Abstract
Hepatitis C virus (HCV) is a major cause of chronic hepatitis worldwide, but the study of HCV infection has been hampered by the lack of an in vitro or in vivo small animal model. The tree shrew Tupaia belangeri is susceptible to infection with a variety of human viruses in vivo, including hepatitis viruses. We show that primary Tupaia hepatocytes can be infected with serum- or plasma-derived HCV from infected humans, as measured by de novo synthesis of HCV RNA, analysis of viral quasispecies evolution, and detection of viral proteins. Production of infectious virus could be demonstrated by passage to naive hepatocytes. To assess whether viral entry in Tupaia hepatocytes was dependent on the recently isolated HCV E2 binding protein CD81, we identified and characterized Tupaia CD81. Sequence analysis of cloned Tupaia cDNA revealed a high degree of homology between Tupaia and human CD81 large extracellular loops (LEL). Cellular binding of E2 and HCV infection could not be inhibited by anti-CD81 antibodies or soluble CD81-LEL, suggesting that viral entry can occur through receptors other than CD81. Thus, primary Tupaia hepatocytes provide a potential model for the study of HCV infection of hepatocytes.
Introduction
Hepatitis C virus (HCV) is a major cause of posttransfusion and community-acquired hepatitis in the world (1–4). The majority of HCV-infected individuals develop chronic hepatitis that may progress to liver cirrhosis and hepatocellular carcinoma (5). Treatment options for chronic HCV infection are limited, and no vaccine to prevent HCV infection is available (3).
HCV has been tentatively classified in a separate genus (Hepaci virus) of the Flaviviridae family (6–8). The virion contains a positive-strand RNA genome of approximately 9.5 kb. The genome consists of a highly conserved 5′ untranslated region (UTR), followed by a long open reading frame of 9,030 to 9,099 nucleotides that is translated into a single polyprotein of 3,010 to 3,030 amino acids (6–8). The polyprotein is posttranslationally processed by host and viral proteases. The HCV structural proteins comprise the putative nucleocapsid or core protein and the two envelope glycoproteins E1 and E2 (6–8). E2 has been shown to interact with CD81 (9, 10), a member of the tetraspanin family (11). CD81 has been suggested as an HCV receptor candidate (10). Viral replication occurs via a negative-strand RNA intermediate, and is dependent on the viral nonstructural proteins, including the RNA-dependent RNA polymerase NS5B (6, 12). In vivo analysis of viral replication and protein expression in blood and liver from infected individuals requires the use of highly sensitive detection methods, such as strand-specific RT-PCR for viral RNA, and immunofluorescence for viral proteins (6, 8).
Although a detailed analysis of the viral genetic organization has led to the identification of various elements, the study of HCV infection has been hampered by the inability to propagate the virus efficiently in vitro and by the limited animal tropism of the virus. The chimpanzee (Pan troglodytes) is the only nonhuman host serving as a model for HCV infection (13–15). Since the availability of these animals is limited, the development of new model systems is pivotal for the analysis of HCV infection. The tree shrew Tupaia belangeri a species closely related to primates (16), has been shown to be susceptible to a variety of human viruses, including herpes simplex, hepatitis B (HBV), and rotavirus (17–21). Recent studies have demonstrated that primary Tupaia hepatocytes can be efficiently infected with HBV (22), allow the study of HBV replication and virion synthesis (23). A recent pilot study has demonstrated that T. belangeri can be infected with HCV in vivo (16). In this study, we demonstrate that primary Tupaia hepatocytes represent a novel cell culture model for the study of HCV infection that allows the functional assessment of HCV receptor candidate CD81.
Methods
Plasmids, antibodies, proteins, and cell lines.
Plasmid pCDHCV.S1b, containing the cDNA for the HCV structural proteins (HCV-J strain; genotype 1b) under the control of the CMV promoter, was obtained by ligating the EcoRI-XbaI fragment of pFastbacHCV.S (24–26) into pCDNA3 (Invitrogen Corp., Carlsbad, California, USA). pEGFP-N1 was obtained from CLONTECH Laboratories Inc. (Palo Alto, California, USA). pCVH77C, containing the cDNA to generate infectious full-length HCV genome (15), was a generous gift of J. Bukh and R. Purcell, of the Hepatitis Viruses Section, NIAID, NIH (Bethesda, Maryland, USA). pGEMT-H77UTRC was generated by ligation of a PCR-amplified DNA fragment coding for the 5′ UTR and amino terminal core region (HCV nt’s 1–410) of pCVH77C into pGEMT (Promega Corp., Madison, Wisconsin, USA), as recommended by the manufacturer. Mouse monoclonal anti-CD81 antibodies 5A6 and 1D6 have been described (10, 11, 27). The mouse monoclonal anti-E2 antibody 3E5 and recombinant E2 protein were generously provided by M. Houghton (Chiron Corp., Emeryville, California, USA). Mouse monoclonal anti-core c1/c2 (24) and anti-NS3 antibodies were generous gifts of H.B. Greenberg (Stanford University, Palo Alto, California, USA) and G. Baccala (Biomerieux Inc., Lyon, France), respectively. Monoclonal mouse anti-galactosidase (anti-LacZ) antibody was obtained from Boehringer Mannheim Biochemicals Inc. (Mannheim, Germany). The human hepatoma cell line HuH-7 has been described (28).
Isolation and culture of primary Tupaia and rat hepatocytes.
T. belangeri were obtained from the German Primate Center, Göttingen, Germany. Sprague-Dawley rats were obtained from the Zentralinstitut für Versuchstierzucht, Hannover, Germany. The animals were bred and maintained at the animal facilities of University Hospital Freiburg, in accordance with institutionally approved protocols and the NIH guidelines for the use of experimental animals. Primary hepatocytes were isolated from adult animals (male and female, 10–12 weeks old, 180–200 g body weight) as described in detail (20, 22). Freshly isolated Tupaia hepatocytes were seeded at a density of 2 × 105 to 3 × 105 cells/ml medium (2 ml per well) on collagen-coated six-well plates (Becton Dickinson Co., Bedford, Massachusetts, USA). Confluence after plating was 80–90%, with hepatocyte viability of greater than 90% as assessed by Trypan blue exclusion. After plating, hepatocytes were maintained as described (22). Medium was changed daily. Primary rat hepatocytes from 10- to 12-week-old female Sprague-Dawley rats were isolated and cultured in a manner similar to that previously described (22, 29).
HCV-positive sera and infection of hepatocytes.
For infection of primary Tupaia hepatocytes, serum samples containing high-titer HCV RNA were obtained from patients with acute and chronic hepatitis C being followed at the Department of Medicine II, University of Freiburg, Germany. Plasma sample H77 7-12-77 (dilution 10–1; described in refs. 13, 15, 30) was a generous gift of R. Purcell. Serum ES-2 was a generous gift of R.S. Ross and M. Roggendorf (Institute for Virology, University of Essen, Germany). Chronic hepatitis C was diagnosed by detection of anti-HCV antibodies (HCV EIA 3.0 RIBA HCV 3.0 SIA; Chiron Corp.), HCV RNA (Amplicor HCV Monitor; Roche Diagnostic Corp., Raritan, New Jersey, USA) and persistently elevated transaminases. HCV RNA was quantitated using Amplicor HCV Monitor (Roche Diagnostic Corp.). HCV genotyping was performed using INNO-LiPA HCV II (Innogenetics Corp., Gent, Belgium). Coinfection with HBV and HIV was excluded by serology. Anti-HCV antibody profile was determined by ELISA (as described in ref. 26) using insect cell–derived HCV-like particles (HCV-LPs, 1 μg/assay) (25, 26) recombinant core (50 ng/assay; G. Baccala, Biomerieux Inc.), E2 (50 ng/assay; M. Houghton, Chiron Corp.), and NS3 proteins (50 ng/assay; Mikrogen GmbH, Munich, Germany) as capture antigens. Anti-HCV immunoreactivity was determined by endpoint titration (26), using a serum panel (n = 10) from anti-HCV–negative healthy volunteers as control. Statistical analyses were performed using the Student t test. All serum samples were aliquoted and stored at –80°C until use.
Hepatocytes were infected with HCV on days 1–5 after hepatocyte plating, by incubation for 6–16 hours with 0.5–50 μl of freshly thawed serum in 1 ml Biocoat medium per well. At various timepoints after infection, hepatocytes and medium were collected and processed as described below. For virus passage medium of six wells of HCV-infected hepatocytes was pooled, concentrated by low-speed centrifugation using Millipore Ultrafree spin columns (Millipore Corp., Bedford, Massachusetts, USA), and added to freshly prepared hepatocytes 1 day after plating.
Analysis of HCV RNA by strand-specific RT-PCR.
At various timepoints after infection, cell culture medium was collected. The cells were washed three times with PBS. Total RNA from cells and medium was isolated using RNeasy from QIAGEN GmbH, Hilden, Germany; and the ULTRASPEC RNA Isolation System from BIOTECX Laboratories Inc., Houston, Texas, USA. Positive- and negative-strand HCV RNA was then analyzed using 1 μl of 50 μl of total RNA by strand-specific RT-PCR using rTth polymerase (31) and two different sets of 5′ UTR–specific primers (31, 32). For the synthesis of control HCV positive- and negative-strand RNA, pCVH77C and pGEMT-H77UTRC were linearized by digestion with XbaI (15) or SpeI, respectively. In vitro transcription and purification of HCV RNA from linearized gel-purified cDNAs (10 μg) was performed as described (15). The absence of residual cDNA template was confirmed by PCR using HCV-specific primers. Quantitation of HCV RNA was performed using the Amplicor HCV Monitor system (Roche Diagnostic Corp.). Ethidium bromide staining of 18S and 28S RNAs after agarose gel electrophoresis, and quantitation of β-actin mRNA were used to assess the quality of purified RNA from Tupaia hepatocytes. Tupaia β-actin mRNA was analyzed by RT-PCR of cellular RNA. Reverse transcription of mRNA was performed using M-MLV reverse transcriptase (Life Technologies Inc., Gaithersburg, Maryland, USA) and random primers (Promega Corp.) according to the manufacturers’ protocols. cDNA amplification was performed using Taq polymerase (Boehringer Mannheim Biochemicals Inc.) and human β-actin–specific primers (nt’s 68–87 and nt’s 843–821 according to GenBank accession no. AB004047; sense primer: 5′-GGCTTCGC-GGGCGACGATGC-3′; antisense primer: 5′-GTAGTTTC-ATGGATGCCACAGGA-3′). Each round of PCR (25 cycles) consisted of denaturation at 94°C for 45 seconds (in first cycle 4 minutes), annealing at 65°C for 30 seconds, and elongation at 72°C for 45 seconds (in last cycle 7 minutes). Amplified products were analyzed by agarose gel electrophoresis, and the identity of the PCR product was confirmed by Southern blot analysis using a 32P-labeled human β-actin–specific cDNA probe. To avoid false-positive RT-PCR results, the procedures suggested by Kwok and Higuchi (33) were followed. Mock-infected hepatocytes served as negative controls in all experiments. For sequence analysis of HCV hypervariable region-1 (HVR-1), HCV RNA purified from inoculum H77 07-12-77 or from infected Tupaia hepatocytes (harvested on day 5 after infection) was amplified by reverse transcription and nested PCR using H77C–HVR-1–specific primers: inner sense primer 5′-GCCATATAAGGCGTCATCGCATGGC-3′ (HCV nt’s 1273–1297 according to GenBank accession no. AF011751); inner antisense primer 5′-GGTGTTAAGGCTTTCATTGCAATT-3′ (nt’s 1646-1623); outer sense primer: 5′-GCATGGGATATGATGATGAACTGGTC-3′ (nt’s 1296–1321); and outer antisense primer 5′-TCTCAGGACAGCCTGAAGAGTTGAA-3′ (nt’s 1704–1680) as described recently (34). RT-PCR products were gel purified and ligated into pCR2.1 vector using the TA cloning kit from Invitrogen BV (Groningen, The Netherlands). Clones were analyzed by cycle sequencing using a 377 DNA sequencer (Perkin-Elmer Life Sciences, Weiterstadt, Germany). Sequences were aligned using MegAlign software (DNAStar Inc., Madison, Wisconsin, USA), and compared with H77C consensus sequence (GenBank accession no. AF011751).
Detection of HCV-encapsidated RNA.
On day 5 after infection, medium was harvested, and hepatocytes were lysed and homogenized in a buffer containing 50 mM Tris-HCl, 50 mM NaCl, and 0.1% NP-40, pH 7.5 (24). Lysates were cleared from nuclei and cellular debris by low-speed centrifugation (15 minutes at 4°C and 10,000 g). The supernatants were layered on 30% (wt/vol) sucrose (in 20 mM Tris-HCl and 150 mM NaCl, pH 7.4) and centrifuged for 4 hours at 4°C and 150,000 g. Pelleted HCV particles were digested with 24 μg/ml Staphylococcus aureus nuclease (Boehringer Mannheim Biochemicals Inc.) and 50 μg/ml of RNase A (QIAGEN GmbH) in 20 mM Tris-HCl, 2 mM NaCl, and 5 mM CaCl2 (pH 8.8) for 2 hours at 37°C to eliminate any nonincorporated nucleic acids (24, 28, 35). Particle-associated RNA was isolated using an ULTRASPEC RNA Isolation System; (BIOTECX Laboratories Inc.), and subjected to HCV-specific RT-PCR as described above.
Immunofluorescence analyses of HCV protein expression.
At various timepoints after infection, cells were fixed with a 1:1 mixture of methanol/acetone (vol/vol) and incubated for 1 hour with serum from a panel of HCV-infected individuals containing high-titer anti-HCV antibody (dilution 1:100 in PBS with 1% BSA) or serum from an anti-HCV–negative individual (dilution 1:100 in PBS with 1% BSA), followed by a 1-hour incubation with Cy3-conjugated anti-human secondary antibody (Jackson ImmunoResearch Laboratories Inc., West Grove, Pennsylvania, USA; dilution 1:250 in PBS with 1% BSA). Between steps, plates were washed five times with PBS. Prior to analysis by immunofluorescence assay, all human sera were preabsorbed twice by a 3 hour incubation on methanol/acetone-fixed uninfected primary Tupaia hepatocytes to remove nonspecific antibodies (25). Preabsorbed sera of noninfected individuals and noninfected Tupaia hepatocytes were used as negative controls in all experiments. To assess specificity and sensitivity of viral protein analysis, primary Tupaia hepatocytes were transfected with the cDNA plasmid pCDHCV.S1b, a control plasmid (pCDNA3; Invitrogen Corp.), and a plasmid expressing the green fluorescent protein (pEGFP-N1; CLONTECH Laboratories Inc.). Tupaia hepatocytes in six-well plates were transfected with pEGFP-N1 and 1 μg plasmid DNA (pCDHCV.S1b or pCDNA3, at a 5:1 ratio) using Lipofectamine Plus (Life Technologies Inc.) in a 3-hour incubation. HCV protein expression was analyzed as described above. GFP expression was analyzed using a rabbit anti-GFP antibody (CLONTECH Laboratories Inc.; dilution 1:100 in PBS with 1% BSA) and an FITC-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories Inc.; dilution 1:500 in PBS with 1% BSA).
The anti-HCV serum panel has been described in detail in immunofluorescence and immunogold affinity labeling studies of HCV structural proteins expressed in insect cells (24, 25). Anti-HCV antibody immunoreactivity of the anti-HCV serum panel was neutralized by coincubation of diluted serum with HCV recombinant proteins at a final concentration of 20 μg/ml prior to use in immunofluorescence. HCV proteins included core, E1, and E2, expressed as HCV-LPs (24–26); recombinant core protein (G. Baccala, Biomerieux Inc.); recombinant E2 (M. Houghton, Chiron Corp.); and purified recombinant nonstructural proteins NS3, NS4, and NS5 (NS3, Mikrogen GmbH and Biomerieux Inc.; NS4 and NS5, Mikrogen GmbH).
Analysis of Tupaia CD81.
Tupaia CD81 cDNA was cloned using human CD81–specific primers covering the large extracellular loop (LEL) domain (nt’s 575–591 and nt’s 841–821 according to GenBank accession no. M33680; sense primer: 5′-GAATTCCACCATGTTTGTCAACAAGGACCA-3′; antisense primer: 5′-TCTAGATCACTTCCCGGAGAAGAGGTCATC-3′). RT-PCR of Tupaia CD81 mRNA was performed on cellular Tupaia RNA as described above, except that the PCR annealing temperature was 60°C. Tupaia CD81 PCR products were purified with the QIAquick PCR purification kit (QIAGEN GmbH) and cloned into plasmid pCR-XL-TOPO (Invitrogen BV). Five clones of the resulting plasmid (pXLTCD81EC2.1-5) were analyzed by automated sequencing (ABI 3700, Applied Biosystems Inc., Weiterstadt, Germany) using CD81-specific primers. For the analysis of CD81 expression, HuH-7 cells and Tupaia and rat hepatocytes were lysed in a buffer containing 50 mM Tris-HCl, 1% NP-40, 50 mM NaCl, and 5 mM EDTA, pH 7.4. After low-speed centrifugation, lysates were subjected to SDS-PAGE under nonreducing conditions on a 12% polyacrylamide gel. Proteins were detected by immunoblot using mouse monoclonal anti-CD81 antibody (1D6; dilution 1:1,000 in PBS containing 1% Tween 20 and 1% BSA). Expression of CD81 in freshly isolated Tupaia hepatocytes was analyzed by FACS using anti-CD81 antibodies (5A6 and 1D6; 10 μg/ml) and phycoerythrin-conjugated (PE-conjugated) anti–mouse IgG antibody (10 μg/ml; Jackson ImmunoResearch Laboratories Inc.) as described recently (10). For the analysis of CD81-dependent viral entry, Tupaia hepatocytes were incubated with anti-CD81 (5A6 and 1D6) under saturating conditions (10 μg/ml) at 37°C for 1 hour (10). Subsequently, the cells were infected by the addition of serum as described above. Six hours later, the medium containing serum and anti-CD81 antibodies was removed, and the cells were cultured and processed as described above.
Binding of recombinant E2 to primary Tupaia hepatocytes.
Binding of recombinant E2 to Tupaia hepatocytes was performed as described recently for MOLT-4 cells (36–38). Primary rat hepatocytes and human hepatoma HuH-7 cells served as negative and positive controls, respectively. Freshly isolated hepatocytes (from rat or Tupaia) or human hepatoma HuH-7 cells (106 cells per assay) were incubated with recombinant E2 protein in PBS (final volume 200 μl) at various concentrations for 1 hour at 4°C. Nonbound E2 protein was removed by three centrifugations in PBS at 200 g for 5 minutes at 4°C. Hepatocytes were subsequently incubated for 30 minutes at 4°C with anti-E2 (3E5) or mouse monoclonal isotype control antibody. The cells were washed four times in PBS at 4°C, suspended in 200 μl PBS, and incubated with PE-conjugated anti–mouse IgG antibody (Dianova AB, Hamburg, Germany). After removing unbound antibodies by a final washing (four times in PBS), cell-bound fluorescence was analyzed with a FACScan flow cytometer using Lysys II software (Becton Dickinson Immunocytometry Systems). Lysys II produces histograms of each sample and calculates the mean fluorescence intensity (MFI) of the cell population, which directly relates to the surface density of PE-labeled E2 bound to hepatocytes (36). MFI values of cells with or without bound E2 protein, and with isotype control antibody or anti-E2, are compared. The threshold for positivity is set for each experiment by flow cytometric analysis of hepatocytes without E2 protein, that have been incubated with anti-E2 and PE-conjugated secondary antibody (36). To study whether binding of E2 can be blocked by recombinant soluble CD81-LEL (expressed as glutathione-S-transferase fusion protein), E2 (10 μg/ml) was preincubated with CD81-LEL (50 μg/ml for 1 hour at 25°C; E2 binding to cell lines was assessed by FACS as described above. To assess whether binding could be blocked by anti-CD81, hepatocytes were incubated for 1 hour at 25°C with anti-CD81 (5A6 and 1D6) at 10 μg/ml (a concentration previously shown to saturate CD81), prior to the addition of E2. E2 binding in the presence of anti-CD81 was detected using a biotinylated anti-E2 antibody and streptavidin-PE.
Results
Infection of primary Tupaia hepatocytes with serum-derived HCV.
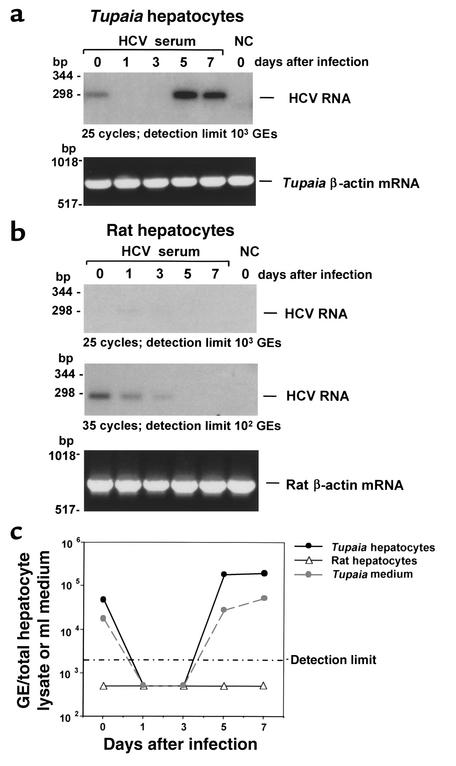
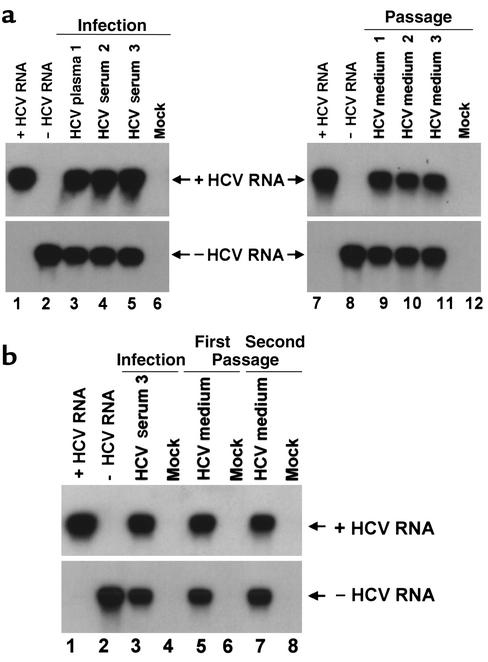
To study whether primary Tupaia hepatocytes can be infected with serum-derived HCV, hepatocytes were incubated with serum from individuals chronically infected with HCV. Analysis of HCV RNA in primary Tupaia hepatocytes by strand-specific RT-PCR, using HCV-specific primers, demonstrated a time-dependent increase of HCV RNA after incubation of cells with HCV-positive serum, indicating successful infection of the hepatocytes (Figure 1a). Quantitation of hepatocyte-derived HCV RNA using a commercially available HCV RNA detection assay (Figure 1c) revealed a time-dependent increase of HCV RNA levels, from less than 2 × 103 genomic equivalents (GEs) per 105 hepatocytes on days 1 and 3, to approximately 104–105 GEs per 105 hepatocytes on days 5 and 7 after incubation. HCV RNA could be detected until day 14 after plating. The rapid decline in hepatocyte viability 14 days after plating precluded a meaningful analysis of HCV RNA at later timepoints. Hepatocytes were susceptible to infection until day 5 after plating. In addition to the de novo synthesis of positive-strand RNA (Figure 1, a and c), infection and replication of HCV in primary Tupaia hepatocytes could be confirmed by detection of negative-strand RNA in hepatocytes (see Figure 2 and Figure 7). Beginning on day 5 after incubation, HCV RNA was detected in culture medium (Figure 1c), indicating secretion of viral particles. HCV RNA in medium reached levels of up to 104 GEs/ml. If the newly synthesized HCV RNA was from replicating virus, RNAs should be packaged in enveloped virions and resistant to degradation by ribonuclease. Indeed, HCV RNA in hepatocytes and medium was resistant to ribonuclease under conditions that completely degraded in vitro synthesized naked RNA (data not shown). Virus could be passaged to uninfected primary Tupaia hepatocytes (Figure 2, a and b). Viral passage was demonstrated by the detection of positive- and negative-strand HCV RNA using two independent RT-PCR methods, as well as analysis of viral quasispecies evolution. Enrichment of virus from cell culture supernatants and the use of freshly prepared hepatocytes were crucial for successful viral passage.
Figure 1.
(a) HCV infection of primary Tupaia hepatocytes. Primary Tupaia hepatocytes were incubated with 50 μl HCV RNA–positive serum 1 day after plating. At various timepoints after plating (days 0–7, as indicated), hepatocyte RNA was isolated, and positive-strand HCV RNA was analyzed by strand-specific RT-PCR using HCV-specific primers as described in Methods. The negative control (NC) consisted of primary Tupaia hepatocytes incubated with HCV-negative control serum harvested and analyzed on day 0 (day of incubation). To control for RNA integrity, cellular Tupaia β-actin mRNA was amplified by RT-PCR as described in Methods. The detection limit of the RT-PCR (25 cycles) was approximately 103 GEs/assay. (b) Lack of HCV infection in primary rat hepatocytes. Primary rat hepatocytes were incubated with the same HCV RNA–positive serum described in a, and HCV RNA was analyzed as described above. To control for RNA integrity, cellular rat β-actin mRNA was amplified by RT-PCR. The detection limit of the RT-PCR was approximately 103 GEs/assay when 25 cycles were used, and increased to approximately 102 GEs/assay when 35 cycles were used. (c) Quantitative analysis of HCV RNA synthesis in primary Tupaia and rat hepatocytes. Primary Tupaia and rat hepatocytes were incubated with HCV RNA–positive serum as described above. At various timepoints, positive-strand HCV RNA was quantified using the Amplicor HCV Monitor assay from Roche Diagnostic Corp. Cellular and medium HCV RNA levels are indicated as GEs/ml of hepatocyte lysate and medium. The detection limit of this assay was 2 × 103 GEs/ml.
Figure 2.
Passage of HCV in primary Tupaia hepatocytes. (a) Passage of HCV to naive cells. Primary Tupaia hepatocytes were incubated with HCV RNA–positive plasma or serum (1: 10 μl H77 7-12-77, 10–1 dilution; 2: 10 μl ES-2; or 3: 50 μl A5387) 1 day after plating. 5 days later, HCV infection was determined by RT-PCR of cellular positive- and negative-strand HCV RNA, on day 5 after incubation with serum-derived HCV, as described above. Strand-specificity of RT-PCR was assessed using 10 fg of in vitro–synthesized positive- (lanes 1 and 7) and negative-strand HCV RNA (lanes 2 and 8). On day 5 after infection, medium was transferred to naive hepatocytes, and infection was assessed 5 days later as described above. (b) Serial passage. Primary Tupaia hepatocytes were infected with serum 3 as described above. On day 5 after infection, medium was transferred to naive hepatocytes from a separate preparation (first passage). Eight days later, medium was used to infect a third preparation of hepatocytes (second passage). Infection of hepatocytes was assessed as described above.
Figure 7.
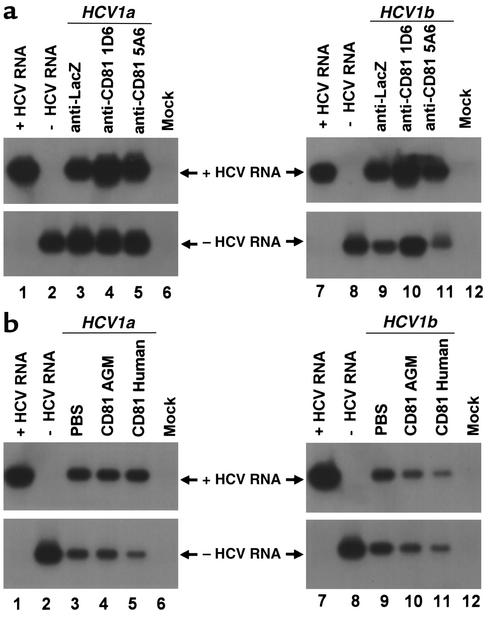
HCV infection of primary Tupaia hepatocytes in the presence of anti-CD81 and soluble CD81-LEL. (a) Hepatocytes were incubated with anti-CD81 antibodies (5A6 and 1D6) at saturating conditions (10 μg/ml) or with control antibody (anti-LacZ, 10 μg/ml) 1 hour prior to the addition of HCV serum as described in Methods. HCV infection was determined by RT-PCR of cellular positive- and negative-strand HCV RNA on day 5 after incubation with RNA-positive serum HCV of genotypes 1a (50 μl, moi 0.2, lanes 3–5) and 1b (50 μl, moi 0.1, lanes 9–11) as described above. Strand specificity of RT-PCR was assessed by RT-PCR of 10 fg of in vitro–synthesized positive- (lanes 1 and 7) and negative-strand HCV RNA (lanes 2 and 8). (b) Serum-derived HCV was mixed with PBS (lanes 3 and 9), soluble CD81-LEL derived from African green monkey (AGM) (50 μg/ml; lanes 4 and 10), or human CD81-LEL (50 μg/ml; lanes 5 and 11), and incubated for 1 hour in PBS. Subsequently, HCV-CD81 complexes were added to the hepatocytes as described in Methods. Infection of hepatocytes was assessed as described above.
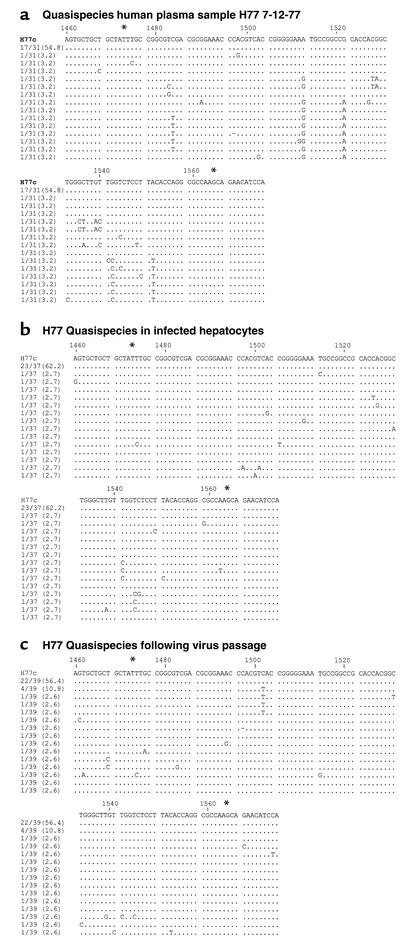
To study the evolution of viral infection in Tupaia hepatocytes, we assessed viral quasispecies in hepatocytes infected with plasma H77 7-12-77. H77 quasispecies evolution has been characterized in detail in humans and chimpanzees (13–15, 30, 39). Sequence analysis of the HCV E2 HVR-1 of the H77 genome in the inoculum and in Tupaia hepatocytes before and after viral passage revealed a marked difference in HCV quasispecies (Figure 3, a–c). HVR-1 quasispecies analysis of H77 plasma (Figure 3a) demonstrated two predominant viral populations: a major population consisting of H77C consensus sequence, and a minor population characterized by defined mutations at nt 1484 (C to T), nt 1513 (A to G), nt 1522 (G to A), nt 1542 (G to C), and nt 1551 (A to T). By contrast, analysis of quasispecies in infected Tupaia hepatocytes (Figure 3b) demonstrated an increase of H77C consensus sequence genomes (to 62.2% of total clones), and a complete disappearance of the second predominant population present in H77 7-12-77 plasma. Following viral passage, a novel population of viral genomes containing a C-to-T mutation at nt 1502 emerged in 7 of 39 clones (Figure 3c). These data are consistent with selection of specific viral genomes and generation of novel mutations in HCV-infected hepatocytes.
Figure 3.
Comparative sequence analysis of HVR-1 from HCV H77 genomes in the inoculum and in infected Tupaia hepatocytes. Primary Tupaia hepatocytes (5 × 105 cells/well) were incubated with HCV RNA–positive plasma (10 μl H77 7-12-77; 10–1 dilution) as described above. On day 5 after infection, hepatocyte RNA was isolated for detection of HCV RNA. Medium was transferred to naive hepatocytes, and infection of passaged virus was assessed as described in Figure legend 2. Plasma- and hepatocyte-extracted HVR-1 sequences of HCV genomes were amplified and sequenced as described in Methods. Comparisons of plasma and hepatocyte HCV (before and after virus passage) HVR-1 sequences with the consensus sequence of functional clone H77C (15) are shown (GenBank accession no. AF011751). The pair of numbers at the left of each line, one before and one after the slash, indicates the fraction of clones and the percentage of each sequence within the spectrum obtained, respectively. *Limits of HVR-1 (HCV nt’s 1473–1565).
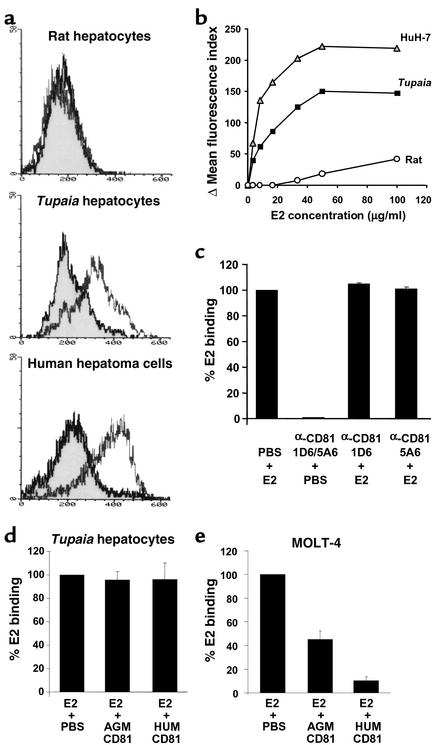
Infection was dependent on specific inocula and required a minimal virus dose. Only 6 of 14 serum samples from HCVinfected patients resulted in infection of Tupaia hepatocytes. HCV viral load, genotype, and anti-HCV antibody profile were not significantly different (P > 0.05) between infectious and noninfectious sera (Table 1). Coincubation of infectious and noninfectious serum resulted in successful infection of Tupaia hepatocytes. This observation indicates that antibodies from noninfectious sera did not inhibit cellular entry of virus from infectious sera. Purification of noninfectious sera by gradient ultracentrifugation (22, 25) did not result in these sera becoming infectious. Serial dilution of six different infectious HCV sera, with subsequent analysis of HCV infection, revealed minimal infectious doses between 2 × 104 and 2 × 105 GEs per 5 × 105 hepatocytes (Table 1). These doses correspond to a multiplicity of infection (moi) of 0.04–0.4. The presence or absence of infection was reproduced in Tupaia hepatocyte preparations from at least three different animals, indicating that the difference in serum infectivity appeared to depend primarily on viral factors. Using specific sera and cell culture conditions (cell viability > 90%), HCV infection of Tupaia hepatocytes was highly reproducible.
Table 1.
Profiles of HCV RNA–positive human serum and plasma samples used for HCV infection of primary Tupaia hepatocytes
To make certain that HCV RNA detected in Tupaia hepatocytes does not represent residual input virus, primary rat hepatocytes were incubated using the same infectious HCV serum and conditions summarized in Figure 1. In contrast to the results obtained using Tupaia hepatocytes, incubation of primary rat hepatocytes did not reveal any evidence of HCV infection. Serial analysis of HCV RNA in similarly incubated rat hepatocytes did not show any detectable HCV RNA (Figure 1b, upper panel). By increasing the detection limit using more sensitive RT-PCR conditions, a low-level signal corresponding to HCV RNA could be detected on the day of incubation and on days 1 and 3 after infection. These signals most likely correspond to input HCV RNA nonspecifically bound to hepatocytes. In contrast to de novo HCV RNA synthesis in primary Tupaia hepatocytes, no HCV RNA could be detected beyond day 3, indicating a lack of viral RNA synthesis and HCV infection in rat hepatocytes. These findings were confirmed by quantitative RT-PCR of HCV RNA (Figure 1c).
De novo synthesis of viral proteins during HCV infection.
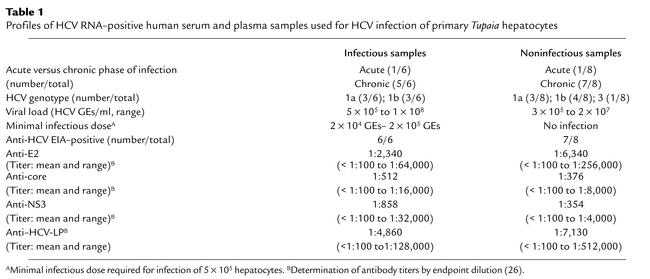
To study viral protein expression in infected Tupaia hepatocytes, infected cells were stained with anti-HCV antibodies. A previously characterized pool of human serum containing high-titer anti-HCV antibodies (24, 25) was used to identify newly synthesized HCV proteins in primary Tupaia hepatocytes. Prior to the analysis of HCV protein expression, the serum pool containing anti-HCV antibodies was preabsorbed on naive primary Tupaia hepatocytes, to remove antibodies binding nonspecifically to the hepatocytes. The specificity of the anti-HCV antibodies present in the serum pool was then confirmed by immunofluorescence of primary Tupaia hepatocytes that had been transfected with a cDNA for HCV structural proteins (Figure 4, a–d), and by inhibition of anti-HCV immunoreactivity using HCV recombinant proteins (Figure 4h). Analysis of HCV proteins in infected primary Tupaia hepatocytes demonstrated a time-dependent increase in HCV protein expression (Figure 4, e and f) that was confined to the cytoplasm (Figure 4, f and g). Staining was positive in only 1–5% of Tupaia hepatocytes, indicating that only a small fraction of hepatocytes synthesize detectable levels of HCV proteins. A strong inhibition of HCV immunoreactivity could be achieved by neutralization of antibodies using HCV recombinant proteins prior to immunofluorescence (Figure 4h). Monoclonal antibodies were less sensitive for the detection of HCV proteins than were anti-HCV antibodies in sera from HCV-infected individuals. Low-level staining was observed using two monoclonal antibodies directed against HCV core and NS3 proteins (data not shown). Viral protein expression could not be detected by immunoblot. Under light microscopy, HCV-infected primary Tupaia hepatocytes did not show any cytopathic changes compared with control hepatocytes incubated with HCV-negative serum.
Figure 4.
HCV protein synthesis in infected primary Tupaia hepatocytes. (a–d) Immunofluorescence of transfected primary Tupaia hepatocytes. To assess specificity and sensitivity of anti-HCV antibodies for immunofluorescence, primary Tupaia hepatocytes were cotransfected with a plasmid (pCDHCV.S1b) expressing the HCV structural proteins, and with a GFP-expression construct (pEGFP-N1) (a and b), or with pEGFP-N1 and pcDNA3 (c and d). Forty-eight hours after transfection, the cells were fixed with a 1:1 mixture of methanol/acetone (vol/vol) and incubated with a well-characterized serum panel (24, 25) from HCV-infected individuals containing high-titer anti-HCV antibody (diluted 1:100 in PBS with 1% BSA), followed by Cy3-conjugated anti–human IgG secondary antibody (b and d). To identify and locate transfected individual cells, the cells were coincubated with anti-GFP antibody, followed by an FITC-conjugated anti–rabbit IgG antibody. (a and c). (e–h) Immunofluorescence of infected Tupaia hepatocytes. For HCV infection, primary Tupaia hepatocytes were incubated with HCV RNA–positive serum 1 day after plating. On days 1 (e) and 5 (f and g) after infection, viral protein expression was analyzed by immunofluorescence using anti–HCV-specific antibodies as described above. Staining was positive in 1–5% of Tupaia hepatocytes (f and g). Anti-HCV immunoreactivity (g) could be inhibited by incubation of anti-HCV antibodies with recombinant HCV proteins prior to use in immunofluorescence (h). Magnification: ×40 (a–f) and ×10 (g and h).
Isolation and characterization of Tupaia CD81.
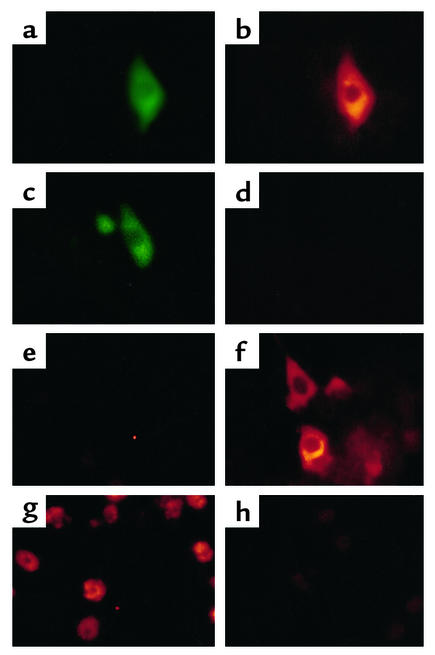
To assess CD81-dependent viral entry into Tupaia hepatocytes, we identified and characterized Tupaia CD81. The LEL of CD81 has been shown to interact specifically with the HCV E2 protein (9, 10), and CD81 has been proposed as an HCV receptor candidate that mediates viral entry (10). E2-CD81 interaction can be blocked by monoclonal anti-CD81 antibodies and soluble CD81-LEL fusion proteins (10, 27, 38). To study whether primary Tupaia hepatocytes express CD81, lysates of primary Tupaia hepatocytes were subjected to SDS-PAGE and immunoblot using monoclonal anti–human CD81 antibodies. Monoclonal anti–human CD81 antibodies 5A6 and 1D6 specifically detected CD81 as a 26-kDa protein in Tupaia hepatocytes and in the human hepatoma cell line HuH-7 (Figure 5a). FACS analysis revealed strong expression of CD81 on the cell surface of freshly isolated Tupaia hepatocytes (Figure 5b). CD81 saturation was achieved at concentrations similar to those described recently for human cell lines. By contrast, anti–human CD81 antibodies did not show any interaction with rat hepatocyte CD81 (Figure 5a). To analyze Tupaia CD81 primary structure, CD81 cDNA was cloned from Tupaia hepatocyte mRNA using human CD81–specific primers. Sequence analysis revealed a strong homology between human and Tupaia CD81, with six amino acid changes in each of their LEL domains (Figure 5c).
Figure 5.
Characterization of Tupaia CD81. (a) CD81 expression in human hepatoma cells and in Tupaia and rat hepatocytes. Cells (5 × 105) grown in monolayer culture were lysed, and lysates were subjected to SDS-PAGE and immunoblot using a mouse monoclonal anti–human CD81 antibody (1D6), as described in Methods. CD81 protein is indicated on the right. Molecular masses (in kDa) are indicated on the left. (b) Analysis of CD81 expressed on the surface of freshly isolated Tupaia hepatocytes. Tupaia hepatocytes (106 cells) were incubated with anti-CD81 or isotype control antibody (10 μg/ml). Staining and flow cytometry were performed as described in Methods. X axis and y axis show mean fluorescence intensity and relative number of stained cells, respectively. (c) Alignment of amino acid sequences of human, chimpanzee, Tupaia, tamarin, African green monkey, and rat LEL domain of CD81. Tupaia CD81-LEL cDNA was cloned and sequenced as described in Methods. CD81-LEL amino acid sequences of other species are depicted according to previous reports (11, 48).
Binding of HCV glycoprotein E2 to Tupaia hepatocytes.
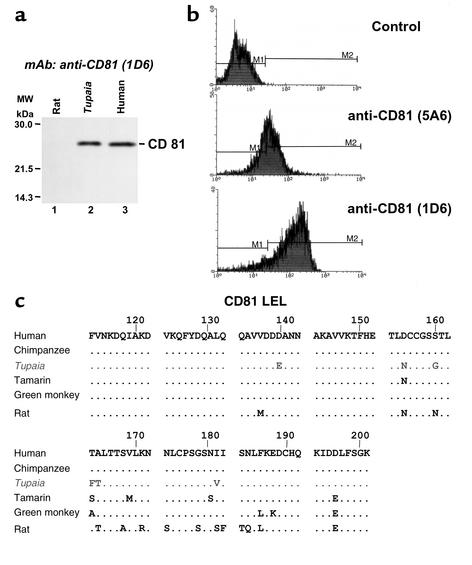
To analyze whether Tupaia hepatocytes interact with E2, binding of recombinant E2 protein to freshly isolated hepatocytes was analyzed by FACS (Figure 6a). Recombinant E2 protein, previously shown to interact with human lymphoma cells (36, 37), did bind to freshly isolated Tupaia hepatocytes. Binding was dose-dependent and saturable, reaching a plateau at an E2 concentration of approximately 50 μg/ml (Figure 6b). Binding of E2 to primary Tupaia hepatocytes was then compared with binding to human hepatoma cells and rat hepatocytes. While human hepatoma cells exhibited an E2 binding profile similar to that of primary Tupaia hepatocytes, primary rat hepatocytes did not demonstrate saturable E2 binding (Figure 6, a and b). E2 binding to primary Tupaia hepatocytes could not be blocked by anti-CD81 antibodies or soluble CD81-LEL fusion proteins (Figure 6, c and d). In contrast, coincubation of E2 with soluble CD81-LEL fusion proteins resulted in a partial (for African green monkey CD81) or almost complete (for human CD81) inhibition of E2 binding to MOLT-4 lymphoma cells (Figure 6e). These data indicate that binding of E2 to hepatocytes is CD81-independent.
Figure 6.
Binding of E2 to Tupaia hepatocytes. (a) Flow cytometry of hepatocyte-bound E2. Primary Tupaia and rat hepatocytes or human hepatoma HuH-7 cells were incubated with PBS (shadowed curve, black line) or E2 protein (17.5 μg/ml; nonshadowed curve, gray line). Staining and flow cytometry of hepatocytes incubated with PBS or E2 protein were performed as described in Methods. X axis and y axis show mean fluorescence intensity and relative number of stained cells, respectively. (b) Dose-dependent binding of E2. Rat and Tupaia primary hepatocytes or HuH-7 hepatoma cells were incubated with E2 at the concentrations indicated. The net MFI values for each E2 concentration were calculated as described in Methods. (c) E2 binding in the presence of anti-CD81 antibodies. Primary Tupaia hepatocytes were incubated with anti-CD81 antibodies (5A6 and 1D6) (10 μg/ml) 1 hour prior to the addition of recombinant E2 protein. E2 binding was determined by FACS using a biotinylated anti-E2 antibody and streptavidin-PE. (d and e) E2 binding in the presence of CD81-LEL. To study whether binding of E2 can be blocked by CD81-LEL, E2 was preincubated with an excess of soluble CD81-LEL as described in Methods. E2-CD81 complexes were added to hepatocytes, and E2 binding to Tupaia hepatocytes (d) or MOLT-4 lymphoma cells (e) was then assessed by FACS as described in a. Data are shown as percent binding compared with binding of E2 in the presence of PBS (100%). Binding was calculated as net mean MFI (as described above) ± SD of three representative experiments. AGM, African green monkey; hum, human.
HCV infection and CD81.
To study whether HCV infection of primary Tupaia hepatocytes was mediated by the LEL of CD81, HCV infection was analyzed in the presence of anti-CD81 antibodies and soluble CD81-LEL. Monoclonal anti–human CD81 antibodies binding to Tupaia CD81 were not able to block viral infection of Tupaia hepatocytes at concentrations recently shown to inhibit E2 binding to target cells (Figure 7a). The functional properties of the antibodies had been assessed by their ability to bind Tupaia hepatocyte CD81 (Figure 5b). Furthermore, HCV infection was observed in the presence of soluble CD81-LEL (Figure 7b). To confirm that soluble fusion proteins were functionally intact, we had studied their ability to block binding of recombinant E2 to MOLT-4 lymphoma cells (Figure 6e), as described previously (9, 10, 37). Taken together, these findings indicate that viral entry of HCV into primary Tupaia hepatocytes appears to be independent of CD81, and suggest that HCV may use a receptor (or receptors) other than CD81 for viral entry.
Discussion
In this study we demonstrate that primary Tupaia hepatocytes are susceptible to HCV infection. Infection is demonstrated by several lines of evidence: (a) a time-dependent, de novo synthesis of HCV RNA in hepatocytes incubated with HCV serum, detected by two independent methods using three sets of HCV-specific primers; (b) detection of negative-strand HCV RNA in infected hepatocytes, using a highly strand-specific RT-PCR method; (c) detection of newly synthesized HCV RNA in culture medium; (d) nuclease resistance of viral RNA in medium, and hepatocytes indicating packaging of newly synthesized HCV RNA into viral particles; (e) formation of new viral genomes during infection, as demonstrated by comparative analysis of viral quasispecies in serum and infected hepatocytes; (f) de novo synthesis of HCV proteins in infected Tupaia hepatocytes; and (g) passage of HCV from infected to naive hepatocytes.
In contrast, no evidence for HCV infection of primary rat hepatocytes was observed, applying the same criteria for infection. Similar to in vivo findings in humans, HCV replication required the use of highly sensitive methods, such as RT-PCR and immunofluorescence, for the detection of viral RNA and proteins. HCV RNA detected in cell culture medium was nuclease-resistant, indicating the presence of packaged viral RNA, consistent with the presence of HCV virions. HCV RNA levels in culture medium reached levels of up to 104 GEs/ml, which is similar to reported HCV RNA serum levels in Tupaia in vivo (1 × 104 to 3.5 × 105 GEs/ml).
Under optimal cell culture conditions, HCV could be passaged (Figure 2 and Figure 3), indicating the production of infectious virus in primary Tupaia hepatocytes. Hepatocytes did not show any cytopathic changes, compared with control hepatocytes, at the light microscopy level. Electron microscopy revealed the presence of putative viral structures in infected Tupaia hepatocytes (data not shown). However, at the present time, the identification of these structures as HCV virions is impaired by the lack of well-defined, reproducible positive control virions from infected human or chimpanzee samples, and by the low abundance of particles in infected cells. Studies are underway to characterize these structures by immunogold affinity antibody labeling.
Analysis of HCV quasispecies in Tupaia hepatocytes infected with a well-characterized serum (13, 15, 30) revealed a selection of genomes consisting of H77C consensus sequence (15). These data suggest that clones of H77C consensus recently shown to be infectious in chimpanzees (15) may have the ability to replicate in primary Tupaia hepatocytes. Indeed, preliminary studies in our laboratory have indicated evidence for replication of transfected full-length H77C HCV RNA (40). Following viral passage, a defined minority population containing a novel HVR-1 mutation could be detected (Figure 3). These data are consistent with recent longitudinal analyses of H77 quasispecies evolution in humans and chimpanzees demonstrating time-dependent development of new mutations (30, 39).
Both virus- and host-specific factors may be important for successful HCV infection in Tupaia in vitro and in vivo. Infection of hepatocytes appeared to be dependent on defined inocula, and required a minimal infectious dose (Table 1). Since only one sample (H77 7-12-77) was tested for infectivity in chimpanzees (13, 15), we do not know whether infectivity of Tupaia hepatocytes correlates with infectivity in chimpanzees. Infectivity of HCV serum samples was not associated with genotype or HCV antibody profile (Table 1). There was no correlation between infectivity and the absence or presence of antibodies against the envelope proteins (Table 1). Noninfectious HCV RNA–positive sera did not inhibit entry of virus present in infectious sera, and purification of virus from noninfectious sera using gradient centrifugation (22, 25) did not result in these sera becoming infectious. These observations indicate that viral entry and infection appear not to be inhibited by antibodies or other factors present in noninfectious sera. Our data suggest that in fact only certain HCV isolates have the ability to infect and replicate in Tupaia hepatocytes. Recent studies resulting in the isolation of infectious full-length or replication-competent subgenomic HCV RNA have demonstrated that only certain HCV RNAs are infectious in vivo (14, 15) or are able to replicate in hepatoma cells (12, 41). Isolate-specific viral factors may include a specific virus envelope protein structure that allows efficient viral entry into Tupaia hepatocytes, or structural or regulatory features of the viral replicative complex that allow efficient viral replication. This hypothesis is corroborated by the results of viral quasispecies analysis in infected cells that demonstrate a selection of certain variants and the suppression of others following viral infection (Figure 3). Further characterization of the infectious isolates may allow development of Tupaia-tropic inocula. The detailed analysis of intracellular HCV RNA after successful infection and passage may result in the isolation of full-length HCV RNA that efficiently replicates in primary Tupaia hepatocytes.
Furthermore, the results of our study may be helpful for the development of Tupaia as an in vivo model for HCV infection. In vivo HCV infection of Tupaia has been described as transient with low viral levels, and occurred in only some of the inoculated animals (16). Our findings in primary Tupaia hepatocytes suggest that the use of Tupaia as an in vivo model could depend on the selection of Tupaia-tropic HCV strains.
Several in vitro models for the study of HCV infection have been proposed. These models include primary hepatocytes from humans (42) or chimpanzees (31), human biliary epithelial cells (43), and human lymphocytic cell lines (44). Limitations of these models are the very low level of HCV replication, requiring the use of ultrasensitive nested RT-PCR methods, and the variable quality of host cells, such as human hepatocytes obtained from surgical specimens. In contrast to other reported human cell culture models, viral replication in primary Tupaia hepatocytes reached levels that could be easily and reproducibly detected by using a strand-specific RT-PCR requiring only one step of cDNA amplification. Since Tupaia can be adapted to a laboratory environment and bred in captivity, primary hepatocytes can be isolated under elective, optimal experimental conditions, resulting in the availability of high-quality hepatocytes.
To gain novel insight into the biology of HCV infection, we studied the functional role of the recently discovered HCV E2–binding molecule CD81 in the establishment of viral infection. CD81 is a member of the tetraspanin family, and has been shown to play an important role in signal transduction and adhesion in the immune system (11). The LEL of CD81 has been shown to interact specifically with HCV E2 protein (9, 10), suggesting a potential role of CD81 as an HCV receptor candidate. CD81-E2 interaction may play an important role in lymphocyte activation (45).
Our studies demonstrate that Tupaia and human CD81 both exhibit a high degree of homology at the amino acid level in the E2-binding domain of h CD81-LEL. This finding is consistent with the close genetic relationship between the two species. Tupaia CD81 exhibited a total of six amino acid changes in the LEL domain. In contrast to African green monkey CD81, which exhibits a mutation at amino acid position 186 (F to L) that reduces E2 binding (27, 38) (Figure 5), amino acid 186 was preserved in Tupaia CD81. Monoclonal anti–human CD81 antibodies showed strong binding to Tupaia CD81 in immunoblots and flow cytometry. Binding of monoclonal anti-CD81 antibody 1D6 to Tupaia CD81 is of particular interest, since this antibody has been used to map the minimal epitope required for CD81-E2 interaction by applying its property to block this interaction (27).
Our data indicate that recombinant E2 binds to Tupaia hepatocytes, but not to rat hepatocytes (Figure 6). Binding reached a plateau at an E2 concentration of approximately 50 μg/ml. Although data are available for binding of E2 to various cell lines (10, 36), no data have been published on E2 binding to primary human hepatocytes. Only one study has demonstrated indirect evidence that E2 may interact with hepatocytes present in fixed liver tissue sections from surgical specimens (38). In conjunction with the observation that Tupaia hepatocytes are susceptible to HCV infection, our results are consistent with the hypothesis that the interaction of E2 protein with the hepatocyte membrane may be important for viral entry (36). Furthermore, our data suggest that primary Tupaia hepatocytes may serve as a model to study binding of HCV E2 protein.
Although both of the anti-CD81 antibodies used in this study interact strongly with Tupaia CD81, and have been shown to inhibit interaction of E2 with CD81 (10, 27, 46), these antibodies failed to block both cellular binding of E2 and HCV infection of Tupaia hepatocytes under CD81-saturating conditions. Furthermore, soluble CD81 did not block E2 binding or inhibit HCV infection at concentrations shown to inhibit E2 binding to MOLT-4 lymphoma cells (Figure 6). These results indicate that both cellular binding of E2 and HCV infection of Tupaia hepatocytes seem to be CD81-independent. Our data suggest that HCV may use an additional receptor or receptors for viral attachment and entry.
Our findings are corroborated by recent observations providing indirect evidence for the lack of CD81 as a receptor mediating viral entry: (a) the molecule is expressed ubiquitously on the surface of various cell types, not limited to HCV-susceptible cells (11); (b) binding of sucrose gradient–purified virions from HCV-infected individuals to lymphoma cells and foreskin fibroblasts does not require HCV-CD81 interaction (47); and (c) binding of E2 to CD81 in various species does not predict susceptibility to HCV infection (48, 49).
In summary, our study demonstrates that primary Tupaia hepatocytes can be infected with HCV independently of CD81. This hepatocyte-based cell culture model should be useful for study of important steps of the viral life cycle, including the assessment of HCV receptor candidates and virus neutralization.
Acknowledgments
The authors thank M. Houghton, S. Abrignani, J.A. McKeating, R. Bartenschlager, and J. Köck for helpful discussions; S. MacNelly and P. Schürmann for excellent technical assistance; S. Wellnitz for initial support of the study; and J. Rasenack and his laboratory for support in HCVgenotyping and HCV RNA quantitation. The generous gifts of recombinant E2 protein and anti-E2 antibody 3E5 (M. Houghton), anti-core antibody (H.B. Greenberg), recombinant NS3 protein and anti-NS3 antibody (G. Baccala), recombinant NS3, NS4, and NS5 proteins (H. Diepolder), and human plasma (R. Purcell) and serum samples (S. Ross and M. Roggendorf) are gratefully acknowledged. This work was supported in part by grants from the Wilhelm Sander-Stiftung, Munich, Germany (99041.1); the European Union, Brussels, Belgium (QLRT-PL1999-00356); and the Bundesministerium für Bildung und Forschung, Bonn, Germany (01KI-9951).
Footnotes
Zhen-Ya Tang’s present address is: Molecular Oncology Laboratory, Alton Ochsner Medical Foundation, New Orleans, Louisiana, USA.
Parts of this study were presented in abstract form at the 6th and 7th International Meetings on Hepatitis C and Related Viruses (Bethesda, Maryland, USA, June 6–9, 1999, and Gold Coast, Australia, December 3–7, 2000).
Xiping Zhao and Zhen-Ya Tang contributed equally to this work.
References
- 1.Choo Q-L, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Alter HJ, et al. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 3.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 4.Alter MJ, et al. The prevalence of hepatitis C virus infection in the United States 1988-1994. N Engl J Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 5.Tong MJ, El-Farra NS, Reikes AR, Co RL. Clinical outcomes after transfusion-associated hepatitis C. N Engl J Med. 1995;332:1463–1466. doi: 10.1056/NEJM199506013322202. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 7.Rice, C.M. 1996. Flaviviridae: the viruses and their replication. In Fields virology. B.N. Fields, D.M. Knipe, and P.M. Howley, editors. Lippincott-Raven Publishers. Philadelphia, Pennsylvania, USA. 931–959.
- 8.Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 9.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 10.Flint M, et al. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy S, Todd SC, Maecker HT. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 12.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 13.Farci P, et al. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 15.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z-C, et al. Transmission of hepatitis C virus infection to tree shrews. Virology. 1998;244:513–520. doi: 10.1006/viro.1998.9127. [DOI] [PubMed] [Google Scholar]
- 17.Darai G, Schwaier A, Komitowski D, Munk K. Experimental infection of Tupaiabelangeri (tree shrews) with herpes simplex virus types 1 and 2. J Infect Dis. 1978;137:221–226. doi: 10.1093/infdis/137.3.221. [DOI] [PubMed] [Google Scholar]
- 18.Park US, et al. Mutations in the p53 tumor suppressor gene in tree shrew hepatocellular carcinoma associated with hepatitis B virus infection and intake of aflatoxin B1. Gene. 2000;251:73–80. doi: 10.1016/s0378-1119(00)00183-9. [DOI] [PubMed] [Google Scholar]
- 19.Pang QF, Liu JC, Wan XB, Qiu FX, Xu AY. Experimental infection of adult Tupaiabelangeri yunalis with human rotavirus. Chin Med J (Engl) 1983;96:85–94. [PubMed] [Google Scholar]
- 20.Walter E, Keist R, Niederöst B, Pult I, Blum HE. Hepatitis B virus infection of Tupaiahepatocytes in vitro and in vivo. Hepatology. 1996;24:1–5. doi: 10.1002/hep.510240101. [DOI] [PubMed] [Google Scholar]
- 21.Yan RQ, et al. Human hepatitis B virus and hepatocellular carcinoma. I. Experimental infection of tree shrews with hepatitis B virus. J Cancer Res Clin Oncol. 1996;122:283–288. doi: 10.1007/BF01261404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köck J, et al. Efficient infection of primary Tupaiahepatocytes with purified human and woolly monkey hepatitis B virus. J Virol. 2001;75:5084–5089. doi: 10.1128/JVI.75.11.5084-5089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren S, Nassal M. Hepatitis B virus (HBV) virion and covalently closed circular DNA formation in primary Tupaiahepatocytes and human hepatoma cell lines upon HBV genome transduction with replication-defective adenovirus vectors. J Virol. 2001;75:1104–1116. doi: 10.1128/JVI.75.3.1104-1116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumert TF, Ito S, Wong D, Liang TJ. The hepatitis C virus structural proteins assemble into virus-like particles in insect cells. J Virol. 1998;72:3827–3836. doi: 10.1128/jvi.72.5.3827-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumert TF, et al. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology. 1999;117:1397–1407. doi: 10.1016/s0016-5085(99)70290-8. [DOI] [PubMed] [Google Scholar]
- 26.Baumert TF, et al. Antibodies against hepatitis-C viruslike particles and viral clearance in acute and chronic hepatitis C. Hepatology. 2000;32:617–624. doi: 10.1053/jhep.2000.9876. [DOI] [PubMed] [Google Scholar]
- 27.Higginbottom A, et al. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol. 2000;74:3642–3649. doi: 10.1128/jvi.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumert TF, Rogers SA, Hasegawa K, Liang TJ. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest. 1996;98:2268–2276. doi: 10.1172/JCI119037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumert TF, Huber M, Mayer D, Keppler D. Ethanol-induced inhibition of leukotriene degradation by ω-oxidation. Eur J Biochem. 1988;182:223–229. doi: 10.1111/j.1432-1033.1989.tb14821.x. [DOI] [PubMed] [Google Scholar]
- 30.Ogata N, Alter HJ, Miller RH, Purcell RH. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 32.Chan SW, et al. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 33.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 34.Enomoto N, Kurosaki M, Tanaka Y, Marumo F, Sato C. Fluctuation of hepatitis C quasispecies in persistent infection and interferon treatment revealed by single-strand conformation polymorphism analysis. J Gen Virol. 1994;75:1361–1369. doi: 10.1099/0022-1317-75-6-1361. [DOI] [PubMed] [Google Scholar]
- 35.Baumert TF, Marrone A, Vergalla J, Liang TJ. Naturally occurring mutations define a novel function of the hepatitis B virus core promotor in core protein expression. J Virol. 1998;72:6785–6795. doi: 10.1128/jvi.72.8.6785-6795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosa D, et al. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heile JM, et al. Evaluation of hepatitis C virus glycoprotein E2 for vaccine design: an endoplasmic reticulum-retained recombinant protein is superior to secreted recombinant protein and DNA-based vaccine candidates. J Virol. 2000;74:6885–6892. doi: 10.1128/jvi.74.15.6885-6892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petracca R, et al. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol. 2000;74:4824–4830. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Major ME, et al. Long-term follow-up of chimpanzees inoculated with the first infectious clone for hepatitis C virus. J Virol. 1999;73:3317–3325. doi: 10.1128/jvi.73.4.3317-3325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, Z.Y, et al. 1999. Primary Tupaia hepatocytes as a model to study hepatitis C virus infection and replication. Sixth International Meeting on Hepatitis C and Related Viruses. Bethesda, Maryland, USA. Presentation 4.1 (Abstr.)
- 41.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1975. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 42.Rumin S, et al. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro. J Gen Virol. 1999;80:3007–3018. doi: 10.1099/0022-1317-80-11-3007. [DOI] [PubMed] [Google Scholar]
- 43.Loriot M-A, et al. Permissiveness of human biliary epithelial cells to infection by hepatitis C virus. Hepatology. 1999;29:1587–1595. doi: 10.1002/hep.510290527. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu Y, Iwamoto A, Hijikata M, Purcell RH, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wack A, et al. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur J Immunol. 2001;31:166–175. doi: 10.1002/1521-4141(200101)31:1<166::aid-immu166>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 46.Flint M, et al. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J Virol. 2000;74:702–709. doi: 10.1128/jvi.74.2.702-709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wunschmann S, Medh JD, Klinzmann D, Schmidt WN, Stapleton JT. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74:10055–10062. doi: 10.1128/jvi.74.21.10055-10062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meola A, et al. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J Virol. 2000;74:5933–5938. doi: 10.1128/jvi.74.13.5933-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allander T, Forns X, Emerson SU, Purcell RH, Bukh J. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology. 2000;277:358–367. doi: 10.1006/viro.2000.0617. [DOI] [PubMed] [Google Scholar]