Abstract
Mutations in PRKAG2, the gene for the γ2 regulatory subunit of AMP-activated protein kinase, cause cardiac hypertrophy and electrophysiologic abnormalities, particularly preexcitation (Wolff-Parkinson-White syndrome) and atrioventricular conduction block. To understand the mechanisms by which PRKAG2 defects cause disease, we defined novel mutations, characterized the associated cardiac histopathology, and studied the consequences of introducing these mutations into the yeast homologue of PRKAG2, Snf4. Although the cardiac pathology caused by PRKAG2 mutations Arg302Gln, Thr400Asn, and Asn488Ile include myocyte enlargement and minimal interstitial fibrosis, these mutations were not associated with myocyte and myofibrillar disarray, the pathognomonic features of hypertrophic cardiomyopathy caused by sarcomere protein mutations. Instead PRKAG2 mutations caused pronounced vacuole formation within myocytes. Several lines of evidence indicated these vacuoles were filled with glycogen-associated granules. Analyses of the effects of human PRKAG2 mutations on Snf1/Snf4 kinase function demonstrated constitutive activity, which could foster glycogen accumulation. Taken together, our data indicate that PRKAG2 mutations do not cause hypertrophic cardiomyopathy but rather lead to a novel myocardial metabolic storage disease, in which hypertrophy, ventricular pre-excitation and conduction system defects coexist.
Introduction
Dominant mutations in the γ2 regulatory subunit (PRKAG2) of AMP-activated protein kinase, an enzyme that modulates glucose uptake and glycolysis (1, 2), have been recently reported (3, 4) to cause hypertrophic cardiomyopathy (HCM). In addition to cardiac hypertrophy, individuals with PRKAG2 mutations frequently manifest electrophysiologic abnormalities, particularly preexcitation (Wolff-Parkinson-White syndrome; WPW), atrial fibrillation, and progressive development of atrioventricular conduction block (3, 4). Although atrial fibrillation is common in HCM patients and becomes increasingly prevalent with disease duration, neither accessory pathways nor conduction system disease are typical features of HCM.
An autosomal dominant disorder, HCM is clinically recognized by unexplained ventricular hypertrophy and a distinctive histopathology that includes myocyte enlargement, myocyte disarray, and increased interstitial fibrosis (reviewed in ref. 5). Previous studies have identified ten different disease genes that are mutated in HCM; despite this genetic heterogeneity, a unified mechanism for disease has been postulated because each of the disease genes encodes a sarcomere protein (reviewed in ref. 6). The discovery of PRKAG2 mutations appears to challenge the hypothesis that HCM is a disease of the sarcomere.
To understand the mechanisms by which PRKAG2 defects cause disease we defined additional, novel mutations and examined the clinical manifestations found in affected individuals. A previously unrecognized and unusual histopathology was identified in hearts with PRKAG2 defects, which prompted biochemical analyses of the functional consequences of human mutations on Snf4, the yeast homologue of the γ2 protein kinase subunit. Collectively our data indicate that PRKAG2 defects do not cause HCM, but rather a novel glycogen storage disease of the heart.
Methods
Clinical studies.
All studies were carried out in accordance with the institutional guidelines for human research. Review of medical records, clinical evaluation, and electrocardiogram and echocardiogram studies were performed as described (7, 8). Detailed clinical characteristics for families AS and MAX were previously reported (8, 9). Cardiac hypertrophy was diagnosed on the basis of left ventricular wall thickness ≥ 13 mm. WPW was diagnosed based on documented spontaneous ventricular preexcitation, positive adenosine test, or demonstration of an accessory pathway on electrophysiologic study. Pathological specimens from members of families A (explanted heart), MF and SS (endomyocardial biopsies), and AS (necropsy specimens from two affected individuals) were examined using hematoxylin and eosin (H&E), Masson Trichrome, periodic acid Schiff (PAS), and PAS plus diastase stains. Samples for electron microscopic examinations were obtained from the paraffin-embedded tissue. After paraffin removal, tissue was fixed in 2% glutaraldehyde and postfixed in osmium tetroxide. Thin sections were stained with uracil acetate and lead citrate.
Genetic studies.
Genomic DNA was extracted from whole blood or Epstein-Barr virus–transformed lymphocytes, as previously described (10). DNA from a deceased individual (Family A) was extracted from the pathological specimen. PRKAG2 cDNA sequence was blasted against the genomic sequence of the three bacterial artificial chromosomes (GenBank accession numbers: CAB65116; and AC006966, AC006358.5, AC074257, respectively) containing exons 1 and 4–16 to generate intronic primers. Genomic sequence encoding exons 2 and 3 was obtained from the Celera database (Celera Genomics, Rockville, Maryland, USA). Exons were PCR-amplified and sequenced using an ABI Prism 377 or ABI Prism 3700 DNA Analyzer (Applied Biosystems, Foster City, California, USA). Primers sequences are available at http://genetics.med.harvard.edu/∼seidman. Mutations were confirmed and families genotyped by restriction enzyme digestion. Sequence variants C1289A and A1553T, encoding Thr400Asn and Asn488Ile substitutions, respectively (Figure 1b), introduce a Tru9I site. To confirm G995A encoding Arg302Gln, a BsrBI restriction site was introduced into exon 7 PCR product using the reverse primer 5′-CTACAAAACTTTGTTTTTTACTCTCCCACAGTGGCGCCGCT-3′. G995A abolishes this BsrBI restriction site. Haplotypes flanking G995A were characterized using single-nucleotide polymorphisms (SNPs) in introns 5 and 6, identified from the Celera database (Celera Genomics) using multiplex PCR (cv2667942-4, cv2667950-2, cv1222232-4) and sequence analyses. Intragenic tandem repeats (poly–CA/GT) were identified from the PRKAG2 genomic sequence. Haplotypes were determined from PCR-amplified tandem repeats using fluorescent-labeled primers.
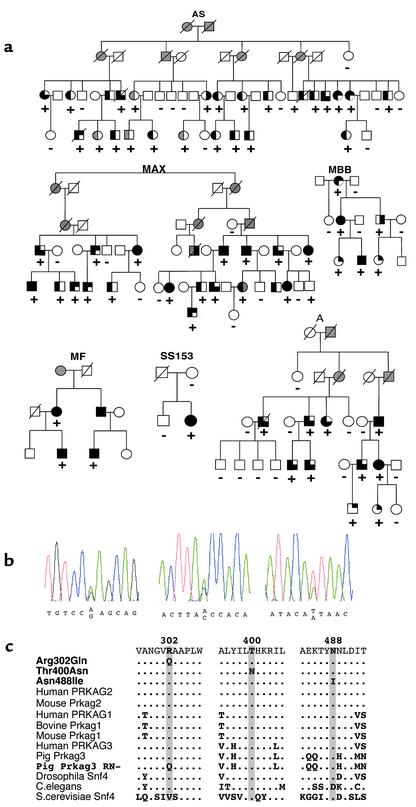
Figure 1.
Identification of three PRKAG2 mutations inherited in six families and the clinical consequences of these mutations. (a) Pedigrees indicate clinical findings of cardiac hypertrophy (left half filled), WPW (right upper quadrant filled), or conduction system disease (right lower quadrant filled) in individuals with a PRKAG2 mutation (+). Open symbols denote unaffected individuals, and shading denotes uncertain clinical status. (b) Sequence traces demonstrating G995A substitution (exon 7), C1289A (exon 11), and A1553T (exon 14), encoding Arg302Gln, Thr400Asn, and Asn488Ile substitutions, respectively. (c) Comparison of PRKAG protein sequences demonstrates evolutionary conservation of residues altered by mutation. Note that human PRKAG2 Arg302Gln mutation is homologous to R200Q mutation in RN– pigs.
Yeast studies.
Growth media were prepared as described (11). Plasmids pPS48 (encoding Snf4) and pGBT9-Snf1 and pPS50 (encoding GAD-Snf4) were generously provided by K.M. Arndt (12). Human mutations Thr400Asn and Asn488Ile were introduced into homologous locations 166 and 251 in Snf4 by standard mutagenesis. Saccharomyces cerevisiae strain FY454a his4-912δ, lys2-128δ, ura3-52, leu2Δ1, snf4Δ1 were transformed with pPS48 (wild-type), pPS48Thr400Asn, pPS48Asn488Ile, or pRS416 (snf4-deficient), grown on SC-Ura selection media. Colonies were then replica-plated on yeast extract peptone dextrose medium (YPD) with 2% glucose, glycerol, or raffinose as a carbon source. Raffinose plates contained 1 μg/ml antimycin A to inhibit mitochondrial phosphorylation. For two-hybrid studies, plasmids (12) (pGBT9-Snf1 + pPS50; pGBT9-Snf1 + pPS50Thr400Asn; pGBT9-Snf1 + pPS50Asn488Ile) and controls (pGBT9 + pGAD10; pGBT9-Snf1 + pGAD10; pGBT9 + pPS50; pGBT9 + pPS50Thr400Asn; pGBT9 + pPS50Asn488Ile) were cotransformed into S. cerevisiae Y190 (CLONTECH Laboratories Inc., Palo Alto, California, USA) and selected on synthetic dropout (SD) media depleted of Trp and Leu (SD-Trp-Leu). For quantitative assay, cells were grown to a mid-log phase in SD-Trp-Leu liquid media containing either Glu2% or galactose 2%/raffinose 1%. β-Galactosidase assays using chlorophenol-red-β-D-galactopyranoside as substrate and calculations were performed according to the manufacturer’s instructions (CLONTECH Laboratories Inc.). β-Galactosidase activity did not exceed 0.5 units in any controls (data not shown).
Statistics.
Two-point lod scores were calculated using an MLINK computer program assuming allele frequency 0.1% and 95% disease penetrance. The χ-square and Student’s t tests were used for statistical comparisons.
Results
The PRKAG2 gene from 18 probands with cardiac hypertrophy and WPW, or isolated, familial WPW, was screened for mutations by nucleotide sequence analyses. Sequence variants were found in six DNA samples from probands with both cardiac hypertrophy and WPW (Figure 1); four sequence variants occurred in families (A, AS, MAX, MBB) with disease linked to the PRKAG2 locus (8, 9). No mutations were identified in 12 samples (Table 1) including three from probands of families with isolated WPW. Retrospective linkage analyses excluded the PRKAG2 locus on chromosome 7q3 as the cause of disease in four kindreds, including two with isolated familial WPW (data not shown). Furthermore, PRKAG2 mutations were not found in 20 unrelated probands with HCM unaccompanied by WPW or in 36 probands with familial dilated cardiomyopathy with or without conduction system disease.
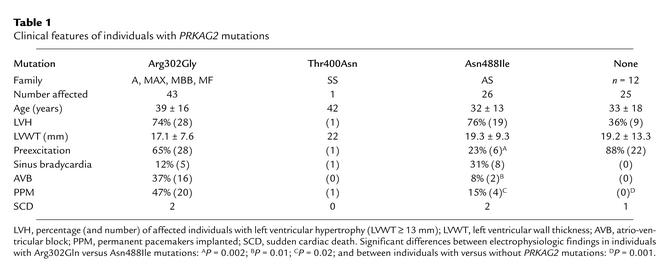
Table 1.
Clinical features of individuals with PRKAG2 mutations
All PRKAG2 sequence variants encoded missense mutations. The previously reported (4) Arg302Gln substitution was found in four samples; analyses of flanking SNPs indicated that this defect arose independently in at least three of four samples (data not shown). Two novel mutations, Thr400Asn and Asn488Ile, were also identified (Figure 1b). All missense mutations were confirmed by restriction enzyme digestion and were absent in more than 200 control samples. Missense mutations Arg302Gln and Asn488Ile cosegregated with affection status in study families (Figure 1a; lod scores = 12.1 [MAX, A, MBB, MF] and 8.4 [AS]; θ = 0). Thr400Asn was found in a proband with no other affected family members and may be a de novo mutation. Each of these three missense mutations altered highly conserved amino acid residues encoded in PRKAG2 (Figure 1c).
The clinical consequences of three different PRKAG2 mutations included cardiac hypertrophy, ventricular preexcitation (WPW), and progressive dysfunction of the conduction system. The presence of conduction system disease was the most significant clinical parameter delineating families with PRKAG2 mutations among kindreds with WPW with or without hypertrophy (Table 1). WPW usually manifested early in life with tachyarrhythmias (atrial fibrillation and other supraventricular arrhythmias) and sometimes caused syncope. Electrophysiologic studies in ten affected individuals demonstrated one or more accessory atrioventricular pathways. With increasing age, affected individuals progressively developed slower heart rates. Sinus bradycardia and/or variable degrees of atrioventricular block resulted in pacemaker implantation in 24 (35%) affected individuals. Left ventricular function markedly deteriorated in five affected adult individuals and necessitated cardiac transplantation in one; sudden death occurred in four. Few differences were observed between the clinical manifestations of the Asn488Ile and Arg302Gln mutations. Conduction disturbances may be more prevalent and severe in individuals with the Arg302Gln mutation, as reflected by more WPW and implantation of more permanent pacemakers, than in individuals with the Asn488Ile mutation.
To evaluate the effects of PRKAG2 mutations on myocyte morphology, histopathology was examined in five cardiac specimens from affected individuals, ages 22–55. Comparable findings were observed in specimens with each of the three different PRKAG2 missense mutations (Figure 2 and data not shown). Marked ventricular hypertrophy was evident on gross inspection, and myocytes were enlarged (Figure 2a), but myofiber disarray, the characteristic feature of HCM, was not detected in any sample. Interstitial fibrosis was minimal and focal. Surprisingly, all five specimens demonstrated isolated, large cytosolic vacuoles in cardiomyocytes (Figure 2b). These vacuoles contained inhomogeneous granular material that stained strongly with PAS (Figure 2c) and was diastase-resistant, a pattern that is characteristic of polyglucan (13). Electron microscopy of specimens revealed densely packed fine granular and fibrillar, electron-dense material within vacuoles (Figure 2, d and e), features that are characteristic of amylopectin, a nonsoluble product of glycogen metabolism. Such findings are typical of type IV glycogenosis and polyglucosan body disease and have also been found in hearts of patients with adult-onset Pompe disease (14–16).
Figure 2.
Histopathology of left ventricular sections obtained from individuals with PRKAG2 mutations. (a) Longitudinal section of left ventricular myocardium from a 26-year-old individual with PRKAG2 mutation Asn488Ile who died suddenly. Note vacuolated myocytes, and lack of myofiber disarray or fibrosis (H&E; bar = 100 μm). (b) High-power magnification of an endomyocardial biopsy (H&E; bar = 100 μm) from a 39-year-old individual with PRKAG2 mutation Thr400Asn shows profound vacuolization (arrows). (c) Homogenous inclusions within vacuoles (arrows and inset) stained positive with PAS are mostly diastase-resistant. (d) Electron micrograph (uranyl acetate and lead citrate) of a sample described in a (bar = 1 μm). Note the large, irregular sarcoplasmic inclusion (arrows) within a large vacuole, and normal-appearing sarcomeres (arrowhead). (e) Higher magnification (bar = 1 μm) demonstrates that the inclusion is composed of a central core of homogenous, electron-dense droplets surrounded by osmiophilic granular and fibrillar material (star).
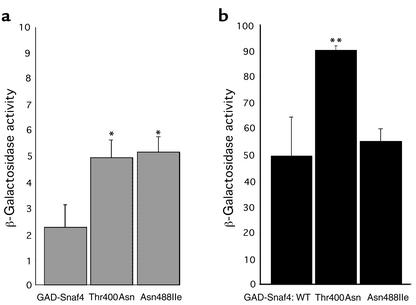
Mammalian AMP-activated protein kinase is a heterotrimeric molecule with regulatory (γ) and enzymatic (α) subunits that are homologous to yeast Snf4 and Snf1 (1, 17), respectively. Two of three amino acid residues altered by human mutations (Asn488 and Thr400; Figure 1c) are conserved in Snf4. To examine the effects of these disease-causing PRKAG2 mutations on γ2 functions, we introduced Asn488Ile and Thr400Asn substitutions into Snf4 from S. cerevisiae. Yeast strains lacking Snf4 require glucose for growth (18). However, Snf4 engineered to contain either Thr400Asn or Asn488Ile allowed yeast growth comparable to that of strains containing wild-type Snf4 (Table 2), indicating that these mutant subunits were functional. Interactions between wild-type Snf4 and Snf1 are normally inhibited by glucose (12, 19), such that kinase activity is very low. However, in the presence of glucose, mutant Snf4Thr400Asn or Snf4Asn488Ile exhibited significantly stronger interaction with Snf1 than did wild-type Snf4 (Figure 3a). In glucose-deprived media, Snf1 interactions with Snf4 mutants was preserved or further increased (Figure 3b). We interpret these data to indicate that Thr400Asn and Asn488Ile mutations produce nonphysiologic, constitutive activation of AMP kinase.
Table 2.
Effects of human PRKAG2 mutations in Snf4 on yeast growth
Figure 3.
Snf1-Snf4 two-hybrid interactions in the presence and absence of glucose. β-Galactosidase activity was measured in cell cultures grown in SD glucose (a) or galactose/raffinose (b) media. The activity from three cultures from different colonies is reported in units (hydrolysis of 1 μmol CPRG/min/cell), mean ± SD. WT, wild-type. *P < 0.02, **P < 0.005 versus wild-type Snf4.
Discussion
We demonstrate that missense mutations in the human γ2 regulatory subunit of AMP-activated protein kinase cause cardiac hypertrophy and electrophysiologic deficits, but not HCM. PRKAG2 mutations produce a distinctive cardiac histopathology characterized by enlarged myocytes with vacuoles containing glycogen derivatives; notably, myocyte disarray is absent and interstitial fibrosis is minimal. Human PRKAG2 defects introduced into the yeast homologue Snf4 result in constitutive enzyme activity. Collectively these data indicate that PRKAG2 mutations cause a novel form of cardiomyopathy associated with polysaccharide storage in the heart.
Persistent activity of AMP kinase in the heart could account for dominant expression of a heterozygous mutation. Nonphysiologic activation of cardiac AMP kinase by these mutations should increase glucose uptake (by stimulating translocation of the glucose transporter GLUT-4 to the plasma membrane) and increase hexokinase activity, thereby leading to glycogen accumulation (20, 21). Polysaccharide storage and vacuole formation in myocytes containing a PRKAG2 mutation (Figure 2) is therefore highly compatible with inappropriately increased base-line AMP kinase activity.
Support for the hypothesis that human PRKAG2 missense mutations cause an increase in AMP kinase activity and stimulate carbohydrate accumulation also comes from the analyses of RN– pigs (22, 23). RN– pigs produce “acid meat” that is of inferior quality due to increased muscle glycogen content. Recent studies defined the RN– mutation to be a missense defect in γ3, a skeletal muscle AMP kinase subunit encoded by the PRKAG3 gene. There is considerable homology between porcine PRKAG3 and human PRKAG2, with 62% identity in a 316–amino acid region. The porcine RN– mutation is analogous to the human cardiac PRKAG2 mutation Arg302Gln (Figure 1c).
Although human PRKAG2 mutations have previously been reported to cause HCM and/or WPW (3, 4), our genetic, biochemical, and histopathologic data indicate that these defects actually produce a novel cardiac glycogen storage disease, manifested by cardiac hypertrophy and conduction system disease. Excess glycogen storage and water in cardiomyocytes appear to account for increased cardiac mass in affected individuals (24). Glycogen accumulation in conductive tissue is also likely to cause sinus and atrioventricular node dysfunction, electrophysiologic abnormalities that are atypical of HCM but are the distinguishing clinical features of PRKAG2 mutations (Table 1). Electrophysiologic disturbances are frequent cardiac manifestations of other glycogen storage disorders such as Pompe disease and Danon disease (24–27). WPW has also been observed in several patients with glycogen storage disease (25, 26); however, neither electrophysiologic nor anatomical evidence of accessory pathways has been obtained in these patients (28). How glycogen accumulation accounts for the presence of atrioventricular accessory pathways associated with PRKAG2 mutations remains an open question. Two mechanisms appear possible: either embryonic atrioventricular connections that normally regress during heart development persist in individuals with PRKAG2 mutations, or metabolic deposits activate quiescent accessory pathways. Although WPW that occurs without cardiac hypertrophy does not appear to be caused by PRKAG2 mutations, understanding this mechanism may well provide insights into other etiologies of WPW.
We conclude that management strategies and prognostic information for individuals with PRKAG2 mutations should not be predicated on HCM. We suggest that therapies directed toward reduction of AMP kinase activity may provide therapeutic benefit and alter the disease progression in these patients.
Acknowledgments
We appreciate the technical assistance of Lisa A. Pacella, Patrik Burgon, Susan Bartlett, Cathy Duffy, and Howard Mulhern. The assistance of John P. DiMarco and Barbara McDonough in collecting clinical material is greatly appreciated. We thank Margaret K. Shirra and Karen M. Arndt for plasmids used in yeast studies, Marian Carlson for helpful suggestions, and Fred Winston for reagents and invaluable advice.
References
- 1.Cheung PC, Salt IP, Davies SP, Hardie DG, Carling D. Characterization of AMP-activated protein kinase gamma-subunit isoforms and their role in AMP binding. Biochem J. 2000;346:659–669. [PMC free article] [PubMed] [Google Scholar]
- 2.Kemp BE, et al. Dealing with energy demand: the AMP-activated protein kinase. Trends Biochem Sci. 1999;24:22–25. doi: 10.1016/s0968-0004(98)01340-1. [DOI] [PubMed] [Google Scholar]
- 3.Blair E, et al. Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10:1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 4.Gollob MH, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 5.Fatkin, D., Seidman, J.G., and Seidman, C.E. 2000. Hypertrophic cardiomyopathy. In Cardiovascular medicine. J.T. Willerson and J.N. Cohn, editors. W.B. Saunders Co. Philadelphia, Pennsylvania, USA. 1055–1074.
- 6.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 7.Niimura H, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 8.MacRae CA, et al. Familial hypertrophic cardiomyopathy with Wolff-Parkinson-White syndrome maps to a locus on chromosome 7q3. J Clin Invest. 1995;96:1216–1220. doi: 10.1172/JCI118154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehdirad AA, et al. Electrophysiologic characteristics of accessory atrioventricular connections in an inherited form of Wolff-Parkinson-White syndrome. J Cardiovasc Electrophysiol. 1999;10:629–635. doi: 10.1111/j.1540-8167.1999.tb00239.x. [DOI] [PubMed] [Google Scholar]
- 10.Siu BL, et al. Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation. 1999;99:1022–1026. doi: 10.1161/01.cir.99.8.1022. [DOI] [PubMed] [Google Scholar]
- 11.Rose, M.D., Winston, F., and Hieter, P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 177–187.
- 12.Shirra MK, Arndt KM. Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics. 1999;152:73–87. doi: 10.1093/genetics/152.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosai J, Lascano EF. Basophilic (mucoid) degeneration of myocardium: a disorder of glycogen metabolism. Am J Pathol. 1970;61:99–116. [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrans VJ, Buja LM, Jones M. Ultrastructure and cytochemistry of glycogen in cardiac diseases. Recent Adv Stud Cardiac Struct Metab. 1973;3:97–144. [PubMed] [Google Scholar]
- 15.Greene GM, et al. Juvenile polysaccharidosis with cardioskeletal myopathy. Arch Pathol Lab Med. 1987;111:977–982. [PubMed] [Google Scholar]
- 16.van der Walt JD, Swash M, Leake J, Cox EL. The pattern of involvement of adult-onset acid maltase deficiency at autopsy. Muscle Nerve. 1987;10:272–281. doi: 10.1002/mus.880100311. [DOI] [PubMed] [Google Scholar]
- 17.Mitchelhill KI, et al. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 18.Celenza JL, Eng FJ, Carlson M. Molecular analysis of the SNF4 gene of Saccharomyces cerevisiae: evidence for physical association of the SNF4 protein with the SNF1 protein kinase. Mol Cell Biol. 1989;9:5045–5054. doi: 10.1128/mcb.9.11.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 20.Bergeron R, et al. Effect of AMPK activation on muscle glucose metabolism in conscious rats. Am J Physiol. 1999;276:E938–E944. doi: 10.1152/ajpendo.1999.276.5.E938. [DOI] [PubMed] [Google Scholar]
- 21.Holmes BF, Kurth-Kraczek EJ, Winder WW. Chronic activation of 5′-AMP-activated protein kinase increases GLUT-4, hexokinase, and glycogen in muscle. J Appl Physiol. 1999;87:1990–1995. doi: 10.1152/jappl.1999.87.5.1990. [DOI] [PubMed] [Google Scholar]
- 22.Milan D, et al. A mutation in PRKAG3 associated with excess glycogen content in pig skeletal muscle. Science. 2000;288:1248–1251. doi: 10.1126/science.288.5469.1248. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton SR, et al. An activating mutation in the gamma1 subunit of the AMP-activated protein kinase. FEBS Lett. 2001;500:163–168. doi: 10.1016/s0014-5793(01)02602-3. [DOI] [PubMed] [Google Scholar]
- 24.Verloes A, et al. Nosology of lysosomal glycogen storage diseases without in vitro acid maltase deficiency. Delineation of a neonatal form. Am J Med Genet. 1997;72:135–142. [PubMed] [Google Scholar]
- 25.Bulkley BH, Hutchins GM. Pompe’s disease presenting as hypertrophic myocardiopathy with Wolff-Parkinson-White syndrome. Am Heart J. 1978;96:246–252. doi: 10.1016/0002-8703(78)90093-5. [DOI] [PubMed] [Google Scholar]
- 26.Francesconi M, Auff E. Cardiac arrhythmias and the adult form of type II glycogenosis. N Engl J Med. 1982;306:937–938. doi: 10.1056/NEJM198204153061515. [DOI] [PubMed] [Google Scholar]
- 27.Danon MJ, et al. Lysosomal glycogen storage disease with normal acid maltase. Neurology. 1981;31:51–57. doi: 10.1212/wnl.31.1.51. [DOI] [PubMed] [Google Scholar]
- 28.Bharati S, et al. The conduction system in Pompe’s disease. Pediatr Cardiol. 1982;2:25–32. doi: 10.1007/BF02265613. [DOI] [PubMed] [Google Scholar]