Abstract
CBF1 is a member of the CSL family of DNA binding factors, which mediate either transcriptional repression or transcriptional activation. CSL proteins play a central role in Notch signaling and in Epstein–Barr virus-induced immortalization. Notch is a transmembrane protein involved in cell-fate decisions, and the cytoplasmic domain of Notch (NotchIC) targets CBF1. The Epstein–Barr virus-immortalizing protein EBNA2 activates both cellular and viral gene expression by targeting CBF1 and mimicking NotchIC. We have examined the mechanism of CBF1-mediated repression and show that CBF1 binds to a unique corepressor, CBF1 interacting corepressor (CIR). A CIR homolog is encoded by Caenorhabditis elegans, indicating that CIR is evolutionarily conserved. Two CBF1 mutants that were unable to bind CIR did not function as repressors, suggesting that targeting of CIR to CBF1 is an important component of repression. When expressed as a Gal4 fusion protein, CIR repressed reporter gene expression. CIR binds to histone deacetylase and to SAP30 and serves as a linker between CBF1 and the histone deacetylase complex.
Epstein–Barr virus (EBV) immortalizes B cells and is associated with human malignancies including Burkitt’s lymphoma, nasopharyngeal carcinoma, Hodgkin’s disease, and lymphoproliferative disease in immunosuppressed patients (1). Two major cell-signaling pathways are constitutively activated by EBV during immortalization. The EBV-latency membrane protein LMP-1 has effects on B cells similar to those that occur after activation of CD40, a membrane protein that is a member of the tumor necrosis factor receptor (TNFR) superfamily (2). LMP-1 functions as a constitutively activated TNFR. The cytoplasmic carboxyl terminus of LMP-1 binds to the TNFR-associated factors TRAF1, TRAF2, TRAF3, and TRADD, leading to activation of the transcription factor NF-κB (3–5). The EBV-encoded nuclear proteins EBNA2, EBNA3A, and EBNA3C each interact with the DNA binding factor, CBF1 (also called RBPJk and Jk) (6–11). EBNA-LP has recently been shown to cooperate with EBNA2 by binding to the EBNA2 activation domain (12) and hence indirectly targets CBF1.
CBF1 belongs to a family of highly conserved CSL proteins with homologs in Drosophila (Suppressor of Hairless, [Su(H)] (13) and Caenorhabditis elegans (Lag-1; ref. 14). CBF1 binds to the DNA sequence GTGGGAA (15, 16) and functions in both transfection assays and in vitro transcription assays as a transcriptional repressor (17, 18). EBNA2 binds to the transcriptional repression domain of CBF1, and relief of repression combined with the effects of the EBNA2 transcriptional activation domain induces expression of repressed genes (18). Interaction of EBNA3A and EBNA3C with CBF1 prevents DNA binding by CBF1, and the interaction of the EBNA3 proteins and EBNA2 is mutually exclusive (9, 10, 19). These properties suggest that the EBNA3 proteins may modulate the effects of EBNA2 in a regulatory partnership. Drosophila also encodes a protein with a function analogous to the EBNA3s. The Drosophila Hairless protein interacts with the Drosophila CSL protein Su(H) and prevents it from binding to DNA (20).
In both Drosophila and C. elegans, there is persuasive genetic evidence that the CSL proteins play a role in Notch signaling (21). Several genes, such as Drosophila enhancer of split and its mammalian homolog HES-1, have been identified that contain CSL binding sites in their regulatory regions and are downstream participants in Notch signaling (22). It was demonstrated in Drosophila (23) and subsequently in mammalian cells (24, 25) that the intracellular domain of Notch directly interacts with CSL proteins. Notch receptors are large transmembrane proteins. Their extracellular domains contain tandem epidermal growth factor repeats, and the intracellular domain contains six ankyrin repeats and a carboxyl-terminal PEST sequence. Notch proteins participate in intercellular signaling events that mediate cell-fate specification (21, 26). There are four mammalian Notches—Notch1, Notch2, Notch3 and Notch4/Int-3. Notch is expressed in uncommitted proliferative cells during development and is believed to function in the adult to maintain the proliferative capacity of immature cells (27, 28). Disruption or disregulation of Notch signaling has been associated with human neoplastic disease (29, 30), cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencepholopathy (CADASIL, ref. 31), and Alagille syndrome (32).
The common targeting of CBF1 by both EBV EBNA2 (6–8, 11, 33) and the intracellular domain of Notch (NotchIC) (24, 25) and the mechanistic similarities of their interaction established a linkage between the early steps in EBV-induced immortalization and Notch signaling. Both EBNA2 and NotchIC bind to the repression domain of CBF1; both proteins abolish CBF1 repression activity and each activates transcription of responsive promoters through a combination of abolition of repression and the positive effects of an endogenous activation domain (17, 18, 24, 34). CBF1 contains a transferable repression domain (18), but the mechanism of CBF1-mediated repression was unknown. Indirect evidence has implicated a corepressor in this activity (35, 36). The importance of CBF1-mediated transcriptional repression in regulating immortalization and developmental pathways led to the initiation of a yeast two-hybrid screen to help identify the hypothetical corepressor.
MATERIALS AND METHODS
Plasmids.
SG5-Gal4(1–95)(pJH385), was generated by moving the sequences for Gal4(1–95) as a PCR-generated BglII fragment into the BglII site of SG5 (Stratagene). CIR–Gal4 fusion plasmids contained either the complete CIR sequence (pJH394–2) or segments of CIR generated as BglII/BamHI fragments by PCR amplification and ligated into the BamHI site of the vector: CIR(1–350), pJH409; CIR(1–304), pJH408; CIR(1–252), pJH407; CIR(1–150), pJH437; and CIR(1–121), pJH406. SG5-CIR-myc, pJH402, had a myc epitope added to the carboxyl terminus. SG5–Gal4–CBF1(1–500)(pJH93) (18), SG5–Gal4–CBF1(1–500)(EEF-233AAA) (pJH111) (18), SG5–CBF1(1–500)(KLV-249AAA)(pJH287) (34), SG5–Flag–CBF1(pJH282) (24), SG5–EBNA2 (pPDL151) (8), 5xGal4TKCAT, and TKLuc (18) have been described. All yeast Gal4DBD fusions were constructed in a pAS1–CYH2 background, and yeast Gal4ACT fusions were constructed in a pACTII background (37). Yeast plasmids generated were: pJH137, Gal4DBD–CBF1(1–500); pJH49, CIR(1–450)–Gal4DBD; pCJC441, Gal4DBD–CMV IE2(290–542); pJH346, Gal4ACT–CBF1(1–500); pJH178, Gal4ACT–CIR(1–240); pJH421, Gal4ACT–CIR(1–121); and pJH442, Gal4ACT–CIR(1–150). pACTII–CBF1(1–500)(EEF-233AAA)(pJH347) was derived from pJH111 (18); pACTII–CBF1(1–500)(KLV-249AAA) (pJH428) was derived from pJH287, (34); and pACTII–EBNA2 (252–476)(pYW163) was prepared by using PCR amplification of EBNA2 sequences from pPDL151 (8). pSZ1, Gal4ACT–mHDAC2 (1–489) and pSZ2, Gal4ACT–mHDAC2 (286–489) were derived from Flag–mHDAC2, a gift from W.-M. Yang (H. Lee Moffit Cancer Center and Research Institute, University of South Florida, Tampa, FL; ref. 38). Sap30(41–221)–ACT was isolated as part of a yeast two-hybrid screen. All clones were sequenced and tested for protein expression by using Western blot analysis.
Immunofluorescence.
Assays were performed as described (34) in Vero cells plated at 0.8 × 105 cells per well onto 2-well glass chamber slides (Nalge Nunc). A total of 1.5 μg of DNA was transfected per well by using the calcium phosphate procedure. Mouse anti-Flag mAb (Kodak) and rabbit anti-CIR antibody were used at a 1:1,000 dilution. The rabbit anti-peptide CIR antibody was raised against the epitope ETRKRAQRNPGEEQSRR. Secondary anti-mouse Ig and anti-rabbit Ig antibodies (Chemicon) were used at a dilution of 1:100. Processed slides were mounted in Mowiol medium (Calbiochem) and viewed by using confocal optics.
Yeast Two-Hybrid Assay.
Saccharomyces cerevisiae Y190 and the yeast plasmids pAS1-CYH2 and pACT2 were a gift from S. Elledge (Howard Hughes Medical Institute, Baylor College of Medicine, Houston). A human lymphocyte cDNA library in pACT was purchased from CLONTECH. Yeast assays were performed according to the CLONTECH manual. A total of 1.5 million transformants were tested for interaction with CBF1 by using growth in His− medium in the presence of 50 mM 3-amino triazole as the first selection and induction of β-galactosidase activity as the second selection. From 65 clones that were positive in these assays, the two showing the strongest CBF1 interaction were selected. These two cDNAs were isolated and sequenced. The cDNAs were then again transformed with pAS1-CBF1 to reconfirm interaction. β-Galactosidase activity was measured by using 2-nitrophenyl β-d galactopyranoside substrate. The amount of liberated 2-nitrophenol was determined as A420 after incubation for a period of 2–4 hours.
Expression Assays.
Transient expression and Northern blot analyses were performed as described (34).
DNA Sequence.
The human CIR (hCIR) cDNA sequence has been submitted to GenBank.
RESULTS
Identification of CIR.
The CBF1 ORF was fused to the Gal4 DNA-binding domain in pAS1CYH2, and a yeast two-hybrid screen for CBF1-interacting proteins was performed by using a B cell cDNA library cloned in pACT. Outgrowth on selective Trp−, Leu−, His− medium followed by selection for yeast colonies expressing β-galactosidase led to the identification of two cDNA clones whose protein products showed strong interaction with CBF1. A database search revealed that both cDNAs encoded uncharacterized proteins. In subsequent experimental analyses, one of the proteins exhibited the properties of a corepressor. This novel 450-aa corepressor was designated CBF1 Interacting coRepressor (CIR).
In the database search, the predicted protein sequence of CIR isolated from the human B cell library matched the amino-terminal 135 amino acids encoded by a previously cloned and uncharacterized gene named recepin (gb U03644), but the predicted protein sequences diverged after amino acid 135. However, the cDNA sequences encoding the carboxyl terminus of hCIR completely matched other sequences in the expressed sequence tag database. Inspection of the recepin sequence revealed that the discrepancy was likely the result of inaccuracies in the recepin sequence that brought about a change of reading frame in the carboxyl-terminal portion of the predicted recepin protein. The 450-aa hCIR protein has a highly charged, serine-rich carboxyl terminus. The database search also revealed that a closely related protein is encoded by C. elegans (designated CeCIR; yk187f6.5). The 561-aa CeCIR shows striking sequence homology in the amino-terminal half of the protein, particularly over the first 150 amino acids. The carboxyl terminus, although diverged in primary sequence, retains the characteristic of being highly charged and serine-rich (Fig. 1A). Analysis of RNA expression indicated that, like CBF1, hCIR transcripts have a widespread tissue distribution, with strong expression in heart, skeletal muscle, and pancreas (Fig. 1B).
Figure 1.
(A) Alignment of the predicted protein sequences of hCIR and a C. elegans homolog (designated CeCIR) that was identified in a database search (National Center for Biotechnology Information accession no. 1707048; ref. 70). Identical amino acids are indicated by the symbol ∗ and conservative changes by (.). Spaces introduced to maximize alignment are shown as –. The amino acid numbers are provided on the right. (B) Northern blot analysis of hCIR expression performed by using a multiple-tissue RNA blot (CLONTECH) and a CIR DNA probe 32P-labeled by using random priming. After exposure to autoradiographic film, the blot was stripped and rehybridized with an actin DNA probe (actin). he, heart; br, brain; pl, placenta; lu, lung; li, liver; sm, skeletal muscle; ki, kidney; and pa, pancreas.
Intracellular Localization of CIR.
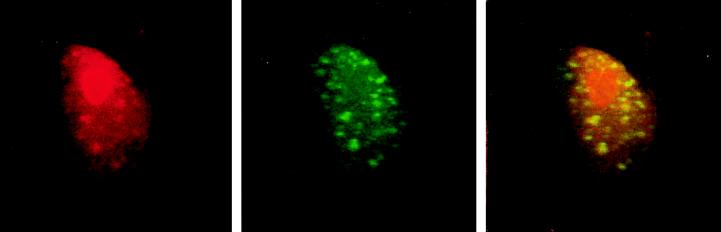
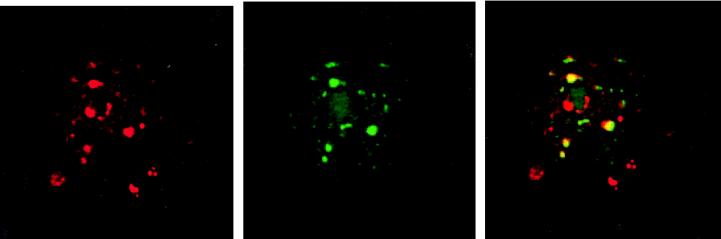
CBF1 is a transcription factor that binds specifically to GTGGGAA motifs in responsive promoters. Complexes of factors involved in RNA synthesis and processing can, in some circumstances, be visualized as discrete intranuclear assemblies that appear as speckles or punctate spots in indirect immunofluorescence assays (reviewed in ref. 39). An example of such an assembly is the array of intranuclear speckles seen on staining for the SC35 splicing factor (40). We used indirect immunofluorescence to examine the intranuclear localization of CBF1 and CIR in transfected cells. Flag–CBF1 gave diffuse intranuclear staining with an underlying pattern of irregular strongly staining speckles plus strong staining in the nucleolus (Fig. 2 Left). Transfected CIR showed a similar pattern minus the nucleolar staining (Fig. 2 Center). Superimposition of the confocal images from cotransfected cells revealed coincidence between the majority of the CIR and CBF1 speckles (Fig. 2 Right). The immunofluorescence data show that CIR is a nuclear protein. The existence of speckles containing both CBF1 and CIR reinforces the suggestion from the yeast two-hybrid screen that these two proteins are capable of interaction and is consistent with both proteins contributing to a common transcriptional complex.
Figure 2.
Colocalization of CIR and CBF1 in transfected Vero cells. In indirect immunofluorescence assays, Flag-CBF1 (Left, red) and CIR (Center, green) each show a mixture of diffuse nuclear staining and strongly staining, irregular nuclear speckles. In the merged images (Right), colocalized staining appears yellow. The pattern indicates colocalization between the majority of CBF1 and CIR nuclear speckles. Primary antibodies were mouse anti-Flag mAb and rabbit anti-peptide CIR antibody. Secondary antibodies were rhodamine-conjugated anti-mouse Ig (red) and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit Ig (green).
CIR Acts as a Repressor.
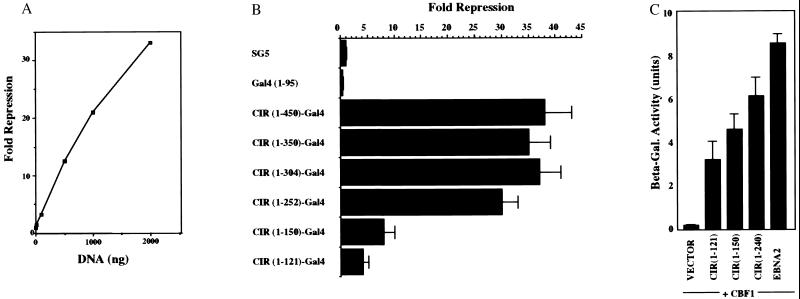
CIR was next tested for transcriptional regulatory activity. Gal4 DNA-binding domain-CIR fusions were constructed in a mammalian expression vector. Cotransfection into HeLa cells of the CIR–Gal4 constructions with a CAT-reporter carrying upstream Gal4 binding sites revealed that CIR efficiently repressed reporter gene expression in a dose-dependent manner in this assay (Fig. 3A). A series of carboxyl-terminal deletions were introduced into CIR to locate the repression domain. When the resulting Gal4-fusion constructions were tested in the repression assay, the data indicated that wild-type levels of repression required CIR amino acids 1–252 (Fig. 3B). The fusion protein containing CIR amino acids 1–121 showed weak activity, whereas CIR(1–150) retained significant repression activity (8- to 10-fold). An examination of the requirements for CIR interaction with CBF1 by using the yeast interaction assay found that the CBF1 interaction domain was located within CIR(1–121) (Fig. 3C). The strongest signal was obtained with CIR(1–252) but CBF1 interaction was readily detectable with both CIR(1–150) and CIR(1–121) placing the CBF1 interaction domain also within the amino-terminal region of CIR. The amino-terminal 150 amino acids represent the region of highest sequence conservation between hCIR and CeCIR and the ability of CIR(1–150) to mediate repressive activity and CBF1 interaction implies that CeCIR is likely to function in a manner similar to hCIR.
Figure 3.
Mapping CIR domains required for repression and CBF1 interaction. (A) Transient-expression assay performed in a dose-response format in HeLa cells cotransfected with a 5xGal4BS–TKCAT reporter (5 μg), a TK-Luc control (1 μg), and increasing amounts of a CIR(1–450)-Gal4(1–95) expression vector. CIR represses reporter-gene expression in a dose-responsive manner. CAT, chloramphenicol acetyltransferase (B) The repression domain is located within CIR amino acids 1–252. Transient-expression assay in HeLa cells cotransfected with 5xGal4BS–TKCAT reporter (5 μg), TK-Luc control (1 μg), and 2 μg of plasmids expressing the indicated CIR–Gal4(1–95) fusion constructions. The results shown are an average of three experiments, with the SD indicated. (C) The CBF1 interaction domain is also located within an N-terminal domain. Yeast two-hybrid assay using β-galactosidase induction as a measure of interaction. CIR(1–121), CIR(1–150), CIR(1–252), and EBNA2 (252–425) were expressed as Gal4ACT fusions, and CBF1 was expressed as a Gal4DBD fusion. The EBNA2–CBF1 pairing served as a positive control for a strong interaction. The results shown are an average of three experiments, with the SD indicated.
Tethering of CIR Is Necessary for CBF1-Mediated Repression.
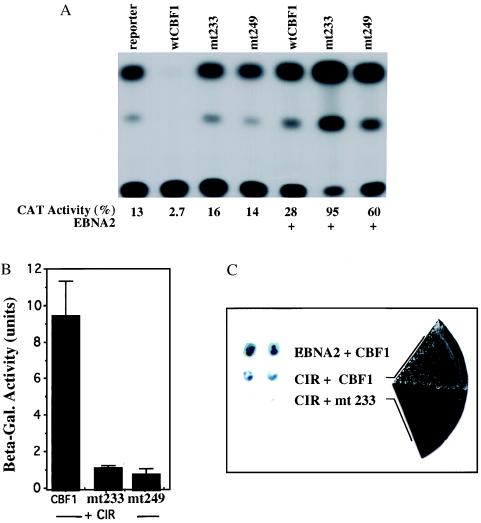
We had previously generated two CBF1 triple-alanine mutants altered at amino acids 233–235 from EEF to AAA and 249–251 from KLV to AAA [CBF1(EEF233AAA) and CBF1(KLV249AAA)] that showed loss of repression activity. The loss of repression did not reflect a global loss of functionality as each mutant retained interaction with EBNA2 (refs. 18 and 34; Fig. 4A). An equivalent mutation to CBF1(EEF233AAA) in Suppressor of Hairless, the Drosophila homolog of human CBF1, displays a hypomorphic hairless phenotype (41) correlating the activity measured in the transient expression assay with a developmental phenotype. The behavior of these two mutants implies that the region of CBF1 between amino acids 233 and 249 forms a key part of the CBF1 repression domain.
Figure 4.
CIR has the properties of a corepressor. (A) Representative transient-expression assay showing loss of repression by the CBF1 mutants CBF1(EEF233AAA) and CBF1(KLV249AAA). The assay was performed in HeLa cells by using a 5xGal4BS–TKCAT reporter (5 μg), TK-Luc control (1 μg), and the indicated CBF1 constructions expressed as Gal4–DBD fusions. The relative specificity of the mutations is indicated by the continued ability of the mutant proteins to mediate EBNA2 activation. (B) CBF1(EEF233AAA) and CBF1(KLV249AAA) show diminished interaction with CIR in a yeast assay that uses quantitative β-galactosidase production as a measure of interaction. The wild-type CBF1 and the CBF1 mutants were expressed as fusions with the Gal4DBD and CIR(1–252) as a fusion with Gal4ACT. The results shown are an average of three experiments, with the SD indicated. (C) Illustration of the results obtained with CBF1(EEF233AAA) by using a qualitative assay for induction of β-galactosidase (Left) or growth on selective His− medium (Right) as a measure of interaction.
If CIR functions as a corepressor for CBF1, then the CBF1 EEF233AAA and KLV249AAA mutations would be expected to have an impact on CBF1–CIR interaction. The two CBF1 mutants and wild-type CBF1 were tested for interaction with CIR in the yeast interaction assay. The two CBF1 repression minus mutants showed a severely diminished ability to interact with CIR as indicated by a quantitative assay for β-galactosidase induction (Fig. 4B). The same loss of interaction was observed by using growth on selective His− medium as a measure of interaction as illustrated for the EEF233AAA mutant in Fig. 4C which also shows a qualitative β-galactosidase induction assay. These results correlate CIR interaction with the ability of CBF1 to function as a repressor.
CIR Links an HDAC Complex to CBF1.
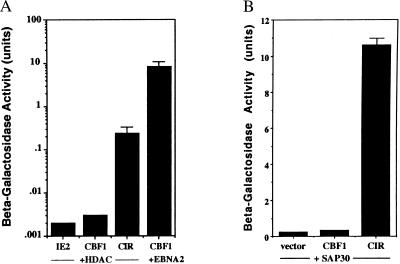
Several mechanisms for transcriptional repression have been described, including DNA binding site competition, DNA bending, and destabilization of the TFIID–promoter complex. An important recent advance in the understanding of transcriptional repression has been the realization that repressor proteins are frequently linked—either directly or indirectly through corepressors such as N-CoR, SMRT, and mSin3—to HDAC (reviewed in refs. 42 and 43). To address the mechanism of CBF1-mediated repression, a yeast two-hybrid assay was used to test for interaction with HDAC. The HDAC2–ACT construction used in this assay contained the same region of HDAC2 (amino acids 286–489) that was originally detected in a yeast two-hybrid screen as an interacting partner for YY1, a well characterized transcriptional repressor that interacts with the adenovirus E1A regulatory protein (38). CBF1 showed no interaction with HDAC2(286–489) (Fig. 5A) or with intact HDAC2 (data not shown). However, the combination of HDAC2–ACT and CIR–Gal4DBD led to induction of β-galactosidase synthesis, indicating interaction (Fig. 5A).
Figure 5.
CIR, but not CBF1, interacts with HDAC2 and SAP30. Interaction between CIR and HDAC2 (A) and CIR and SAP30 (B) was examined in a yeast assay. CBF1 and CIR were each expressed as Gal4DBD fusions and tested in yeast for interaction with HDAC2 (286–489) and SAP30 (41–221) expressed as a Gal4ACT fusions. The negative control in A was the CMV IE2 transcriptional regulatory protein plus HDAC, and the positive control for high-affinity interaction was EBNA2 (252–425) plus CBF1. The results shown are an average of three experiments, with the SD indicated.
HDAC has been found previously to be brought into repression complexes through interactions with the corepressors N-CoR and mSin3. A subset of mSin3 complexes also includes SAP30 (44, 45). SAP30 was described only recently, and because CIR had not previously been recognized as a component of repression complexes, we considered the possibility that CIR may interact with SAP30. We tested CIR for interaction with SAP30 by using a SAP30 construction that was isolated from a B cell library in a separate two-hybrid screen and contained SAP30 amino acids 41–221. The CIR–Gal4 DBD protein showed a strong interaction with SAP30–ACT. In contrast, there was no interaction between SAP30 and CBF1 in this assay (Fig. 5B). The interaction between CIR and HDAC2 and CIR and SAP30 implies that CIR forms a linker between CBF1 and the HDAC–mSin3–SAP30 complex and that CBF1-mediated transcriptional repression involves chromatin modification.
To confirm that the intranuclear localization of CIR and HDAC in mammalian cells was consistent with the interactions observed in yeast, Vero cells were cotransfected with CIR and Flag–HDAC, and the proteins were visualized by indirect immunofluorescence using rabbit anti-CIR polyclonal antibody to detect CIR and mouse anti-Flag mAb to detect Flag–HDAC. Both CIR and HDAC produced a pattern of irregular intranuclear speckling (Fig. 6). In the merged image, there was partial colocalization between CIR and HDAC.
Figure 6.
Partial colocalization of CIR and HDAC2 in the nucleus of transfected cells. In an indirect immunofluorescence assay, Flag-HDAC2 (Left, red) and CIR (Center, green) colocalize in transfected Vero cells in a subset of the intranuclear speckles. In the merged image on the Right, colocalized staining appears yellow. The immunological reagents used were the same as for Fig. 2.
DISCUSSION
The importance of histone deacetylation in mediating transcriptional repression has become increasingly clear with a recent example being the association between the retinoblastoma protein and HDAC (46–48). The involvement of histone deacetylation in repression has been most extensively characterized for the Mad and nuclear hormone receptor family proteins where the corepressor complex includes SMRT or N-CoR, mSin3A or 3B, SAP30, SAP18, RbAp46 and RbAp46 in addition to HDAC1 and HDAC2 (49–54). In these examples, N-CoR or the related SMRT protein interacts with the DNA bound hormone receptor or MAD/MAX proteins. mSin3 in turn binds to N-CoR and brings in HDAC and the other mSin3 associated proteins SAP18, SAP30 and the histone binding proteins RbAp46 and RbAp48 (44, 45). SAP30 appears to function as a linker between N-CoR and mSin3. The amino terminus of SAP30 binds to N-CoR and the carboxyl lterminus binds to mSin3 (45). Interestingly, SAP30 is present in only a subset of corepressor complexes. SAP30 is required for N-CoR-mediated repression through antagonist bound estrogen receptor and the POU domain protein PIT-1 but not for repression by the unliganded retinoic acid receptor or thyroid hormone receptor complexes (45). Here we describe a newly identified corepressor, CIR, that is a component of the CBF1-mediated repression complex and participates in the recruitment of HDAC to DNA-bound CBF1. CIR interacts strongly with SAP30 suggesting that SAP30 also participates in the CBF1 associated repression complex. Because SAP30 is a component of the mSin3 complex, it seems likely that mSin3 will also be present in this complex although this was not addressed experimentally. The yeast two-hybrid experiments do not distinguish between direct and indirect contacts made through yeast homologs and hence it is also possible that there may be intermediate proteins between CBF1 and CIR. Whether CIR is a general participant in all corepressor complexes containing SAP30 or is present in only a subset of complexes of which CBF1 is representative remains to be investigated. CBF1-mediated repression is multifactorial. In addition to the recruitment of the HDAC complex, repression through destabilization of interactions between the transcription factors TFIID and TFIIA also has been recently described (55).
Overcoming CBF1-mediated repression is a crucial step in EBV immortalization of B cells and plays a role in the transmission of Notch signaling during development. The mechanism by which ligand-activated Notch signaling leads to transcriptional changes in the nucleus has been the subject of controversy. An understanding of the intracellular events that follow Notch signaling initially arose from the phenotypes observed in the presence of truncated Notch receptors. In particular, a truncated Notch composed only of the intracellular domain (NotchIC) translocates to the nucleus and functions as a ligand-activated receptor as judged by its gain-of-function phenotype (21). This activated form of Notch not only diverts cell-fate determination but also is associated with the development of tumors, including both naturally occurring and experimental T-cell neoplasms (30, 56–59). It had been postulated that natural ligand-induced activation of Notch leads to a proteolytic cleavage event that releases NotchIC, but this proposal was not universally accepted. However, the recent identification of a proteolytic cleavage site between Notch1 amino acids G1743 and V1744 and the demonstration that cleavage at this site occurs in response to ligand-binding releasing NotchIC, along with evidence obtained by using a V1744 mutant that proteolytic cleavage at this site is necessary for complete activation of an HES-luciferase reporter (60), supports the model of Notch processing with nuclear translocation of NotchIC and interaction with CSL proteins in the nucleus.
Loss of CBF1(mouse RBPJk; ref. 61) expression in mice is lethal to embryos at 10.5 days of gestation—an earlier stage than observed with Notch1 knockout mice—suggesting that CBF1 does not function solely as a downstream target of Notch but also plays a unique role in development (62). It is interesting to note that the CBF1 binding site, which ends in GGGAA, can overlap with an NFκB binding site, which starts with the sequence GGGA. Repression of genes whose promoters contain this sequence overlap could also be relieved by displacement of CBF1 as a consequence of the availability of increased levels of nuclear NFkB. Cellular promoters carrying CBF1 binding sites that overlap with binding sites for the transcription factor NFkB include interleukin 6, major histocompatibility complex class 1, and interferon β (63–67).
Notch signaling is evolutionarily conserved and has been demonstrated in C. elegans. The C. elegans Notch homologs are LIN-12 and GLP-1 and the CBF1 homolog is LAG-1 (14, 68, 69). The existence of a CIR homolog in C. elegans suggests that the pathways that utilize CIR as part of developmental gene regulation are highly conserved. The contribution of CIR apparently is essential for normal development, because loss of CeCIR function by RNA-mediated interference is embryonic-lethal at an early stage of C. elegans development (J.J.-D.H., A. Godbey, and T. Schedl, unpublished results). Further genetic analysis in C. elegans will provide information on the role of CeCIR in Notch signaling and additional insight into the contribution that transcriptional repression makes to embryonic development. In addition, it will be interesting to determine whether NotchIC and EBNA2 use the same or different mechanisms to overcome the repressive effects of the promoter-bound CBF1 complex.
Acknowledgments
We thank Wen-Ming Yang for the Flag-HDAC2 plasmid, S. Elledge for yeast Y190 and the two-hybrid vectors, Chuang-Jiun Chiou for the CMV IE2 plasmid pCJC441, and Michael Delannoy for assistance with confocal microscopy. This work was supported by Public Health Service Grant CA42245 and the American Cancer Society (MCB-85262).
ABBREVIATIONS
- EBV
Epstein–Barr virus
- CIR
CBF1-interacting corepressor
- HDAC
histone deacetylase
- TNFR
tumor necrosis factor receptor
- EBNA
EBV-encoded nuclear antigen
- IC
intracellular domain
- hCIR
human CIR, TK, thymidine kinase
- luc
luciferase
- CMV
cytomegalovirus
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF098297).
References
- 1.Rickinson A B, Kieff E. In: Fields Virology. Field B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2397–2446. [Google Scholar]
- 2.Izumi K M, Kaye K M, Kieff E D. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 5.Izumi K M, Kieff E D. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henkel T, Ling P D, Hayward S D, Peterson M G. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 8.Ling P D, Rawlins D R, Hayward S D. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B, Marshall D R, Sample C E. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimber-Strobl U, Strobl L J, Meitinger C, Hinrichs R, Sakai T, Furukawa T, Honjo T, Bornkamm G W. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada S, Kieff E. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweisguth F, Posakony J W. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 14.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. Development (Cambridge, UK) 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 15.Ling P D, Hsieh J J-D, Ruf I K, Rawlins D R, Hayward S D. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales L D. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh J J-D, Hayward S D. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 19.Johannsen E, Miller C L, Grossman S R, Kieff E. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Genes Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- 21.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 22.Lecourtois M, Schweisguth F. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 23.Fortini M E, Artavanis-Tsakonas S. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh J J-D, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Nature (London) 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 26.Blaumueller C M, Artavanis-Tsakonas S. Perspect Dev Neurobiol. 1997;4:325–343. [PubMed] [Google Scholar]
- 27.Lardelli M, Williiams R, Lendahl U. Int J Dev Biol. 1995;39:769–780. [PubMed] [Google Scholar]
- 28.Robey E. Curr Opin Genet Dev. 1997;7:551–557. doi: 10.1016/s0959-437x(97)80085-8. [DOI] [PubMed] [Google Scholar]
- 29.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis-Tsakonas S. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 31.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, et al. Nature (London) 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Krantz I D, Deng Y, Genin A, Banta A B, Collins C C, Qi M, Trask B J, Kuo W L, Cochran J, et al. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 33.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh J J-D, Nofziger D E, Weinmaster G, Hayward S D. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Development (Cambridge, UK) 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 36.Waltzer L, Bourillot P Y, Sergeant A, Manet E. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 38.Yang W-M, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer R H, Green M R. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 40.Spector D L. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 41.Furukawa T, Maruyama S, Kawaichi M, Honjo T. Cell. 1992;69:1191–1197. doi: 10.1016/0092-8674(92)90640-x. [DOI] [PubMed] [Google Scholar]
- 42.Wolffe A P. Nature (London) 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 43.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Sun Z-W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 45.Laherty C D, Billin A N, Lavinsky R M, Yochum G S, Bush A C, Sun J-M, Mullen T-M, Davie J R, Rose D W, Glass C K, et al. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 46.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 47.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 48.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 50.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 51.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 52.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 53.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 54.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 55.Olave I, Reinberg D, Vales L D. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Girard L, Hanna A Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 57.Jhappan C, Gallahan D, Stahle C, Chu E, Smith G H, Merlino G, Callahan R. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 58.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohn J L, Lauring A S, Linenberger M L, Overbaugh J. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 61.Amakawa R, Jing W, Ozawa K, Matsunami N, Amaguchi Y, Matsuda F, Kawaichi M, Honjo T. Genomics. 1993;17:306–315. doi: 10.1006/geno.1993.1326. [DOI] [PubMed] [Google Scholar]
- 62.Oka C, Nakano T, Wakeham A, de la Pompa J L, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak T W, et al. Development (Cambridge, UK) 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 63.Shirakata Y, Shuman J D, Coligan J E. J Immunol. 1996;156:4672–4679. [PubMed] [Google Scholar]
- 64.Goodbourn S, Maniatis T. Proc Natl Acad Sci USA. 1988;85:1447–1451. doi: 10.1073/pnas.85.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kannabiran C, Zeng X, Vales L D. Mol Cell Biol. 1997;17:1–9. doi: 10.1128/mcb.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyazawa K, Mori A, Yamamoto K, Okudaira H. J Biol Chem. 1998;273:7620–7627. doi: 10.1074/jbc.273.13.7620. [DOI] [PubMed] [Google Scholar]
- 67.Plaisance S, Vanden Berghe W, Boone E, Fiers W, Haegeman G. Mol Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kimble J, Simpson P. Annu Rev Cell Dev Biol. 1997;13:333–361. doi: 10.1146/annurev.cellbio.13.1.333. [DOI] [PubMed] [Google Scholar]
- 69.Yochem J, Greenwald I. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- 70.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]