancer. Nat Med. 1995;1:348–352. doi: 10.1038/nm0495-348. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki H, Komiya A, Aida S, Akimoto S, Shiraishi T, Yatani R, Igarashi T, Shimazaki J. Microsatellite instability and other molecular abnormalities in human prostate cancer. Jpn J Cancer Res. 1995;86:956–961. doi: 10.1111/j.1349-7006.1995.tb03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herfarth KK, Kodner IJ, Whelan AJ, Ivanovich JL, Bracamontes JR, Wells SA, Jr, Goodfellow PJ. Mutations in MLH1 are more frequent than in MSH2 in sporadic colorectal cancers with microsatellite instability. Genes Chromosomes Cancer. 1997;18:42–49. doi: 10.1002/(sici)1098-2264(199701)18:1<42::aid-gcc5>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 39.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 40.King BL, Peng HQ, Goss P, Huan S, Branson D, Kacinski BM, Hogg D. Repeat expansion detection analysis of (CAG)n tracts in tumor cell lines, testicular tumors, and testicular cancer families. Cancer Res. 1997;57:209–214. [PubMed] [Google Scholar]
- 41.Peltomaki P, Lothe RA, Aaltonen LA, Pylkkanen L, Nystrom-Lahti M, Seruca R, David L, Holm R, Ryberg D, Haugen A, et al. Microsatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 42.Holzman D. Mismatch repair genes matched to several new roles in cancer. J Natl Cancer Inst. 1996;88:950–951. doi: 10.1093/jnci/88.14.950. [DOI] [PubMed] [Google Scholar]
- 43.Davis TW, Wilson-Van Patten C, Meyers M, Kunugi KA, Cuthill S, Reznikoff C, Garces C, Boland CR, Kinsella TJ, Fishel R, Boothman DA. Defective expression of the DNA mismatch repair protein, MLH1, alters G2-M cell cycle checkpoint arrest following ionizing radiation. Cancer Res. 1998;58:767–778. [PubMed] [Google Scholar]
- 44.Friedman HS, Johnson SP, Dong Q, Schold SC, Rasheed BK, Bigner SH, Ali-Osman F, Dolan E, Colvin OM, Houghton P, Germain G, Drummond JT, Keir S, Marcelli S, Bigner DD, Modrich P. Methylator resistance mediated by mismatch repair deficiency in a glioblastoma multiforme xenograft. Cancer Res. 1997;57:2933–2936. [PubMed] [Google Scholar]
- 45.Shellard SA, Hosking LK, Hill BT. Characterisation of the unusual expression of cross-resistance to cisplatin in a series of etoposide-selected resistant sublines of the SuSa testicular teratoma cell line. Biochem Pharmacol. 1994;47:775–779. doi: 10.1016/0006-2952(94)90476-6. [DOI] [PubMed] [Google Scholar]
- 46.Hamaguchi K, Godwin AK, Yakushiji M, PJ OD, Ozols RF, Hamilton TC. Cross-resistance to diverse drugs is associated with primary cisplatin resistance in ovarian cancer cell lines. Cancer Res. 1993;53:5225–5232. [PubMed] [Google Scholar]
- 47.Fritzell JA, Narayanan L, Baker SM, Bronner CE, Andrew SE, Prolla TA, Bradley A, Jirik FR, Liskay RM, Glazer PM. Role of DNA mismatch repair in the cytotoxicity of ionizing radiation. Cancer Res. 1997;57:5143–5147. [PubMed] [Google Scholar]
- 48.Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 49.Kaye SB. Ovarian cancer, from the laboratory to the clinic: challenges for the future. Ann Oncol. 1996;7:9–13. doi: 10.1093/oxfordjournals.annonc.a010488. [DOI] [PubMed] [Google Scholar]
- 50.Wei Q, Bondy ML, Mao L, Gaun Y, Cheng L, Cunningham J, Fan Y, Bruner JM, Yung WK, Levin VA, Kyritsis AP. Reduced expression of mismatch repair genes measured by multiplex reverse transcription-polymerase chain reaction in human gliomas. Cancer Res. 1997;57:1673–1677. [PubMed] [Google Scholar]