Abstract
Interleukin 13 (IL13) binds a receptor that is highly overexpressed in malignant gliomas, IL13Rα2. IL13 protein is composed of four helices: α-helix A, B, C, and D, and we found a new “hot spot” in α-helix D that is crucial for the binding of IL13 to IL13Rα2. Lys-105 plus Lys-106 and Arg-109 represent this hot spot. In the current study, we have made substitutions at these three positions in IL13. We examined both neutralization of an IL13-based cytotoxin's glioma cell killing and direct receptor binding of the new IL13 mutants. We observed that Lys-105 and Arg-109 are critical for IL13 binding to IL13Rα2, indeed. However, new mutants of important properties were identified with regard to tumor targeting. IL13.K105R mutant, in which lysine was substituted by arginine, neutralized the killing of IL13Rα2-positive cells by IL13-based cytotoxin more efficiently than wild-type IL13. However, IL13.K105L or IL13.K105A was deprived of any such activity. Furthermore, IL13.K105R and IL13.R109K competed 77- and 27-fold better, respectively, with the binding of [125I]IL13 to the IL13Rα2 binding sites when compared with wild-type IL13. Thus, we have uncovered the first forms of IL13 of higher avidity toward IL13Rα2. These mutants should prove useful in the further design of anticancer diagnostics/therapeutics.
Keywords: Interleukin 13, IL13Rα2, mutants, brain tumors, tumor-specific
Introduction
Interleukin 13 (IL13) is a cytokine that elicits both proinflammatory and anti-inflammatory immune responses [1,2]. IL13 is structurally homologous to IL4 and contains four (A, B, C, and D) α-helices. IL13 and IL4 bind to IL13/4 receptor (IL13/4R), a heterodimeric receptor complex composed of IL13Rα1 and IL4Rα, that is expressed in many normal organs and in adenocarcinomas [3–5]. However, IL13 binds malignant brain tumor (glioma) cells independently of IL4, which indicated the existence of another binding moiety specific for IL13 [6,7]. Subsequently, IL13 receptor α 2 (IL13Rα2) was cloned [8] and this monomeric protein exhibits affinity for IL13, but not IL4. Our studies revealed that malignant gliomas overexpress this restricted receptor for IL13 and that IL13Rα2 is the molecular entity responsible for the binding of IL13 to glioma tumors [9–13].
We have been analyzing the structure-function relationship for the purpose of redirecting IL13 from its normal tissue receptor, IL13/4R, to the tumor-associated receptor, IL13Rα2 [14]. We demonstrated that residues in α-helix A (Pro-7 to Gln-23), α-helix C (Ala-60 to Cys-72), and α-helix D (Lys-90 to Glu-110) take part in the interaction with both IL13Rα2 and IL13/4R (Figure 1) [15–17]. For example, we discovered glutamic acid at position 13 (α-helix A) to be a “hot spot” for IL13 interaction with IL13/4R, and it contributes to site I of the binding regions in IL13 [17]. Thus, IL13 with mutation of the glutamic acid at position 13 to lysine (IL13.E13K) does not bind nor activate IL13/4R, but it retains avidity toward the glioma-restricted IL13Rα2 [16]. Similarly, IL13.R66D and IL13.S69D mutants (α-helix C) binding to the IL13/4R is altered without a loss of its affinity toward the glioma-restricted receptor [17].
Figure 1.
Primary structure of IL13, a 114-amino-acid cytokine, with secondary structure features underlined [23]. Alpha-helical regions A, B, C, and D of IL13 are in bold face letter whereas interconnecting loops are italicized.
Hypothetically, site II of the IL13Rα2 binding region in IL13 was considered to be located in another helix, α-helix D, at the C-terminal end of the cytokine [15]. Of interest, the first IL13 mutant in D helix, IL13.R109D, showed an impaired interaction with glioma cells [17]. This experimental result strongly supported the notion of α-helix D belonging to the receptor binding site II of IL13. However, no derivatives of IL13 were known with selectively enhanced recognition of the IL13Rα2.
More recently, we performed alanine scanning of the entire α-helix D segment of human IL13 in order to systematically delineate the residues that take part in the binding to IL13Rα2 (Figure 1) [18]. Thus, we found that residues 105, 106, and 109 in the α-helix D region of IL13 form the interacting triad toward the glioma-restricted receptor, a new receptor binding “hot spot” in IL13 [18]. Moreover, we noted that these three residues are the only hydrophilic ones located in the amphipathic middle region of α-helix D, comprising the motif LLLHLKKLFR (hydrophilic residues are in bold) (Figure 1) [18]. In our present investigation, we have substituted amino acids at positions 105, 106, and 109 in the α-helix D region with amino acids other than alanine, based on the charge, chain length, and bulkiness of the residues, and analyzed their functional properties. In the course of this investigation, we have identified the first mutants of IL13 of higher avidity toward the IL13Rα2 when compared with wild-type cytokine.
Materials and Methods
Materials
The mutagenesis primers were synthesized in-house in Macromolecular Core Laboratory, Pennsylvania State University College of Medicine (Hershey, PA). Fast protein liquid chromatography (FPLC), columns and media, and unique site elimination mutagenesis kit were obtained from Amersham Pharmacia Biotech (Piscataway, NJ). Glioblastoma multiforme (GBM) (a grade IV astrocytoma) U-251 MG cell line was from American Type Culture Collection (ATCC; Rockville, MD). G-26 mouse glioma cells were transfected with IL13Rα2 receptors to obtain G-26-IL13Rα2+ cells [11]. The TF-1 cell line was obtained from the ATCC. Tissue culture equipment were from Corning Glass (Corning, NY). 3(4,5-Dimethylthiazol-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt/phenazine methosulfate (MTS/PMS) was purchased from Promega (Madison, WI). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot transferring equipment were from Bio-Rad (Hercules, CA). Antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Super-Signal Substrate for chemiluminescent detection was from Pierce (Rockford, IL). IODO-GEN reagent for 125I labeling was purchased from Pierce.
Methods
Expression, purification, and structural characterization of IL13 α-helix D mutants The plasmid was constructed with T7 promoters and isopropyl-1thio-β-d-galactopyranoside (IPTG)-inducible T7 RNA polymerase gene in IPTG-inducible form [16]. The primers for mutagenesis were designed using Vector NTI suite software and mutagenesis was performed by site-directed mutagenesis using a selection primer and a mutagenic primer. A selection primer 5′-TGAGGATCCGAGCTCTAATCTAGAGGCTG TGAGGATCCGAGCTCTAATCTAGAGGCTG CTAAC-3′ was used to introduce XbaI site in pIL13-CLS plasmid in place of EcoRI site to carry out restriction selection digestion and transformation in the process of mutagenesis.
The Escherichia coli cells transformed with plasmids were allowed to grow until A600 reached 2.0. Then the cells were induced with 250 µM final concentration of IPTG and incubated at 37°C for 2 hours in a 1-I flask with shaking. The inclusion body pellets were isolated and solubilized in 7 M guanidine HCl and renatured in the presence of l-arginine and oxidized glutathione, as described previously [17]. The renatured proteins were dialyzed against 10 mM sodium acetate buffer, pH 5.2, and purified on ion exchange columns SP Sepharose and Mono S (Amersham Pharmacia Biotech) columns.
SDS-PAGE and Western blot analyses The purity and molecular weight of the D-helix mutants were verified by SDS-PAGE under nonreducing conditions. The procedure for SDS-PAGE has been described elsewhere [18]. All SDS polyacrylamide gels were 15%. After electrophoresis, the gel was stained in Coomassie blue. For Western blot analysis, the proteins were transferred to PVDF membrane using standard electroblotting procedures. The PVDF membrane was blocked with 5% nonfat dry milk in phosphate-buffered saline (PBS) with 0.05% Tween 20 at 4°C overnight. Then the membrane was incubated at room temperature in 5% milk containing the polyclonal goat anti-human IL13 antibody (1:1000 dilution) for 45 minutes. The membrane was washed three times with 0.05% Tween 20/PBS followed by 45-minute incubation in 5% milk containing secondary antibody, antigoat IgG conjugated to horseradish peroxidase. Subsequently, the membrane was washed three times with 0.05% Tween 20/PBS. The proteins were detected on the film by enhanced chemiluminescence (ECL) detection system (Pierce).
Circular dichroism (CD) spectrum analysis The protein samples used for CD spectrum analyses were dissolved in PBS (0.1 mg/ml). All the analyses were performed in a quartz cuvette with a path length of 1 mm using a Jasco (Eden Prairie, MN) J-710 spectropolarimeter. The spectra were recorded in the wavelength region of 200 to 260 nm, and the spectra were subtracted from the blank PBS spectrum.
Cytotoxicity neutralization assay using constant concentrations of IL13 mutants The cytotoxicity neutralization experiment was performed as described elsewhere [17]. IL13-PE38QQR is a cytotoxin and fusion protein composed of IL13 and Pseudomonas exotoxin (PE38QQR), which was used in the assay [5]. Briefly, 1 x 103 cells of U-251 MG cells or 2.5 x 103 cells of G-26-IL13Rα2+ cells per well were plated in 96-well culture plates and incubated at 37°C, 5% CO2, and 90% humidity for 24 hours. The receptor sites on the cells were blocked with IL13 or its mutants, at a final protein concentration of 1 µg/ml, for 1 hour at 37°C in the incubator. Subsequently, the cytotoxin, IL13-PE38QQR, was added at increasing concentrations (0.01–100 ng/ml final concentration) and the plates were incubated for 48 hours. The proliferated cells in each well were determined by colorimetric MTS/PMS method, as described [18]. All the experiments were performed in duplicates.
Cytotoxicity neutralization assay using constant concentrations of IL13-based cytotoxin The assay was performed as described above except that the inhibitor's (IL13 or its mutants) concentrations varied, whereas the concentration of cytotoxin was constant. The U-251 MG or G-26-IL13Rα2+ cells plated in 96-well plates were blocked with IL13 or its mutants (0.1–1000 ng/ml final concentration) for 1 hour at 37°C. Then, IL13-PE38QQR at a concentration of 10 ng/ml was added and incubated at 37°C for 48 hours. Following incubation, the proliferated cells in each well were determined by colorimetric MTS/PMS method.
TF-1 cell proliferation (IL13/4R stimulation) assay The proliferation assay was carried out in 96-well culture plates and the cells were allowed to grow in the presence of different concentrations of IL13 and its mutants, and incubated at 37°C, 5% CO2, and 90% humidity for 72 hours. The rate of proliferation was determined by colorimetric MTS/PMS cell proliferation assay and the absorbency of each well at 490 nm was recorded on a microplate reader. Cells treated with high concentrations of cyclohexamide served as background for the assay. The experiment was performed in duplicate.
IL13Rα2 receptor binding analyses In order to establish the number of binding sites for IL13 on glioma cells, we performed a saturation binding experiment. To establish the total, nonspecific, and specific binding curves, we added a range of concentrations of [125I]IL13 or its mutants (1 pM–2 nM) in the presence or absence of unlabeled IL13 at 400 times molar excess, to 0.5 x 106 cells (U-251 MG or G-26-IL13Rα2+ cells). Incubations were carried out at 4°C for 3 hours. Then the cells were harvested in a cell harvester, followed by the radioactivity measurement in a γ-counter.
The second set of binding assays was performed in the form of equilibrium competition assays using increasing concentrations of the unlabeled IL13 or its mutants (10 pM–10 nM) in the presence of a constant concentration of the labeled [125I]IL13 (100 pM). For this assay, 0.2 x 106 cells (G-26-IL13Rα2+cells) were used. They were incubated at 4°C for 3 hours, followed by harvesting in a cell harvester and counting in a γ-counter. Analyses of the binding assays were carried out using the computer program, Graphpad Prism (San Diego, CA). The Ki values were calculated by the program Prism using the equation of Cheng and Prusoff [19]:
The Ki value represents an affinity of the radioligand ([125I]IL13) toward the receptor in the presence of competitive inhibitor ligands. Each experiment was performed in duplicate.
Results and Discussion
Site-Directed Mutagenesis of α-Helix D Residues of Human IL13
α-Helix D of the IL13 molecule is composed of 24 amino acid residues. Based on the results of alanine scanning mutagenesis [18], we have selected residues, which appeared to be of greatest importance for the binding of IL13 to IL13Rα2, and mutated them to charged or neutral residues. A series of single-site mutagenic primers was constructed (Table 1). In particular, residues Lys-105, Lys-106, and Arg-109 were substituted for amino acids of varying charge, chain length, and bulkiness of functional groups. The residues Ala-94 and Gln-95 were mutated as well because they had displayed a partial impairment in their ability to interact with IL13Rα2. We also changed hydroxyproline at position 103 to proline in an intended helix-destabilizing manner.
Table 1.
Mutagenic Primers Used for α-Helix D of IL13 Mutagenesis.
| Position | Oligonucleotide Primers (5′ to 3′) |
| A94R | ACC-AAA-ATC-GAG-GTG-CGC-TTT-GTA-AAA-GAT |
| Q95R | AAA-ATC-GAG-GTG-GCC-CGC-TTT-GTA-AAA-GAT-CTG |
| H103P | AAA-GAT-CTG-CTC-TTA-CCA-TTA-AAG-AAA-CTT |
| K105R | CTG-CTC-TTA-CAT-TTA-CGC-AAA-CTT-TTT-CGC-GAG |
| K105E | CTC-TTA-CAT-TTA-GAG-AAA-CTT-TTT-CGC |
| K105D | CTC-TTA-CAT-TTA-GAC-AAA-CTT-TTT-CGC |
| K105L | TTC-TTA-CAT-TTA-CTG-AAA-CTT-TTT-CGC-GAG |
| K106E | TTA-CAT-TTA-AAG-GAA-CTT-TTT-CGC-GAG |
| K106R | TTA-CAT-TTA-AAG-CGC-CTT-TTT-CGC-GAG |
| R109D | AAG-AAA-CTT-TTT-GAC-GAG-GGA-CGG-TTC |
| R109K | AAG-AAA-CTT-TTT-AAA-GAG-GGA-CGG-TTC-AAC |
| R109L | AAG-AAA-CTT-TTT-CTG-GAG-GGA-CGG-TTC-AAC |
Expression and Purification of Recombinant Interleukins
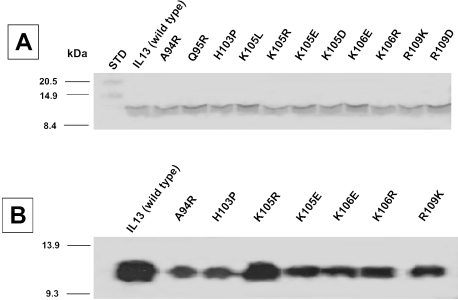
All the expressed proteins in E. coli were localized to the inclusion bodies and, on average, the yield of all the purified proteins was found to be 0.5 to 1 mg/liter culture, except for IL13.K105L, IL13.H103P, and IL13.R109L. IL13 mutant proteins were pure and homogeneous with an approximate molecular mass of ∼ 13 kDa after the purification on SP Sepharose fast flow and Mono S columns (Figure 2A). All the mutant proteins were immunoreactive using an anti-IL13 antibody and Figure 2B demonstrates the Western blot of several representative mutants.
Figure 2.
(A) SDS-PAGE analysis (15%) of α-helix D mutants of IL13. Each well was loaded with 1 µg of protein and stained in Coomassie blue. (B) The Western blot of IL13 α-helix D mutant proteins. The purified recombinant proteins were resolved on 15% SDS-PAGE and electroblotted to PVDF membrane. The proteins were detected by ECL detection system.
CD Spectrum Analysis of α-Helix D Mutants of IL13
The CD spectra of the studied D-helix mutant proteins of IL13 were analyzed according to the reported procedure [18]. All the mutant proteins were folded properly into α-helical enriched structure, as all of them exhibited two spectral minima at 208 and 222 nm, a characteristic property of helical proteins (not shown).
IL13 Mutants Neutralization of Cell Killing by a IL13-Based Cytotoxin
The cytotoxicity neutralization assay was carried out in U-251MG and G-26-IL13Rα2+ cells, which express IL13Rα2 either naturally or as a transgene, respectively (Figure 3). These experiments were performed in order to reveal the importance of studied substitution mutants in the α-helix D region of IL13 in their functional binding toward IL13Rα2. IL13Rα2 is the receptor that is utilized by the IL13-based cytotoxin to bind and kill cells [11]. Most of the substitutions made at Lys-105 in the α-helix D resulted in IL13 mutants with severely impaired cytotoxicity neutralization efficiency when compared with wild-type IL13 (Figure 3, A and B). For example, IL13.K105L and IL13.K105D did not compete for the receptor sites on the cells that are used by IL13-PE38QQR cytotoxin, unlike wild-type IL13 (Figure 3, A and B). This is in line with our earlier study where the replacement of lysine at position 105 with alanine resulted in total loss of cytotoxicity neutralization efficiency of the mutant [18]. Surprisingly, substitution of lysine at position 105 to arginine (IL13.K105R) resulted in a mutant with at least, similar to wild-type IL13, cytotoxicity neutralization efficiency in the two studied cancer cell lines (Figure 3, A and B).
Figure 3.
Cytotoxicity neutralization by IL13 α-helix D mutants in malignant glioma cells. Neutralizing concentrations of the IL13 mutants were constant (1000 ng/ml) and the cytotoxin, IL13-PE38QQR, was serially diluted (0.01–100 ng/ml). Experiments were performed using a colorimetric cell MTS/PMS proliferation assay. (A) U-251 MG, human GBM cells. (B) G-26-IL13Rα2+, mouse glioma cells transfected with IL13Rα2. The IC50 for the cytotoxin alone (no cytokine) was 0.5 and 0.6 ng/ml in U-251 MG and G-26-IL13Rα2+ cells, respectively. A wild-type IL13, or its mutants with preserved competition for the receptor used by the cytotoxin, augmented the IC50 values to > 100 ng/ml (e.g., IL13, IL13.K105R, and IL13.K106R neutralized cytotoxicity). The mutants of IL13 that lost competitive properties did not change or changed little the IC50 (e.g., IL13.R109D, IL13.K105L, and IL13.K105D).
Similar to the phenomenon observed with Lys-105, an anionic glutamic acid substitution at position 106 resulted in partial loss in cytotoxicity neutralization efficiency of IL13.K106E in U-251 MG and G-26-IL13Rα2+ cells (Figure 3, A and B). However, the IL13.K106R mutant was able to neutralize IL13-PE38QQR cytotoxicity and behaved similar to wild-type IL13 (Figure 3, A and B). Furthermore, substitution of arginine at position 109 in α-helix D to other amino acids, such as leucine and aspartic acid, also resulted in mutants with altered cytotoxicity neutralization capacities by displaying a poor neutralization efficiency of IL13-PE38QQR cytotoxicity in glioma cells (Figure 3, A and B). Again, substitution of similarly charged lysine in the place of arginine (IL13.R109K) resulted in an unaltered neutralization efficiency of the mutant when compared with an intact cytokine (Figure 3, A and B). Thus, the results obtained with substitution mutants at positions 105, 106, and 109 are consistent with the results of the alanine scanning mutagenesis studies [18]. Of great interest, we have found evidence that substitution mutants at position 105 and 109, such as IL13.K105R and IL13.R109K, resulted in IL13 of high avidity toward the tumor-associated receptor mutants.
We also made a substitution of alanine at position 94 to arginine. This substitution did not result in any prominent loss of cytotoxicity neutralization potential of the mutant in either U-251 MG cells (Figure 3A) or in G-26-IL13Rα2+ cells (Figure 3B). However, mutation of glutamine at position 95 to arginine resulted in impaired cytotoxicity neutralization efficiency when compared with wild-type IL13 (Figure 3, A and B). Furthermore, the mutant IL13.H103P did not lose all cytotoxicity neutralization efficiency when compared to wild-type IL13, which implies that proline at position 103 may not completely disrupt the helical orientation of α-helix D (Figure 3, A and B).
Titered Cytotoxicity Neutralization Experiment
Next, we performed cytotoxicity neutralization assay in a reversed scenario: a constant concentration of the cytotoxin was used instead of serial dilutions (10 ng/ml), and serial dilutions of IL13 mutants were used instead of constant concentrations (0.1–1000 ng/ml); IL13-PE38QQR cytotoxin efficiently kills malignant glioma cells at 10 ng/ml. The cytotoxicity neutralization potential of the wild-type IL13 and its mutants at variable concentrations of the ligand and at a constant concentration of the cytotoxin is demonstrated in Figure 4.
Figure 4.
Cytotoxicity neutralization by IL13 α-helix D mutants in U-251 MG cells. Wild-type IL13 and neutralizing mutants were serially diluted (0.01–1000 ng/ml) whereas IL13-PE38QQR concentration was constant (10 ng/ml). The experiments were performed using colorimetric cell MTS/PMS proliferation assay. Vertical bars correspond to standard deviation. Higher percentage of cells was protected against IL13-PE38QQR cytotoxicity when, for example, the IL13.K105R mutant was used rather than the wild-type IL13.
In this titered cytotoxicity neutralization experiment, the highest neutralization efficiency was consistently observed for IL13K105R. IL13.K105R neutralized the cytotoxicity by 99% at 100 ng/ml concentration when compared with the mutant's blocking efficiency at 1000 ng/ml. The wild-type IL13 and the IL13.K106R and IL13.R109K mutants exhibited 86.5%, 86.7%, and 80.7% cytotoxicity neutralization efficiency, respectively. We then determined effective neutralization concentration (ENC) as a minimum concentration of the cytokine that would bring about 50% of cytotoxicity neutralization. ENC was found to be lower for IL13.K105R (10 ng/ml) than that for the wild-type IL13 (50 ng/ml) (Figure 4). In addition, IL13.R109K had a strong tendency to neutralize the cytotoxin at lower concentrations that wild-type IL13, but less so than IL13.K105R. The least competitive to neutralize the cytotoxicity of IL13-PE38QQR was IL13.R109K; however, it was still neutralizing better than wild-type IL13 (Figure 4). This experiment underlined the better cytotoxicity neutralizing of IL13.K105R, IL13.R109K, and IL13.K106R, compared to that seen with wild-type IL13. Thus, these mutants exhibited an enhanced ability to compete for the receptor sites used functionally by the IL13-based cytotoxin.
IL13Rα2 Receptor Binding Studies of IL13 D-Helix Mutants
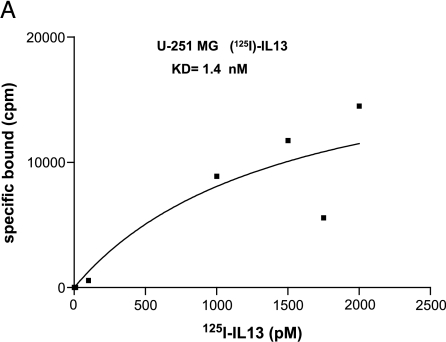
The receptor binding experiments with 125I-labeled IL13 revealed Kd values of 1.4 and 1.3 nM on U-251 MG and G-26-IL13Rα2+ cells, respectively (Figure 5A). The number of binding sites per cell was found be 7325 and 14,550, respectively. Subsequently, we performed studies in which we examined the competition of IL13 α-helix D mutants with 125I-labeled IL13 for the binding to receptors on G-26-IL13Rα2+ cells and the results are depicted in Figure 5B. The binding of [125I]IL13 to the cells was competed for by IL13.K105R and IL13.R109K, with the EC50 of IL13.K105R being 0.027 nM and that of IL13.R109K being 0.073 nM, which is 77- and 27-fold less, respectively, when compared with wild-type IL13 (2.1 nM) (Table 2). The Ki values of competitive ligands, namely IL13.K105R, IL13.K106R, and IL13.R109K, were calculated (Table 2). The results indicate that IL13.K105R and IL13.R109K are potent competitive ligands for the iodo-IL13 binding to IL13Rα2, with the Ki values being 0.025 and 0.068 nM, respectively. However, the Ki value for IL13.K106R was lower than for wild-type IL13 (Table 2). Thus, IL13.K105R and IL13.R109K compete better for the IL13Rα2 than the wild-type IL13 in receptor binding experiments.
Figure 5.
Binding studies of IL13 and its mutants to a glioma-associated receptor for IL13. (A) Saturation curves for [125I]IL13 on U-251 MG cells. (B) Competition for the [125I]IL13 binding sites by α-helix D mutants of IL13 in G26-IL13Rα2+ cells.
Table 2.
EC50 and Ki Values for IL13, IL13.K105R, IL13.K106R, and IL13.R109K Obtained in Receptor Binding Assay in G26-IL13Rα2+ Cells (Figure 5B).
| IL13 (nM) | IL13.K105R (nM) | IL13.K106R (nM) | IL13.R109K (nM) | |
| EC50 | 2.134 | 0.027 | 3.39 | 0.073 |
| Ki | 2.00 | 0.025 | 3.18 | 0.068 |
TF-1 Cell Proliferation Activity of D-Helix Mutants
TF-1 cells are preleukemic human B-cells, which express the shared IL13/4R, but little, if any, of IL13Rα2 [20]. Hence, TF-1 cell proliferation assay directly reflects an ability of the mutants to bind effectively and activate IL13/4R. The TF-1 cell proliferation ability of α-helix D mutants is demonstrated in Figure 6. The mutant proteins like IL13.K105D, IL13.K105L, IL13.K105E, IL13.K106E, and IL13.R109D lacked proliferative activity on TF-1 cells. However, mutants like IL13.K105R, IL13.K106R, and IL13.R109D proliferated TF-1 cells similarly to wild-type IL13. Noteworthy, IL13.K105R, IL13.K106R, and IL13.R109D did not appear to a have clear-cut superagonistic activities toward IL13/4R.
Figure 6.
Proliferative (IL13/4R-stimulating) potency of the α-helix D mutants on TF-1 cells. Experiments were performed using a colorimetric MTS/PMS cell proliferation assay.
Triad of Residues in IL13 That Takes Part in the Interaction with IL13Rα2
Residues Lys-105, Lys-106, and Arg-109 in IL13 are solvent-accessible and are the most likely residues to be involved in receptor binding interactions [18]. Furthermore, these three residues are in close spatial proximity in the three-dimensional structure of IL13. Our results demonstrate that residue Lys-105 in the α-helix D region in IL13 is a receptor contact site as its substitution to uncharged or negatively charged residues (e.g., leucine, alanine, glutamic acid, aspartic acid, etc.) resulted in overall loss in their binding to the glioma-restricted receptor. However, to our surprise, the substitution of this amino acid with another positively charged amino acid, arginine, resulted in a mutant, which binds IL13Rα2 more competitively when compared with wild-type IL13. Thus, substitutions with glutamic acid, leucine, and aspartic acid likely distort the recognition site, resulting in an impaired binding affinity for the receptor, whereas substitution with positively charged arginine results in an enhanced avidity. It is plausible that the contact sites in the IL13Rα2 protein involve negatively charged residue(s) or neutral residue(s). It is also noteworthy that the large residues, such as arginine, contribute to the higher side-chain flexibility of the ligand-receptor interface [21].
Figure 7 indicates the orientation of receptor binding residues in α-helix A, B, and C in IL13. Our previous studies demonstrated that IL13.E13K in A-helix and IL13.R66D and IL13.S69D in C-helix retained an ability to interact with IL13Rα2, but not with the shared IL13/4R, which is composed of IL4Rα and IL13Rα1. Thus, we conclude that the residues Glu-13, Arg-66, and Ser-69 are the contact residues with IL4Rα, and Lys-105 and Arg-109 in the C-terminal region of IL13 are the contact residues with IL13Rα2 (Figure 7). The IL4Rα receptor and the IL13Rα2 interacting residues lie, respectively, at receptor binding site I (yellow) and receptor binding site II (green) in the IL13 molecule, as shown in Figure 7.
Figure 7.
Main structural features of IL13 (α-helix A, C, and D are seen). The residues that contribute to receptor binding site I (Glu-13, Arg-66, and Ser-69) are shown in green (Ser-69 is hidden behind α-helix C). The residues that contribute to receptor binding site II (Lys-105, Lys-106, and Arg-109) are shown in yellow. Sites I and II take part in the binding of IL13 to IL4Rα and IL13Rα protein chains, respectively.
In order to further visualize the function of Lys-105 and Arg-109 residues in IL13 binding to IL13Rα2, we modeled the structural side-chain changes at position 105 upon substitution with various amino acids using Swiss-PDB molecular viewer and the coordinates available from the solution structure for IL13, as presented in Figure 8 [22,23]. The proximity between the side chain -NH2 group (residue 105) and -C (NH2)NH-group (residue 109) and their orientation is shown in Figure 8. It is plausible that such orientation favors the residues to anchor the receptor site coordinately and that this is achieved more optimally in the IL13.K105R version of the cytokine (Figure 8B). All the other mutants, namely IL13.K105E, IL13.K105D, IL13.K105A, and IL13.K105L did not have the extended side chain at position 105 in order to provide hypothetically required optimal space for the interaction with the receptor (e.g., IL13.K105E in Figure 8C). Thus, we propose that IL13 interacts with IL13Rα2 coordinately through positions 105 and 109, providing an ambient anchoring toward IL13Rα2.
Figure 8.
Modeled side-chain orientations of IL13 with substitutions of Lys-105. The pictures were generated using Swiss-PDB. The side chains for the position 105 are underlined in green, and for the position 109 in red. (A) IL13 (wild type). (B) IL13.K105R. Circled are two amino acids that may provide more optimal orientation for IL13 engagement with IL13Rα2: Lys-105 substituted for by Arg and Arg-109. (C) IL13.K105E.
Conclusion
The α-helix D mutants of IL13, IL13.K105R, and IL13.R109K were found to bind IL13Rα2 better than the native protein. The most plausible explanation for this phenomenon is the orientation and alignment of positions 105 and 109 in the α-helix D region toward the receptor engagement. Thus, we identified mutants of IL13 having desired characteristics for future development of tumor targeting molecules while sparing normal organs [15]. Previously, we presented evidence for IL13Rα2 being a unique molecular target in cancer [11]. Several therapeutic approaches that target this receptor, including vaccines [24], gene therapy [25], passive immunotherapy represented by retargeted cytotoxic T cells [26], and new IL13-based cytotoxins [12,27], are being developed preclinically and clinically. New mutant proteins of IL13 will facilitate achieving the goal of developing agents of specific targeting properties to the tumor-associated receptor, IL13Rα2.
Acknowledgements
We thank Ira Ropson (Department of Biochemistry and Molecular Biology, Pennsylvania State University College of Medicine) for assistance in obtaining the CD spectra, and we also extend our thanks to Denise Gibo and Becky Slagle for their excellent technical assistance. We also thank James R. Connor for the use of cell harvester in binding experiments.
Abbreviations
- CD
circular dichroism
- FPLC
fast protein liquid chromatography
- GBM
glioblastoma multiforme
- h
human
- HGA
high-grade astrocytoma
- IL4
interleukin 4
- IL13
interleukin 13
- IPTG
isopropyl-1thio-β-d-galactopyranoside
- m
murine
- MTS
3-(4,5-dimethylthiazol-2yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- PAGE
polyacrylamide gel electrophoresis
- PE
Pseudomonas exotoxin
- PMS
phenazine methosulfate
- SDS
sodium dodecyl sulfate
Footnotes
This work was supported by NIH/NCI grant 2R01 CA 74145.
References
- 1.Minty A, Chalon P, Derocq JM, Dumont X, Guillemot JC, Kaghad M, Labit C, Leplatois P, Miloux B, Minty C, Casellas P, Loison G, Lupker J, Shire D, Ferrara P, Caput D. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;36:248–251. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 2.McKenzie ANJ, Culpepper JR, De Waal Malefyt R, Briere F, Punnonen G, Aversa G, Sato A, Dang W, Cocks BG, Menon S, De Vries JE, Bancherau J, Zurawski G. Interleukin 13, a T-cell derived cytokine that regulates human monocyte and B-cell function. Proc Natl Acad Sci USA. 1993;90:3735–3739. doi: 10.1073/pnas.90.8.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hilton DJ, Zhang J-G, Metcalf D, Alexander WS, Nicola NA, Wilson TA. Cloning and characterization of a binding subunit of the interleukin 13 receptor that is also a component of the interleukin 4 receptor. Proc Natl Acad Sci USA. 1996;93:497–501. doi: 10.1073/pnas.93.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tony HP, Shen BJ, Reush P, Sebal W. Design of human interleukin-4 antagonists inhibiting interleukin-4-dependent and interleukin-13-dependent responses in T-cells and B-cells with high efficiency. Eur J Biochem. 1994;225:659–666. doi: 10.1111/j.1432-1033.1994.00659.x. [DOI] [PubMed] [Google Scholar]
- 5.Debinski W, Obiri NI, Pastan I, Puri RK. A novel chimeric protein composed of interleukin 13 and Pseudomonas exotoxin is highly cytotoxic to human carcinoma cells expressing receptors for interleukin 13 and interleukin 4. J Biol Chem. 1995;270:16775–16780. doi: 10.1074/jbc.270.28.16775. [DOI] [PubMed] [Google Scholar]
- 6.Debinski W, Obiri NI, Powers SK, Pastan I, Puri RK. Human glioma cells overexpress receptors for interleukin 13 and are extremely sensitive to a novel chimeric protein composed of interleukin 13 and Pseudomonas exotoxin. Clin Cancer Res. 1995;1:1253–1258. [PubMed] [Google Scholar]
- 7.Debinski W, Miner R, Leland P, Obiri NI, Puri RK. Receptor for interleukin 13 (IL13) does not interact with IL4 but receptor for IL4 interacts with IL13 on human glioma cells. J Biol Chem. 1996;271:22428–22433. doi: 10.1074/jbc.271.37.22428. [DOI] [PubMed] [Google Scholar]
- 8.Caput D, Laurent P, Kaghad M, Lelias JM, Lefort S, Vita N, Ferrara P. Cloning and characterization of a specific interleukin (IL)-13 binding protein structurally related to the IL-5 receptor α chain. J Biol Chem. 1996;271:16921–16926. doi: 10.1074/jbc.271.28.16921. [DOI] [PubMed] [Google Scholar]
- 9.Debinski W, Gibo DM, Hulet SW, Connor JR, Gillespie GY. Receptor for interleukin-13 is a marker and therapeutic target for human high-grade gliomas. Clin Cancer Res. 1999;5:985–990. [PubMed] [Google Scholar]
- 10.Debinski W, Slagle B, Gibo DM, Powers SK, Gillespie GY. Expression of a restrictive receptor for interleukin 13 is associated with glial transformation. J Neuro-Oncol. 2000;48:103–111. doi: 10.1023/a:1006446426611. [DOI] [PubMed] [Google Scholar]
- 11.Mintz A, Gibo DM, Becky SW, Christensen ND, Debinski W. IL13Ralpha2 is a glioma-restricted receptor for interleukin-13. Neoplasia. 2002;4:388–399. doi: 10.1038/sj.neo.7900234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mintz A, Gibo DM, Madhankumar AB, Debinski W. Molecular targeting with recombinant cytotoxins of interleukin-13 receptor alpha-2-expressing glioma. J Neuro-Oncol. 2003;64:117–123. doi: 10.1007/BF02700026. [DOI] [PubMed] [Google Scholar]
- 13.Gibo W, Debinski DM. Molecular expression analysis restrictive receptor for interleukin 13, a brain tumor-associated cancer/testis antigen. Mol Med. 2000;6:440–449. [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson JP, Debinski W. Retargeting interleukin 13 for radioimmunodetection and radioimmunotherapy of human high-grade gliomas. Clin Cancer Res. 1999;5:3143s–3147s. [PubMed] [Google Scholar]
- 15.Debinski W. An immune regulatory cytokine receptor and glio-blastoma multiforme: an unexpected link. Crit Rev Oncog. 1998;9:255–268. [PubMed] [Google Scholar]
- 16.Debinski W, Gibo DM, Obiri NI, Kealiher A, Puri RK. Novel anti-brain tumor cytotoxins specific for cancer cells. Nat Biotechnol. 1998;16:449–453. doi: 10.1038/nbt0598-449. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JP, Debinski W. Mutants of interleukin 13 with altered reactivity toward interleukin 13 receptors. J Biol Chem. 1999;24:29944–29950. doi: 10.1074/jbc.274.42.29944. [DOI] [PubMed] [Google Scholar]
- 18.Madhankumar AB, Mintz A, Debinski W. Alanine scanning mutagenesis of alpha-helix D segment of interleukin-13 reveals new functionally important residues of the cytokine. J Biol Chem. 2002;277:43194–43205. doi: 10.1074/jbc.M205047200. [DOI] [PubMed] [Google Scholar]
- 19.Cheng Y, Prusoff WH. Relationship between the inhibition constant and the concentration of inhibitor, which causes 50 percent inhibition of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 20.Zurawski SM, Vega F, Jr, Huyghe B, Zurawski G. Receptors for interleukin-13 and interleukin-4 are complex and share a novel component that function in signal transduction. EMBO J. 1993;12:2663–2670. doi: 10.1002/j.1460-2075.1993.tb05927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz GE, Schirmer RH. Principles of Protein Structure. New York: Springer-Verlag; 1979. pp. 1–16. [Google Scholar]
- 22.Eisenmesser EZ, Horita DA, Alteiri AS, Byrd RA. Solution structure of interleukin-13 and insights into receptor engagement. J Mol Biol. 2001;310:231–241. doi: 10.1006/jmbi.2001.4765. [DOI] [PubMed] [Google Scholar]
- 23.Moy FJ, Diblasio E, Wilhelm J, Powers R. Solution structure of human IL-13 and implication for receptor binding. J Mol Biol. 2001;310:219–230. doi: 10.1006/jmbi.2001.4764. [DOI] [PubMed] [Google Scholar]
- 24.Okano F, Storkus WJ, Chambers WH, Pollack IF, Okada H. Identification of a novel HLA-A*0201-restricted, cytotoxic T lymphocyte epitope in a human glioma-associated antigen, interleukin 13 receptor α2 chain. Clin Cancer Res. 2002;8:2851–2855. [PubMed] [Google Scholar]
- 25.Zhou G, Ye G-J, Debinski W, Roizman B. Genetic engineering of a herpes virus 1 vector dependent on the IL13Rα2 receptor for entry into cells: interaction of glycoprotein D with its receptors is independent of the fusion of the envelope and the plasma membrane. Proc Natl Acad Sci. 2002;99:15124–15129. doi: 10.1073/pnas.232588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhlon KS, Mumelak A, Morgan R, Cooper L, Jensen MCV. The IL13 Zetakine chimeric immunoreceptor : a novel approach to genetically engineer T cells for glioma immunotherapy. Neuro-Oncology. 2001;3:315. [Google Scholar]
- 27.Hall WA, Jin N, Todhunter DA, Panoskaltis-Mortari A, Vallera DA. Targeting glioblastoma multiforme with an IL-13/diphtheria toxin fusion protein in vitro and in vivo in nude mice. Prot Eng. 2002;5:419–427. doi: 10.1093/protein/15.5.419. [DOI] [PubMed] [Google Scholar]