Abstract
Cranial neural crest cells differentiate into diverse derivatives including neurons and glia of the cranial ganglia, and cartilage and bone of the facial skeleton. Here, we explore the function of a novel transcription factor of the spalt family that might be involved in early cell-lineage decisions of the avian neural crest. The chicken spalt4 gene (csal4) is expressed in the neural tube, migrating neural crest, branchial arches and, transiently, in the cranial ectoderm. Later, it is expressed in the mesectodermal, but not neuronal or glial, derivatives of midbrain and hindbrain neural crest. After over-expression by electroporation into the cranial neural tube and neural crest, we observed a marked redistribution of electroporated neural crest cells in the vicinity of the trigeminal ganglion. In control-electroporated embryos, numerous, labeled neural crest cells (∼80% of the population) entered the ganglion, many of which differentiated into neurons. By contrast, few (∼30% of the population) spalt-electroporated neural crest cells entered the trigeminal ganglion. Instead, they localized in the mesenchyme around the ganglionic periphery or continued further ventrally to the branchial arches. Interestingly, little or no expression of differentiation markers for neurons or other cell types was observed in spalt-electroporated neural crest cells.
Keywords: Spalt, neural crest, branchial arches, trigeminal ganglia
INTRODUCTION
The neural crest is a migratory cell type that originates from the dorsal neural tube and emigrates shortly after its closure. At cranial levels, neural crest cells migrate beneath the ectoderm and populate many ganglia of the PNS, differentiating into the glia and some of the neurons within these ganglia. In addition to neural derivatives, the neural crest cells that invade the head and the branchial arches give rise to most of the cartilage, bone and connective tissue of the face. How cells within the cranial neural crest population home to the proper structures and, subsequently, make cell-lineage decisions to differentiate into cell types as divergent as cartilage, neurons and glia remain unknown.
In the trunk region, several signaling pathways and transcription factors have been implicated in cell fate decisions of neural crest. For example, in dorsal root ganglia, Notch signaling appears to influence the decision to assume neuronal versus glial fates (Morrison et al., 2000; Wakamatsu et al., 2000). In the autonomic nervous system, bone morphogenetic protein (BMP) biases neural crest precursors toward a neural fate whereas glial growth factor/neuregulin drives cells into the glial lineage (Shah et al., 1994; Shah et al., 1996; Shah et al., 1997). At the transcriptional level, null mutations in Mash-1 lead to selective loss of sympathetic ganglion (Guillemot et al., 1993) whereas overexpression of neurogenin biases neural crest cells toward the sensory lineage (Perez et al., 1999). Transcription factors that affect cell localization and lineage decisions in cranial neural crest cells have yet to be described.
The spalt genes are a family of evolutionarily conserved zinc finger-containing transcription factors, mutations of which lead to homeotic transformations in Drosophila (Kühnlein et al., 1994). Loss of Drosophila spalt transforms prothorax into head and abdominal segments into tail (Jürgens, 1988). Selective deletion of Drosophila spalt in the embryonic PNS causes a cell-fate switch in neuronal precursors and defects in migration (Rusten et al., 2001; Elstob et al., 2001). In the thorax, spalt and spalt-related are involved in positioning of specific proneural clusters (de Celis et al., 1999). In addition, spalt is required for the directed migration of dorsal trunk cells in the trachea (Kühnlein and Schuh, 1996). Thus, this gene family appears to play important roles in cell migration and cell-fate decisions.
Here, we describe a novel chicken transcription factor of the spalt family that might be involved in specification of avian cranial neural crest cell lineages. We have isolated a chicken spalt gene, csal4, which is expressed in the neural tube, migrating neural crest and branchial arches. Both the sequence and expression pattern most closely resemble that of Xenopus xsal-3 (Onuma et al., 1999) and mouse Sall4 (Kolhase et al., 2002). Overexpression of this spalt cDNA in migrating cranial neural crest cells causes a redistribution of neural crest derivatives within the trigeminal ganglia to its periphery; furthermore, these spalt-expressing cells fail to express detectable differentiation markers of neurons or other cell types examined. This suggests that spalt can affect the localization of cranial neural crest cells, which, in turn, might influence the ability to form sensory neurons.
OBJECTIVE
We hypothesized that csal4 might influence cranial neural crest migration and/or lineage decisions, based on its expression in neural crest-derived branchial arches but not sensory ganglia and its known function in cell fate and migration decisions in Drosophila. To test this idea, we overexpressed the transcription factor in migrating neural crest cells, which resulted in altered localization of transfected cells to the periphery but not the core of the trigeminal ganglion where neurons form.
METHODS
Isolation of csal4
csal4 was isolated using RT-PCR from RNA derived from stage-8 embryos using the oligonucleotides OLF UP (5’-ATGTGGAGAACTAGTTTG-3’) and OLF DN (5’-CCACAGATGATGAAGGCA-3’) and the Access RT-PCR system (Promega). DNA fragments of 450 bp were isolated and cloned into pGEM-T (Promega). Two independent clones were identified and used to screen a stage 10 chicken-embryo cDNA library in HYBRIDZAP 2.1 (Stratagene). Several cDNA clones were isolated, including one that contained a 6-kb insert. Sequence analysis of the longest clone showed that it contained all but the most 5’ sequence of the coding region. The 5’ coding sequence was amplified from the stage 10 cDNA library. The deduced amino acid sequence of the entire coding region was compared to that of other spalt genes using the ClustalW program (Thompson et al., 1994).
In situ hybridization
The complete csal4 cDNA and several fragments were used as templates to prepare antisense RNA probes. In situ hybridization was performed as described (Wilkinson, 1992). The embryos were staged according to the criteria of Hamburger and Hamilton (Hamburger and Hamilton, 1951), fixed and embedded in gelatin and sectioned in a cryostat at 20 μm. Gelatin was removed from the sections by soaking for 5 minutes in PBS at 42°C. Some of these sections were used for immunohistochemistry.
Overexpression of csal4 by electroporation
The coding region of csal4 was cloned into the pCIG vector (Megason and McMahon, 2002) downstream of a chicken b-actin promoter and upstream of enhanced green fluorescent protein (EGFP) under the control of an internal ribosomal entry site (IRES) sequence. The plasmid in 10 mM tris was diluted 1:1 with Ringers solution and injected into the lumen of the neural tube of embryos with 3-6 somites. Two pulses of 25 volts and 50 msec duration were delivered through two platinum electrodes, 4 mm apart, placed on top of the vitelline membrane on either side of the embryo (Itasaki et al., 1999). The embryos were placed at 38°C for up to 48 hours of further development. Embryos were collected, fixed in 4% paraformaldehyde overnight at 4°C, embedded in gelatin and sectioned on a cryostat at 10 μm. Sections were then stained with antibodies that recognize green fluorescent protein (GFP) (Abcam), and HNK-1 and TUJ1 (Covance). Anti-GFP antibodies were used to identify cells containing the pCIG plasmid and HNK-1 to identify neural crest cells. TuJ1 (against b3-tubulin) immunostaining were done after electroporation to identify differentiated neurons. They were visualized by Alexa 568-conjugated donkey anti-mouse IgG or Alexa 568-conjugated goat anti-mouse IgG (Molecular Probes).
RESULTS
Isolation of a chick spalt homologue csal4
Using an RT-PCR approach followed by library screening, we isolated a full length cDNA clone for a novel chicken spalt gene, csal4 (GenBank accession number AY342354). The coding region contains three sets of zinc fingers and a single zinc-finger domain at the amino terminus, a glutamine rich region, and the 8 amino acid motif, referred to as the SAL-box, that is common to all spalt genes (Fig. 1A). Comparison with full length sequences of other spalt genes reveals that it is most closely related to Xsal3 (61% identity at the amino acid level) and human SALL4 (57% identity at the amino acid level). A dendogram of the ClustalW data places these three genes in one of four separate spalt groups (Fig. 1B). However, the degree of sequence conservation between csal4, Xsal3 and SALL4 is less than that the relatedness observed within other groups. For example, human SALL1 is 86% identical to chicken csal1 and human SALL3 is 71% identical to chicken csal3. No more closely related genes were uncovered by searches of genomic and EST databases. These results indicate that csal4, Xsal3 and SALL4 are homologues, but we cannot rule out the possibility that undiscovered discovered gene might be more related.
Fig. 1.
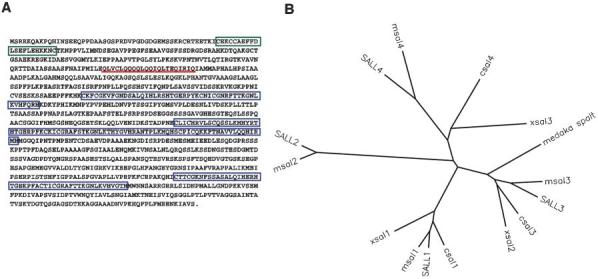
Sequence analysis of csal4. (A) Amino-acid sequence of csal4. The nucleotide sequence has been assigned the GenBank accession number AY342354. The amino terminal CCHC zinc-finger region is boxed in green, the three C2H2 zinc-finger regions are boxed in blue and the glutamine-rich domain is underlined in red. (B) Dendogram of ClustalW analysis of vertebrate spalt genes. There are four paralog groups of spalt. csal4 is most closely related to Xsal3 and human SALL4. The proteins (and their accession numbers) used in this analysis: human SALL1 (Q9N5C2); human SALL2 (Q9Y467); human SALL3 (Q9BXA9); human SALL4 (Q9UJQ4); mouse msal1 (Q9ER74); mouse msal2 (NP_056587); mouse msal3 (XP_129051); mouse msal4 (BAC33598); chicken csal1 (AAG13011); chicken csal3 (AAK38370); Xenopus Xsal1 (AAG45108); Xenopus Xsal2 (T30341); Xenopus Xsal3 (JC7116); and medaka spalt (AAB51127). For clarity, the mouse genes are labeled to be consistent with the naming of the human genes.
Expression pattern of csal4 in chicken embryos
We examined the pattern of csal4 expression by in situ hybridization of chicken embryos from the early neurula (E1) stage to the time of gangliogenesis (E3). As the neural folds are beginning to elevate (3-somite stage), csal4 expression is found throughout the head ectoderm and the neural plate. A few hours later, as the neural tube begins to close at the level of the forebrain/midbrain, csal4 expression is downregulated from the forming neural tube, but remains expressed in more caudal regions of the open neural plate (not shown).
By the 10-somite stage, there is strong staining in the lateral ectoderm and caudal dorsal neural tube. The otic placode, lateral ectoderm, pharyngeal endoderm, early somites and the notochord express csal4. Expression is also seen in migrating neural crest (Fig. 2A,B) and is particularly notable in cells emigrating from the hindbrain of stage-13 embryos (Fig. 2D,E). To verify that these csal4-positive cells are neural crest cells, embryos were sectioned following whole mount in situ hybridization and the sections stained with the HNK-1 antibody, which is a marker for early migrating neural crest cells (Bronner-Fraser, 1986). The csal4 and HNK-1 expression patterns overlapped, which indicates that csal4 is expressed in this migratory population (Fig. 2C,F).
Fig. 2.
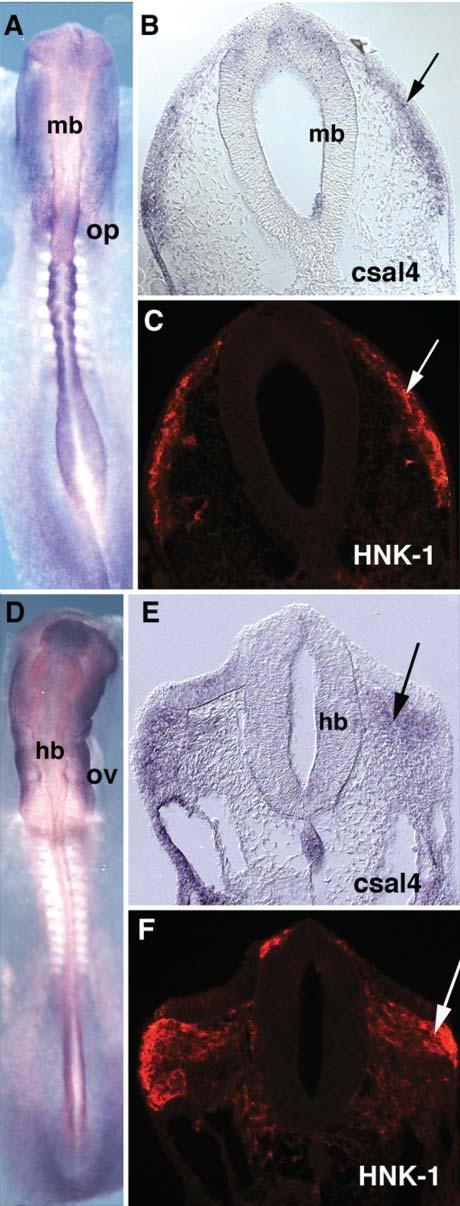
Migrating neural crest cells express csal4. (A) Whole-mount in situ hybridization of a stage-10-chicken embryo with a csal4 RNA probe. (B) Section through the midbrain of the embryo in (A). The arrow points to the migrating neural crest. (C) The same section as B, stained with HNK1 antibody. The arrow points to the migrating neural crest cells. The HNK1 antibody colocalizes with csal4 expression. (D) Whole-mount in situ hybridization of a stage-13-chicken embryo with csal4 probe. (E) Section of the embryo in D. The neural crest migrating from the hindbrain expresses csal4 (arrow). (F) Same section as E, stained with HNK1 antibody. Arrows point to the migrating neural crest. At this level, csal4 expression overlaps with HNK1. Abbreviations: hb, hindbrain; mb, midbrain; op, otic placode; ov, otic vesicle.
The mesoderm in the branchial arches strongly expresses csal4, with highest levels in branchial arch 1 (Fig. 3A,D). Because the migrating neural crest marker, HNK-1, is downregulated as neural crest cells enter the branchial arches, we used a pan-Dlx antibody that recognizes all Dlx proteins (Panganiban et al., 1997) as a marker for neural crest cells within this location (Panganiban and Rubenstein, 2002). We found extensive overlap of csal4 RNA and Dlx protein at stages 14 and 15 in the branchial arches (compare Fig. 3B,C and Fig. 3E,F). However, the overlap was not absolute because there were areas that either expressed csal4 only or Dlx proteins only. For example, during neural crest migration, csal4 was expressed primarily in the ventral portion of the arches whereas Dlx transcripts were also expressed in the dorsal portion (Fig. 3B,C). At later stages, csal4 appeared to be expressed more medially than Dlx (Fig. 3E,F). This indicates that csal4 is expressed in some, but not all, neural crest cells in the branchial arches.
Fig. 3.
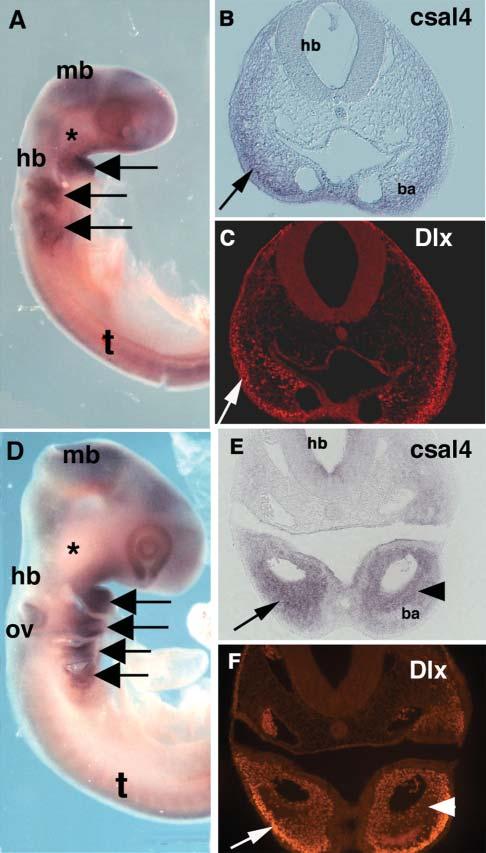
Neural crest cells in the branchial arches express csal4. (A) Whole-mount in situ hybridization of stage-14-chicken embryo with csal4 probe. A strong signal can be seen in the branchial arches (arrows) but there is no detectable signal at site of the trigeminal ganglia (asterisk). (B) Section through the region of the hindbrain of the embryo in A. Neural crest in the branchial arch (arrow) expresses csal4. (C) Same section as in B, stained with pan-Dlx antibody. Although not all Dlx-expressing cells express csal4, there is an overlapping population that expresses both (arrows). (D) Whole mount in situ hybridization of a stage-15-chicken embryo with csal4 probe. All the branchial arches (arrows) but not the trigeminal ganglia (asterisk) express csal4 strongly.(E) Section through the embryo in D showing extensive expression of csal4 throughout the branchial arch (arrow) with highest levels in the core (arrowhead). (F) Section in E stained with pan-Dlx antibody. Although there is an overlapping population that expresses both csal4 and Dlx protein, the highest level of Dlx expression (arrow) is distal to the region in which csal4 expression is highest (arrowhead). Abbreviations: ba, branchial arch; hb, hindbrain; mb, midbrain; ov, otic vesicle; t, trunk.
By stage 15, the ectoderm no longer expresses csal4 but expression continues in the otic vesicle (Fig. 3D). In the neural tube, csal4 transcripts are observed in the forebrain, midbrain and hindbrain but are missing at the boundaries between these brain regions. At spinal cord levels, csal4 is first expressed throughout the neural tube except for the most dorsal region; later csal4 becomes restricted to a band of cells in the dorsal half of the spinal cord with the exception of the dorsal-most (presumptive roof plate) cells. However, csal4 expression was undetectable in the trigeminal ganglia, which is made up of neural crest as well as placode cells (Fig. 3D).
Expression of csal4 in the branchial arches remains strong until stage 25, the last stage examined. Collectively, these results indicate that csal4 is expressed primarily in the branchial arch neural crest but is absent from the neuronal neural crest-derived population that emerges from the same axial level.
In addition to neural and neural crest derivatives, csal4 transcripts are also found in the nephric ducts, notochord, sclerotomal portion of the trunk somites, pharyngeal endoderm and very strongly in the tail (data not shown). Staining was detected in the distal limb buds with faint staining in the somatic mesoderm at the level of the future wing bud (data not shown).
Overexpression of csal4 alters localization of neural crest cells
We tested the possible function of csal4 by overexpressing it in the forming nervous system using electroporation. The coding region of csal4 was cloned into the pCIG expression vector that coexpresses EGFP through an IRES sequence. This plasmid was injected into the lumen of the midbrain and hindbrain prior to neural crest migration and, subsequently, electroporated into the neural tube (Fig. 4A). This allows for transfection of the both neural tube and premigratory neural crest cells. At 1 day after electroporation, at a time when neural crest cells are at late stages of migration in the cranial region, no differences were noted between csal4 and control-transfected cells, which indicates that there was no obvious effect on their early migratory behavior (data not shown). However, differences in the distribution pattern of electroporated experimental versus control neural crest cells were noted in embryos examined 48 hours after electroporation, by which time neural crest-derived cranial ganglia have formed (Fig. 4B). We focused our studies particularly on the trigeminal ganglion, a cranial sensory ganglion that receives a contribution from neural crest cells derived from the midbrain and rostral hindbrain.
Fig. 4.
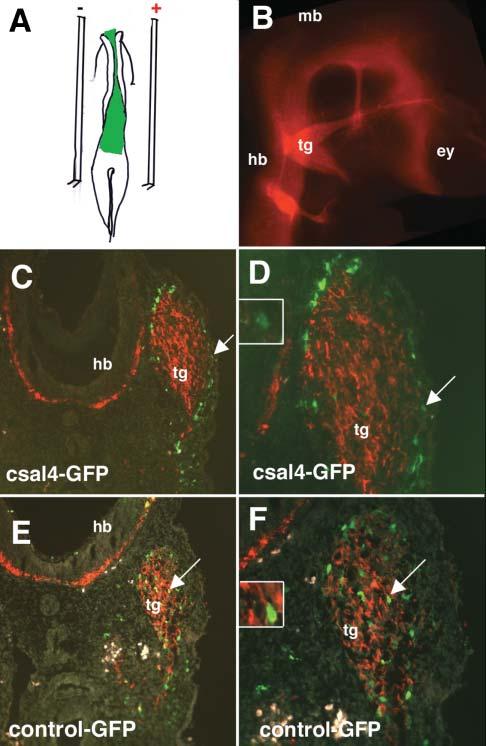
Misexpression of csal4 alters the localization of transfected neural crest cells in the vicinity of the trigeminal ganglia. (A) Schematic diagram illustrating the strategy for overexpressing csal4. Either csal4 or control DNA solution, visualized with fast green, was injected in the lumen of the neural tube and the open neural plate (green). The anode and cathode electrodes are placed on top of the vitelline on either side of the embryo and two 50 msec pulses of 25 mv delivered. Embryos were incubated for an additional 48 hours.(B) Whole-mount immunofluoresence at the level of the head. TUJ1 antibody was used to stain the neurons of a stage-19 embryo to show the location of the cranial ganglia. (C) A transverse section at the level of the trigeminal ganglia through an embryo transfected with pCIG-sal expressing both csal4 and GFP and stained with antibodies to GFP (green) and TUJ1 (red). A large fraction of the GFP-expressing cells (green, arrow) are localized on the periphery of the TUJ1-positive ganglia (red). (D) Higher magnification of C, showing that GFP-positive cells are outside the TUJ1-positive ganglia (see arrow). Inset is a detail of the cell marked by the arrow showing little to no detectable cytoplasmic TUJ1. (E) A transverse section at the level of the trigeminal ganglia through a control embryo transfected with pCIG expressing only GFP and stained with antibodies to GFP (green) and TUJ1 (red). Most of the GFP-positive cells are localized uniformly throughout the TUJ1-expressing ganglia (see arrow). (F) Higher magnification of E showing GFP-positive cells throughout the ganglia. Inset is a detail of the cell marked by the arrow showing cytoplasmic TUJ1 immunoreactivity in a cell with with a GFP-positive nucleus. Abbreviations: ey, eye; hb, hindbrain; mb, midbrain; tg, trigeminal ganglia.
Overexpression of csal4 affected localization of neural crest cells within the trigeminal ganglion. Normally, neural crest cells contribute both neurons and glia of the trigeminal ganglia, with crest-derived neurons distributed throughout the proximal portion of the ganglion and non-neural/glial cells found both in the proximal and distal portions (Noden, 1980). Consistent with this, in control transfected embryos, GFP-labeled neural crest cells were distributed throughout the trigeminal ganglion in a relatively uniform pattern (Fig. 4E,F). By contrast, the bulk of the csal4 expressing/GFP-labeled neural crest cells localized to the periphery of the ganglia and in the mesoderm adjacent to the ganglia (Fig. 4C,D). Only a few csal4-transfected cells were observed in the interior of the ganglion (Fig. 4C,D). In contrast to the trigeminal region, no obvious differences were noted in the distribution of electroporated neural crest cells in the branchial arch mesoderm of control versus csal4-transfected embryos (data not shown).
The number of GFP-labeled cells in the mesoderm adjacent to the trigeminal ganglion and at the periphery of the trigeminal ganglia as well as those inside the ganglion were counted after electroporation of control or csal4 plasmids. A total of 1053 GFP-positive cells were counted in six csal4-GFP transfected embryos and compared with 1666 cells in six control transfected embryos. We noted a significant difference between the percentage of electroporated cells that localized to the periphery of the ganglion versus total number of GFP-positive cells in the region of the ganglia. In csal4-electroporated embryos, 79% of the GFP-positive cells were localized to the periphery whereas in control electroporated embryos only 28% of GFP-positive cells were localized to the periphery (P<0.001). These results demonstrate that overexpression of csal4 alters the localization of neural crest cells.
Neural crest cells overexpressing csal4 fail to express differentiation markers for neurons or other cell types
To assess whether the types of neural crest derivatives were altered after overexpression of csal4, sections through control and csal4-transfected embryos were stained with the neural marker TUJ1 that recognizes b3 tubulin. In control electroporated embryos, numerous GFP-positive cells were present in the trigeminal ganglion. Many of these GFP-expressing cells were also positive for TUJ1 (Fig. 4E,F). By contrast, no TUJ1-positive cells were observed in GFP/csal4-expressing cells. Although the majority of these cells were found in the periphery where neurons do not typically differentiate, none of the labeled cells (28%) found within the ganglion expressed Tuj1 (Fig. 4C,D). In addition, the csal4-expressing cells within or at the periphery of the ganglion did not stain with the Hu antibody, another pan-neuronal marker, nor did they express the transcription factor, neurogenin-1 (data not shown). These results indicate that csal4-electroporated cells do not express detectable markers of neuronal differentiation.
We next looked for other signs of differentiation in the spalt-expressing population. The csal4-GFP cells on the periphery of the ganglion or in the mesoderm did not express smooth muscle actin, indicating that they had not differentiated into smooth muscle (data not shown). Nor did they stain with Dlx 3 antibody, suggesting that they had not assumed properties of branchial arch neural crest (data not shown). At the time of our analysis, glial cells have not yet differentiated so we were unable to assess glial fate in these embryos. Given the lack of differentiation markers in the electroporated cell, an intriguing possibility is that spalt4 maintains cells in an undifferentiated state.
CONCLUSIONS
Chicken spalt4 is expressed in early migrating cranial neural crest cells; later, it is maintained in the population within the branchial arches but absent in the trigeminal ganglion.
Over-expression of csal4 in the cranial neural crest alters their pattern of localization within the trigeminal ganglia but does not affect migration within the branchial arches; spalt4-expressing neural crest cells localize primarily in the periphery rather than in the core of the ganglia.
Neural crest cells over-expressing csal4 do not express detectable neuronal or other differentiation markers.
DISCUSSION
In this study, we have isolated a novel chicken spalt-family member that biases cell localization and might alter fate decisions in the cranial neural crest. Based on sequence similarity and expression pattern, we surmise that csal4, Xsal3 and SALL4 are likely to be homologues. Although the degree of sequence conservation between csal4, Xsal3 and SALL4 is less than that the relatedness observed within other groups, csal4, Xsal3 (Onuma et al., 1999) and mouse Sall 4 (Kollhase et al., 2002) have similar expression patterns in the neural tube and the neural crest, as well as the limbs and other tissues, consistent with these being true homologues.
In the early embryo, strong expression of csal4 was noted in both the neural plate and the ectoderm as early as stage7. At subsequent stages, expression in the neural tube is downregulated but expression in the ectoderm is maintained in some placode regions. Neural tube expression reappears later at variable levels depending upon age and rostrocaudal location. In the brain, csal4 expression declines after neural tube closure, but is re-expressed by stage 13, when it is present in the forebrain, midbrain and hindbrain with the exception of the borders between them. This is in contrast with the expression of another chicken spalt gene, csal3, which is expressed only in the border regions (Farrell et al., 2001). Thus, the two spalt genes have complementary, non-overlapping patterns in the head. At the level of the trunk, csal4 is expressed first throughout the neural plate. Later, expression in the most dorsal cells is reduced and afterwards the expression in the ventral neural tube becomes downregulated, which leaves a stripe of csal4 expression in the dorsal half of the spinal cord.
Neural crest cells migrate from the midbrain and the hind-brain to form the cartilage, bone and connective tissue of the face as well as the glia and many neurons of the cranial ganglia. We find expression of csal4 in the migrating neural crest of the hindbrain as early as stage 10. However, at later stages of development, only a subset of neural crest cells express csal4, as seen by comparing the in situ pattern of csal4 with the antibody pattern of the neural crest marker HNK1. Once the neural crest cells migrate into the branchial arches, csal4 is detected in the arches in many cells that co-express Dlx, although the overlap is not complete. The co-expression of Dlx and in the branchial arches presents an interesting parallel with Drosophila where distalless (the Drosophila Dlx homolog) has been shown to induce spalt in the antenna (Dong et al., 2000).
The expression of csal4 in the branchial arches but absence from the cranial ganglia raises the interesting possibility that csal4 might bias cell fate decisions toward cartilage and bone lineages at the expense of neurons and/or glia. To test for possible effects of csal4 on neural crest cells, we over-expressed it in migrating neural crest cells of the developing chicken embryo via electroporation. The results show that excess spalt affects the localization of neural crest cells within the trigeminal ganglion. The cranial ganglia of embryos that over-express spalt contain significantly fewer GFP-positive cells than in control embryos. Most of the labeled cells failed to incorporate into the core of the ganglia itself and were found predominantly either at its edges or in the adjacent mesoderm.
Given that most of the csal4-electroporated cells localized at the periphery of the trigeminal ganglia whereas they were uniformly distributed throughout the ganglia in controls, an intriguing possibility is that these alterations in cell localization might be due to differences in cell adhesion as a consequence of spalt overexpression. Thus, the primary function of spalt could be to affect cell-cell adhesion, which, in turn, affects cell localization. If this is the case, it is possible that its influences on cell fate are a secondary consequence of the site of localization.
A second, but not mutually exclusive, possibility is that csal4 might be involved in the cell fate decisions that determine whether a neural crest differentiates into branchial arch cartilage or forms neuronal derivatives in the neural crest-derived ganglia. The csal4-electroporated cells failed to express markers of neuronal differentiation, as assayed by TUJ1 staining. Even after 3 days (stage 23), csal4-expressing cells in the periphery failed to express the neuronal marker Hu. These results indicate that downregulation of csal4 might be necessary for proper localization and/or neuronal differentiation within the ganglion.
If overexpression of csal4 alters the distribution and, consequently, the differentiation of electroporated neural crest cells, what is the fate of these cells? One possibility is that many of the electroporated cells switch from neuronal fate to a mesectodermal or glial fate. However, we were unable to detect phenotypic markers of glia, smooth muscle or branchial arch neural crest (which express Dlx genes) in the electroporated cells at the times examined. Given that they fail to express known differentiation markers, it is possible that they remain in an undifferentiated state. If this is the case, csal4 may keep cells in a ‘stem-like’ state.
Spalt was first isolated in Drosophila as a homeotic gene (Jürgens, 1988). It is expressed in two bands, one near the head and the other near the tail. Mutations in spalt cause prothorax structures to develop in the head and structures of the eighth abdominal segment to develop in the tail (Jürgens, 1988). Spalt is also involved during later cell fate decisions in Drosophila development, including eye (Mollereau et al., 2001), trachea (Kühnlein and Schuh, 1996), and wing (de Celis et al., 1996).
Regulation of spalt genes in vertebrates has been examined during medaka brain development (Köster et al., 1997) and chicken limb development (Farrell and Münsterberg, 2000). These experiments show that spalt genes are under the control of a number of different signaling molecules, including sonic hedgehog, and members of the Wnt and FGF family of growth factors. Many of these growth factors are required both at early stages of neural crest development (Garcia-Castro et al., 2002) and for proper patterning and differentiation of the branchial arches (Le Douarin and Kalcheim, 1999). One intriguing possibility is that they might be intimately interlinked with regulation of csal4 in the migration of neural crest from the neural tube to the branchial arches.
ACKNOWLEDGEMENTS
We thank Drs. Tatjana Sauka-Spengler and Vivian Lee for helpful comments on the manuscript. This work is supported by USPHS grants NS42287 and NS41070 to M.B.-F. We would like to thank Jhumku Kohtz for providing the pan-DLX antibody, which was made from a construct from Grace Panganiban.
REFERENCES
- Bronner-Fraser M. Analysis of the early stages of trunk neural crest migration in avian embryos using the monoclonal antibody HNK-1. Developmental Biology. 1986;115:44–55. doi: 10.1016/0012-1606(86)90226-5. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. Regulation of the spalt/spalt-related gene complex and its function during sensory organ development in the Drosophila thorax. Development. 1999;126:2653–2662. doi: 10.1242/dev.126.12.2653. [DOI] [PubMed] [Google Scholar]
- Dong PDS, Chu J, Panganiban G. Coexpression of the homeobox genes Distal-less and homothorax determines Drosophila antennal identity. Development. 2000;127:209–216. doi: 10.1242/dev.127.2.209. [DOI] [PubMed] [Google Scholar]
- Elstob PR, Brodu V, Gould AP. Abstract spalt-dependent switching between two cell fates that are induced by the Drosophila EGF receptor. Development. 2001;128:723–732. doi: 10.1242/dev.128.5.723. [DOI] [PubMed] [Google Scholar]
- Farrell E, Münsterberg AE. csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Developmental Biology. 2000;225:447–458. doi: 10.1006/dbio.2000.9852. [DOI] [PubMed] [Google Scholar]
- Farrell ER, Tosh G, Church E, Münsterberg AE. Cloning and expression of CSAL2, a new member of the spalt gene family in chick. Mechanisms of Development. 2001;102:227–230. doi: 10.1016/s0925-4773(01)00296-9. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of Morphology. 1951;88:49–92. [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, Krumlauf R. ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nature Cell Biology. 1999;1:E203–E207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO Journal. 1988;7:189–196. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhase J, Heinrich M, Liebers M, Fröhlich Archangelo L, Reardon W, Kispert A. Cloning and expression analysis of Sall4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenetic and Genome Research. 2002;98:274–277. doi: 10.1159/000071048. [DOI] [PubMed] [Google Scholar]
- Köster R, Stick R, Loosli F, Wittbrodt J. Medaka spalt acts as a target gene of hedgehog signaling. Development. 1997;124:3147–3156. doi: 10.1242/dev.124.16.3147. [DOI] [PubMed] [Google Scholar]
- Kühnlein RP, Schuh R. Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development. 1996;122:2215–2223. doi: 10.1242/dev.122.7.2215. [DOI] [PubMed] [Google Scholar]
- Kühnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, et al. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO Journal. 1994;13:168–179. doi: 10.1002/j.1460-2075.1994.tb06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C. The Neural Crest. 2nd Edition Cambridge University Press; 1999. [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Mollereau B, Dominguez M, Webel R, Colley NJ, Keung B, de Celis JF, et al. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, et al. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Noden DM. Somatotropic and functional organization of the avian trigeminal ganglion: An HRP analysis in the hatchling chick. Journal of Comparative Neurology. 1980;224:415–427. doi: 10.1002/cne.901900302. [DOI] [PubMed] [Google Scholar]
- Onuma Y, Nishinakamura R, Takahashi S, Yokota T, Asashima M. Molecular cloning of a novel Xenopus spalt gene (Xsal-3) Biochemical and Biophysical Research Communications. 1999;264:151–156. doi: 10.1006/bbrc.1999.1479. [DOI] [PubMed] [Google Scholar]
- Panganiban G, Irvine SM, Lowe C, Roehl H, Corley LS, Sherbon B, et al. The origin and evolution of animal appendages. Proceedings of the National Academy of Sciences of the U.S.A. 1997;94:5162–5166. doi: 10.1073/pnas.94.10.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Cantera R, Urban J, Technau G, Kafatos FC, Barrio R. Spalt modifies EGFR-mediated induction of chordotonal precursors in the embryonic PNS of Drosophila promoting the development of oenocytes. Development. 2001;128:711–722. doi: 10.1242/dev.128.5.711. [DOI] [PubMed] [Google Scholar]
- Shah NM, Marchionni MA, Isaacs I, Stroobant P, Anderson DJ. Glial growth factor restricts mammalian neural crest stem cells to a glial fate. Cell. 1994;77:349–360. doi: 10.1016/0092-8674(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Shah NM, Anderson DJ. Integration of multiple instructive cues by neural crest stem cells reveals cell-intrinsic biases in relative growth factor responsiveness. Proceedings of the National Academy of Sciences of the U.S.A. 1997;94:11369–11374. doi: 10.1073/pnas.94.21.11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu Y, Maynard TM, Weston JA. Fate determination of neural crest cells by NOTCH-mediated lateral inhibition and asymmetrical cell division during gangliogenesis. Development. 2000;127:2811–2821. doi: 10.1242/dev.127.13.2811. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Wholemount in situ hybridization of vertebrate embryos. In: Wilkinson DG, editor. Situ Hybridization: A Practical Approach. IRL Press; 1992. pp. 75–83. [Google Scholar]


