Abstract
Non-invasive imaging of gene expression opens new prospects for the study of transgenic animals and the implementation of genetically based therapies in patients. We have sought to establish a general paradigm to enable whole body non-invasive imaging of any transgene. We show that the expression and imaging of HSV1-tk (a marker gene) can be used to monitor the expression of the LacZ gene (a second gene) under the transcriptional control of a single promoter within a bicistronic unit that includes a type II internal ribosomal entry site. In cells bearing a single copy of the vector, the expression of the two genes is proportional and constant, both in vitro and in vivo. We demonstrate that non-invasive imaging of HSV1-tk gene accurately reflects the topology and activity of the other cis-linked transgene.
Keywords: gene therapy, gene imaging, tumor, HSV1-tk, IRES
Introduction
The investigation of genetically based therapies in patients and transgenic animal models relies heavily on the accurate localization and measurement of transgene activity. The direct analysis of transgene expression typically requires invasive procedures based on tissue sampling to examine and quantitate levels of transgene products. The topology of active transgenes can only be determined through systematic analyses of multiple tissues. Thus, marker gene studies that seek to determine the localization and level of transgene expression in transduced cells and tissues require serial organ harvest, biopsies or blood collection. A complete analysis of all tissues in the body is seldom performed. As yet, there are no general non-invasive methods for quantitative imaging transgene activity in patients undergoing genetically based treatments or in transgenic animals, and tissue-sampling techniques to assess the extent of gene transfer and the pattern of transgene expression are laborious and time consuming. The ability to image transgene activity would greatly facilitate the evaluation and monitoring of gene transfer in human subjects by defining the location, magnitude, and persistence of gene expression over time.
We and others have recently demonstrated that imaging marker gene expression is feasible using quantitative autoradiography, gamma camera, single photon emission tomography and positron emission tomography techniques [1–6]. This was accomplished in an animal model using herpes simplex virus 1 thymidine kinase gene (HSV1-tk) as the marker (reporter) gene. We have shown that tumors retrovirally transduced with HSV1-tk can be imaged following i.v. administration of radiolabeled 5-iodo-2′-fluoro-2′-deoxy-1-β-d-arabino-furanosyl-uracil (FIAU). FIAU is efficiently phosphorylated by the HSV1-tk enzyme, but not by endogenous mammalian thymidine kinase. This results in the selective trapping of FIAU in HSV1-tk-transduced cells. We showed previously that the magnitude of FIAU accumulation in transduced cells is proportional to the level of HSV1-tk mRNA expression and to their sensitivity to the anti-viral drug ganciclovir [1]. We also showed proportional relationships between these independent measures of HSV1-tk expression and the level of radio-iodinated FIAU accumulation measured in vivo by non-invasive gamma camera imaging and positron emission tomography (PET) [2,3].
Most gene products, however, lack appropriate ligands or substrates that can be radiolabeled for imaging purposes. Therefore, it is necessary to resort to indirect imaging strategies. In this report, we describe and validate a paradigm for monitoring the expression of any gene by non-invasive imaging of a cis-linked marker (reporter) gene. This strategy is based on demonstrating that a constant relationship in the coexpression of two genes is established. For this purpose, we investigated whether an internal ribosomal entry site (IRES) element within a single bicistronic transcription unit allows for coexpression that is reliable and quantitative in the context of in vivo imaging. The IRES element enables translation initiation within the bicistronic mRNA, thus permitting gene coexpression by cap-dependent translation of the first cistron and cap-independent, IRES-mediated translation of the second cistron [7]. We utilized a type II IRES element derived from the 5′ untranslated region of encephalomyocarditis virus (EMCV) because of its apparent lack of tissue-specificity relative to other IRES types [8,9]. When coexpression was carefully examined at the single cell level in retrovirally transduced cells, the EMCV IRES has been shown to confer reliable dual gene expression in certain cell types, including chicken embryo [10], myeloid cells [11], and human T lymphocytes [12].
This is the first report to demonstrate that non-invasive imaging of a reporter gene (HSV1-tk) can provide quantitative as well as topographical information related to the expression of a cis-linked transgene.
Methods
Retroviral Vector STLEO
The retroviral vector STLEO was derived from the vector MoT [13] which encodes HSV1-tk gene under the transcriptional control of the Moloney murine leukemia virus long terminal repeat, by incorporating the LacZ/NeoR fusion gene linked by the IRES sequence of ECMV [12]. The expression of HSV1-tk is cap-dependent and that of the LacZ/NeoR fusion gene is IRES-mediated and cap-independent. The LacZ/NeoR fusion gene expresses a chimeric protein that retains both β-galactosidase and neomycin phosphotransferase enzymatic activities. Vesicular stomatitis virus G-glycoprotein (VSV-G)-pseudotyped recombinant virions were generated from the gpg29 packaging cell line [14] as previously described [15].
Cell Cultures and Gene Transduction In Vitro
The RG2 rat glioma cells were kindly provided by Dr. D. Bigner (Duke University Medical Center, Durham, NC). The W256 rat mammary carcinoma cells were obtained from American Type Tissue Culture Collection (Manassas, VA). Both cell lines were grown in MEM supplemented with 10% FCS. The STLEO vector was used to transduce these cell lines. The STLEO retroviral vector producer cells were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). A retroviral suspension was obtained from filtered (0.45 µm) supernatants of the vector producer cell line cultures. In vitro transduction of tumor cells was accomplished by exposing cell monolayers to the retroviruses for 8 hours in the presence of polybrene (8 µg/ml). A selection medium containing G418 (800 µg/ml) was used to isolate G418 resistant colonies. After the bulk cultures of transduced cells RG2STLEO and W256STLEO were established, they were maintained in the same culture media containing G418 (250 µg/ml). Multiple single cell clones of RG2STLEO and W256STLEO cells were obtained by terminal dilution in 96-well plates. On reaching confluence, each clone was duplicated in a 96-well format (duplicate plates) for selection of clones with different levels of transgene expression.
Radiotracer Assay for HSV1-tk Expression In Vitro
The cells were seeded in 150 mmx25 mm culture dishes (at 5000 cells/dish) and grown until 50% confluent. The incubation medium was replaced with 14 ml of medium containing 2-[14C]FIAU (56 mCi/mmol) and [met-3H]TdR (65.4 Ci/mmol) (Moravek Biochemicals, Brea, CA). Radiochemical purity of each compound is routinely checked in our laboratory by HPLC and found to be more than 97% pure. The concentrations of 2-[14C]FIAU and methyl-[3H]TdR were 0.01 and 0.2 µCi/ml, respectively. The cells were harvested using a scraper after various periods of incubation of 10, 30, 60, 90, and 120 minutes, weighed and assayed using a Packard B1600 TriCarb beta spectrometer and standard 3H and 14C dual-channel counting techniques. The medium was counted before and following incubation. The data were expressed as a harvested cell-to-medium concentration ratio (dpm/g cells)/(dpm/ml medium) and plotted against time. The steady-state accumulation rate of FIAU, normalized by that of TdR, was obtained from the slopes of the plot and used as a measure of HSV1-tk gene expression in vitro (FIAU/TdR ratio). We have shown that the FIAU/TdR accumulation ratio correlates highly with the other independent measures of the HSV1-tk gene expression; namely, ganciclovir sensitivity (IC 50) and concentration of HSV1-tk mRNA [1].
Assay for E. Coli β-Galactosidase
The cells were gently scraped in PBS and centrifuged. The cell pellets were weighed (usually 10–20 mg) and homogenized in 1 ml of lysis buffer containing 200 µg/ml of pefabloc SC, 40 µg/ml of aprotinin, and 5 µg/ml of leupeptin (Boehringer Manheim, Indianapolis, IN), in 10 mM Tris-HCL (pH 7.5), 1 mM DTT, and 102 mM 2-mercaptoethanol. The cell lyzate was obtained by centrifugation after brief ultrasonic disruption. One hundred microliters of the lysate was added to the reaction mixture (in the spectrophotometric cuvette) containing 2.6 ml of 100 mM d-galactose in 100 mM sodium phosphate buffer (pH 8), 100 µl of 30 mM MgCl solution, 100 µl of 3.36 mM 2-mercaptoethanol solution. The assay was performed at pH 8 and in the presence of 100 mM d-galactose to further suppress the endogenous mammalian enzyme activity [16]. After equilibration at 37°C, the baseline absorbance at 410 nm was obtained. Thereafter, 100 µl of 68 mM O-nitrophenyl-β-galactoside (ONPG) solution (Sigma Chemical, St. Louis, MO) was added to the reaction mixture and a linear increase in 410 nm absorbance was recorded for 5 to 25 minutes with Beckman DU-65 Spectrophotometer (Beckman Instruments, Columbia, MD). The rate of increase in 410 nm absorbance was converted into the units (U) of β-galactosidase using a standard curve obtained from standards with known concentrations of E. coli β-galactosidase (0.05 to 50 U/ml) (Sigma) prepared with the wild-type tumor cell lyzates (10 mg cells/ml). The results (U/ml) were normalized by the initial cell pellet weight, and β-galactosidase concentration was expressed in terms of U/g of cells.
Generation of Subcutaneous Tumors
Different single cell-derived clones of W256STLEO and RG2STLEO transduced cells were used to produce multiple subcutaneous tumors in Sprague-Dawley (Harlan Sprague Dawley, IN) (n=12) and RNU/rnu rats (n=4), respectively. The animals were anesthetized with a gas mixture containing 5% isoflurane, 70% nitrous oxide and 30% oxygen, and maintained with 1.5% isoflurane. Tumor cells (106 cells in 100 µl) were injected subcutaneously (s.c.) into both flanks of rats weighing 200–250 g. Eight RG2STLEO or seven W256STLEO different transduced clonal cells were injected into multiple s.c. sites into RNU-rnu immunodeficient rats and Sprague-Dawley rats (Harlan Sprague Dawley), respectively. A wild-type s.c. tumor was also produced in each animal and used as a nontransduced tumor control. The multitumor animal model allows for a direct comparison between tumors with different levels of transgene expression during the same imaging session in the same animal. The experimental protocol involving animals has been approved by the Institutional Animal Care and Use Committee of the Memorial Sloan Kettering Cancer Center.
Gamma Camera Imaging
No carrier-added [131I]FIAU was synthesized as described previously [2,3]. Gamma-camera imaging was performed with a dual-headed ADAC Genesys gamma camera (ADAC, Milpitas, CA) equipped with a high-energy, high-resolution (HEHR) collimator. The HEHR collimator characteristics are: hole diameter 3.06 mm, septa thickness 0.152 mm, tunnel thickness 32.2 mm. Reference 131I standards were imaged in parallel for semiquantitative assessment of the images. Radioactivity (µCi) in the tumor was determined from the ratio of the counts in a region of interest (ROI) placed over the tumor divided by the counts in an ROI enclosing the reference standards, multiplied by the known radioactivity (µCi) of the standards. Radioactivity concentration was obtained by dividing the gamma camera measured radioactivity of the tumor by the weight of the tumor measured at time of sacrifice. No correction for attenuation or scatter was applied to these measurements.
Tissue Sampling and Analysis
The animals were sacrificed after gamma-camera imaging. Tumors were removed and assayed by gamma spectroscopy to verify the measures of tissue radioactivity (% dose/g tissue) obtained by gamma-camera imaging. β-galactosidase activity was measured in the same samples. Multiple 10- to 20-mg tissue samples were obtained from different sites of each tumor and each sample was homogenized and processed as described above. Part of the lyzate (before centrifugation) was assayed for radioactivity concentration (% dose/g tissue) by gamma spectroscopy. The remaining lyzate was assayed for lacZ expression (β-galactosidase, U/g tissue) using the ONPG spectrophotometric assay described above.
Results
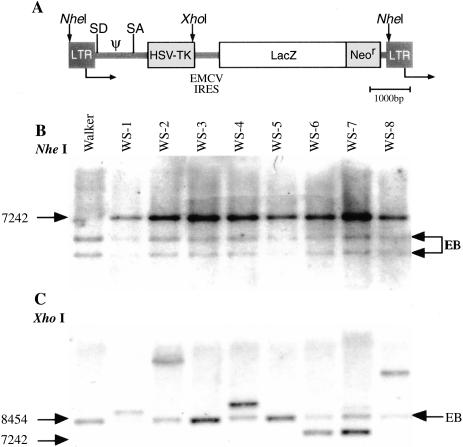
The vector STLEO combines the HSV1-tk gene and a fusion gene encoding the bacterial enzymes β-galactosidase and neomycin phosphotransferase (Figure 1A). The latter is expressed in an IRES-dependent fashion. The bicistronic transcription unit was retrovirally transduced into rat RG2 glioma and W256 mammary carcinoma cells. To investigate the spectrum of transgene coexpression that is achieved in cell populations, we generated two series of clones bearing a single copy of the STLEO vector to examine coexpression of both genes in a strict bicistronic context (Figure 1, B and C). Taking advantage of chromosomal position effects, we obtained a spectrum of expression levels over which we could explore the reliability of measuring coexpression of both genes (Figures 1C and 2).
Figure 1.
Genetic analysis ofsubcloned W256 tumor cells transduced with the retroviral vector STLEO. (A) The retroviral vector STLEO. (B) Southern blot analysis of subclones of STLEO-transduced W256 tumor cells show single-copy integration of the intact proviral structure (EB, endogenous bands; LacZ/NeoR probe). (C) Integration analysis (Xhol digest) shows unique integration sites in each clone (WS 3 and 5, as well as 6 and 7, are distinct after digestion with BgIII, data not shown).
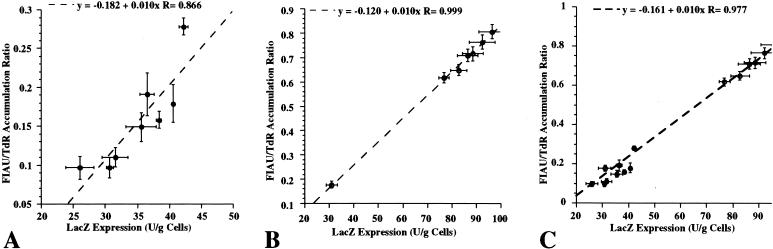
Figure 2.
IRES-mediated coexpression of the HSV1-tk and lacZ genes in vitro. LacZ gene expression (β-galactosidase enzyme activity, U/g cells) was measured using the ONPG spectrophotometric assay for (A) RG2STLEO and (B) W256STLEO transduced single cell clones, and plotted against measurements of the HSV1-tk gene expression (FIAU/TdR accumulation ratio). The relationship between the two measurements was defined by regression analysis. (C) The same relationship was observed when the RG2STLEO and W256STLEO data sets were combined.
The level of HSV1-tk enzyme activity in individual clones was measured by a double-label radiotracer accumulation assay using [3H]TdR and [14C]FIAU. β-galactosidase activity was measured by a spectrophotometric assay using ONPG as the substrate. There was a linear relationship between the levels of lacZ and HSV1-tk gene expression in corresponding clones of retrovirally transduced RG2 glioma cells (RG2STLEO, r=0.86; Figure 2A) and in W256 carcinoma cells (W256STLEO, r=0.99; Figure 2B). Combining the RG2STLEO and W256STLEO data sets yields a similar relationship over a wide range of transgene expression levels (Figure 2C). This suggests that the relationship of IRES-mediated coexpression is independent of cell type. Furthermore, the level and range of HSV1-tk gene expression (as measured by the FIAU/TdR accumulation ratio) in the transduced clones was within the range that is adequate for non-invasive imaging [1–3].
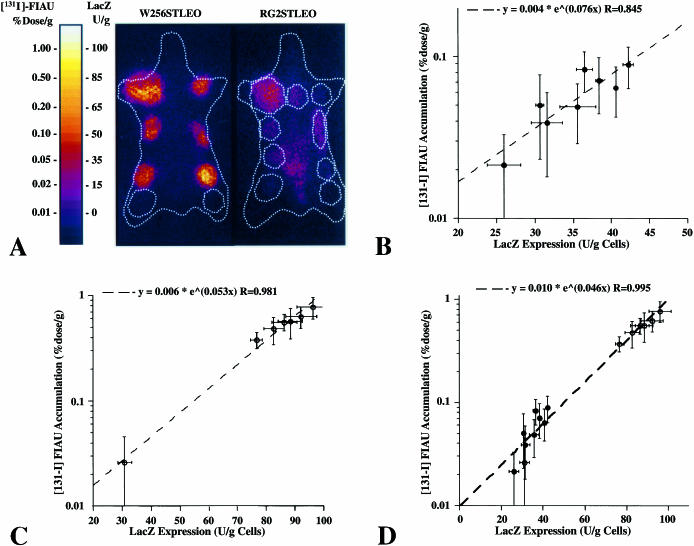
We investigated whether the relationship between lacZ and HSV1-tk expression that was observed in tissue culture would remain proportional in vivo. Multiple clones of transduced RG2 and W256 tumor cells were implanted subcutaneously in single animals so that different levels of transgene expression could be imaged under identical conditions. The range of expression levels in the different clones reflected the variability of expression obtained in polyclonal populations of tumor cells. Wild-type W256 and RG2 tumors served as negative controls. Three weeks after tumor cell inoculation, animals were injected with [131I]FIAU (800 µCi, i.v.) to image HSV1-tk gene expression. Gamma-camera imaging was performed 24 hours after [131I]FIAU administration and demonstrated highly specific localization of radioactivity to the areas of transduced tumors (Figure 3A). Low background levels of radioactivity were observed in the nontransduced wild-type RG2 and W256 tumors. A linear relationship was observed between the levels of HSV1-tk expression ([131I]FIAU accumulation, % dose/g) and levels of lacZ expression (β-galactosidase, U/g) in corresponding explanted RG2STLEO (Figure 3B) and W256STLEO (Figure 3C) clonal tumors. The relationship between expression levels of lacZ and HSV1-tk indicates that the proportionality of IRES-mediated coexpression of the two genes is maintained in vivo. Combining the RG2STLEO and W256STLEO tumor data sets extends the lacZ and HSV1-tk gene expression relationship over a broad range (Figure 3D). Knowing the relationship between [131I]FIAU accumulation in transduced tumors (% dose/g) and corresponding levels of β-galactosidase activity (U/g), a color-coded parametric image and scale bar of lacZ gene expression could be constructed (Figure 3A).
Figure 3.
IRES-mediated coexpression of the HSV1-tk and lacZ genes in vivo. (A) Gamma camera images of [131I]FIAU accumulation in W256SLEO and RG2STLEO subcutaneous tumors derived from clonal cell lines, 24 hours after tracer administration reflect HSV1-tk expression. The white dashed line outlines the contours of the animal and tumors with relatively low activity and wild-type control tumors (lowest tumor on the right thigh). Some residual bladder activity is still present in RG2STLEO tumor-bearing rats. Note the difference in [131I]FIAU accumulation between different tumors. The [131I]FIAU images of HSV1-tk expression were also converted into parametric images of lacZ gene expression based on the relationship defined in panel D, and this is indicated by the color-coded intensity bar showing units of β-galactosidase activity (U/g) in panel A. The levels of HSV1-tk gene expression measured in different tumors (% dose/g [131I]FIAU) were plotted against measures of lacZ gene expression (U/g) obtained in corresponding tumor samples for different (B) W256SLEO and (C) RG2STLEO clonal tumors. The relationship between the two measures was defined by regression analysis. (D) The same relationship was observed when the RG2STLEO and W256STLEO data were combined.
Discussion
The imaging paradigm we describe can be applied to any gene combination, and is based on the stable coexpression of two genes. Strict coexpression of two proteins in equimolar amounts can only be achieved by a fusion gene encoding both gene products [17]. This approach, however, cannot be generalized as many fusion proteins may not yield functional activity or the fusion protein may not localize in the appropriate subcellular compartment. Fusion proteins may also induce immunogenic molecules, poorly suited for in vivo studies. Coexpression of two distinct gene products is commonly achieved by using dual-promoter systems. However, when transgene expression is carefully examined in single cells rather than in cell populations in which total expression is averaged, coexpression is unreliable because of promoter interference or epigenetic transcriptional repression [18–20].
The imaging paradigm we describe is not confined to one type of vector or transgene delivery system and could be extended to microinjected DNA and other viral vectors as well; this is because IRES-mediated coexpression is determined at the translational level. However, the efficacy of translation may depend on protein synthetic activity and cycling of the transduced cells. Indeed, cap-dependent translation is relatively inefficient during mitosis in mammalian cells because of the presence of underphosphorylated elF-4F [21]. Eventually, it will be important to assess which IRES elements are reliable indicators of transgene coexpression in different tissues, taking into account the half-life of each encoded protein. These considerations will be important when using non-invasive imaging to assess organ (tissue) specificity, as well as level and duration of transgene expression.
In this report, we present as “proof of principle” the results of imaging studies with the HSV1-tk gene positioned proximally in the IRES-based coexpression cassette (HSV1-tk-IRES-geneX). Previously, we studied the range of HSV1-tk expression levels when the HSV1-tk gene was positioned distal to the IRES in combination with genes other than lacZ (geneX-IRES-HSV1-tk). For example, when the HSV1-tk gene was positioned distal to the IRES element and the gene encoding for the mutant low affinity nerve growth factor receptor (mLNGFR) was positioned proximal (mLNGFR-IRES-HSV1-tk), the average level of HSV1-tk expression in a mixed population of transduced cells was two to three times lower [22] compared to the expression levels observed in the current studies. Similar results were obtained with the HSV1-tk (distal) and GM-CSF (proximal) using the GMCSF-IRES-HSV1-tk expression cassette [23]. Nevertheless, the range of HSV1-tk expression levels is more than adequate for non-invasive imaging in vivo when the HSV1-tk gene is positioned distal to the IRES. It is important to note that the expression of the IRES-linked genes was driven by the same promoter, retroviral LTR, in this and all of our previous studies. The level of expression of the both IRES-linked genes can be further increased by using promoters stronger than the retroviral LTR.
In most gene-therapy protocols, appropriate reporter substrates for direct imaging of transgene expression are not available, and their development and validation would require considerable time and effort. Furthermore, developing specific tracers for direct imaging would have to be repeated for each therapeutic transgene, and this is simply not feasible. An alternative approach is to use “indirect” imaging based on proportional coexpression of a reporter gene, such as HSV1-tk. This report demonstrates the feasibility of indirect reporter gene imaging to monitor the expression of therapeutic genes of interest by constructing appropriate IRES-based gene expression cassettes. For example, non-invasive imaging could facilitate the assessment of the intratumoral delivery of genes encoding different cytokines and costimulatory molecules (e.g., IL2, IL4, IL12, GM-CSF, INF-γ, B7, etc.), prodrug activating genes that can not be imaged directly (e.g., cytosine deaminase), tumor suppressor genes (e.g., p53). Non-invasive imaging could be applied to monitor the delivery and expression of angiogenic genes (e.g., VEGF) in cardiovascular gene therapy; genes that correct genetic deficiencies such as the Duschene muscular distrophy (dystrophin gene). Non-invasive imaging could also be applied to monitor gene expression and to track the genetically modified cells in immunotherapy (e.g., tumor vaccines), adoptive cell therapies (e.g., infusion of antigen-specific T-lymphocytes), and stem cell transfer (e.g., hematopoietic or muscle stem cells). Thus, the demonstration of proportional coexpression of a cis-linked transgene over a wide range of expression levels and the ability to image this range of coexpression non-invasively represents an important and essential step in development of the imaging paradigms discussed above.
In summary, we demonstrate that non-invasive imaging of a marker gene (HSV1-tk) can provide quantitative as well as topographical information related to the expression of a cis-linked transgene. This imaging paradigm could be applied to investigate the activity of specific promoter and enhancer elements in transgenes or genes targeted by homologous recombination. Furthermore, non-invasive imaging of HSV1-tk could be used to investigate and monitor a wide range of clinical gene therapy trials involving cis-linked transgenes; it could be readily applied to several ongoing and newly developing clinical gene therapy protocols by defining the location, magnitude and persistence of transgene expression over time.
Acknowledgements
We thank I. Riviere for critical review of the manuscript.
Abbreviations
- EMCV
encephalomyocarditis virus
- FCS
fetal calf serum
- HSV1-tk
herpes simplex virus type one thymidine kinase
- IRES
internal ribosomal entry site
- PET
positron emission tomography
- s.c.
subcutaneous
Footnotes
This work was supported by NIH RO1 CA59350, NIH RO1 CA60706, NIH RO1 CA69769, NIH RO1 CA76117, DOE 86ER60407, the James McDonnell Foundation Scholars Award, and the Gershel Foundation.
References
- 1.Tjuvajev JG, Stockhammer G, Desai R, Uehara H, Watanabe K, Gansbacher B, Blasberg RG. Imaging the expression of transfected genes in vivo. Cancer Res. 1995;55:6126–6132. [PubMed] [Google Scholar]
- 2.Tjuvajev JG, Finn R, Watanabe K, Joshi R, Oku T, Kennedy J, Beattie B, Koutcher J, Larson S, Blasberg RG. Noninvasive imaging of herpes virus thymidine kinase gene transfer and expression: a potential method for monitoring clinical gene therapy. Cancer Res. 1996;56:4087–4095. [PubMed] [Google Scholar]
- 3.Tjuvajev JG, Avril N, Oku T, Sasajima T, Miyagawa T, Joshi R, Safer M, Beattie B, DiResta G, Daghighian F, Augensen F, Koutcher J, Zweit J, Humm J, Larson SM, Finn R, Blasberg RG. Imaging herpes virus thymidine kinase gene transfer and expression by positron emission tomography. Cancer Res. 1998;58:4333–4341. [PubMed] [Google Scholar]
- 4.Morin KW, Knaus EE, Wiebe LI. Non-invasive scintigraphic monitoring of gene expression in a HSV-1 thymidine kinase gene therapy model. Nucl Med Commun. 1997;18:599–605. doi: 10.1097/00006231-199707000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Gambhir SS, Barrio JR, Wu L, Iyer M, Namavari M, Satyamurthy N, Bauer E, Parrish C, MacLaren DC, Borghei AR, Green LA, Sharfstein S, Berk AJ, Cherry SR, Phelps ME, Herschman HR. Imaging of adenoviral-directed herpes simplex virus type 1 thymidine kinase reporter gene expression in mice with radiolabeled ganciclovir. J Nucl Med. 1998;39:2003–2011. [PubMed] [Google Scholar]
- 6.Gambhir SS, Barrio JR, Phelps ME, Iyer M, Namavari M, Satyamurthy N, Wu L, Green LA, Bauer E, MacLaren DC, Nguyen K, Berk AJ, Cherry SR, Herschman HR. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc Natl Acad Sci USA. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 8.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 10.Ghattas IR, Sanes JR, Majors JE. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medin JA, Migita M, Pawliuk R, Jacobson S, Amiri M, Kluepfel-Stahl S, Brady RO, Humphries RK, Karlsson S. A bicistronic therapeutic retroviral vector enables sorting of transduced CD34+ cells and corrects the enzyme deficiency in cells from Gaucher patients. Blood. 1996;87:1754–1762. [PubMed] [Google Scholar]
- 12.Gallardo HF, Tan C, Sadelain M. The internal ribosomal entry site of the encephalomyocarditis virus enables reliable coexpression of two transgenes in human primary T lymphocytes. Gene Ther. 1997;4:1115–1119. doi: 10.1038/sj.gt.3300506. [DOI] [PubMed] [Google Scholar]
- 13.Barba D, Hardin J, Sadelain M, Gage FH. Development of anti-tumor immunity following thymidine kinase-mediated killing of experimental brain tumors. Proc Natl Acad Sci USA. 1994;91:4348–4352. doi: 10.1073/pnas.91.10.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riviere I, Sadelain M. Methods for the construction of retroviral vectors and the generation of high titer producers. In: Robbins P, editor. Methods in Molecular Biology, Gene Therapy Protocols. Totowa, NJ: Humana Press; 1997. pp. 59–78. [DOI] [PubMed] [Google Scholar]
- 16.Hendrikx PJ, Martens AC, Visser JW, Hagenbeek A. Differential suppression of background mammalian lysosomal beta-galactosidase increases the detection sensitivity of LacZ-marked leukemic cells. Anal Biochem. 1994;222:456–460. doi: 10.1006/abio.1994.1516. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs A, Dubrovin M, Hewett J, Sena-Esteves M, Tan C, Slack M, Sadelain M, Breakfield X, Tjuvajev JG. Functional coexpression of HSV1 thymidine kinase and green fluorescent protein: implications for noninvasive imaging of transgene expression. Neoplasia. 1999;1:154–161. doi: 10.1038/sj.neo.7900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emerman M, Temin HM. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984;39:449–467. [PubMed] [Google Scholar]
- 19.Emerman M, Temin HM. Quantitative analysis of gene suppression in integrated retrovirus vectors. Mol Cell Biol. 1986;6:792–800. doi: 10.1128/mcb.6.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowtell DD, Cory S, Johnson GR, Gonda TJ. Comparison of expression in hemopoietic cells by retroviral vectors carrying two genes. J Virol. 1988;62:2464–2473. doi: 10.1128/jvi.62.7.2464-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pause A, Belsham GJ, Gingras AC, Donzo O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 22.Tjuvajev J, Gallardo H, Joshi A, Joshi R, Blasberg R, Sadelain M. Bi-cistronic co-expression of a cell surface marker gene and the HSV1-tk gene in Jurkat cells: implications for noninvasive imaging in vivo. Cancer Gene Ther. 1997;4:S44. [Google Scholar]
- 23.Tjuvajev J, Sharma S, Miller P, Paul R, Joshi R, Dubinett S, Blasberg R. Monitoring the expression of therapeutic genes linked to a marker gene by an IRES sequence. J Neuro-Oncol. 1996;30:101. [Google Scholar]