Abstract
The ability of actin to both polymerize into filaments and to depolymerize permits the rapid rearrangements of actin structures that are essential for actin’s function in most cellular processes. Filament polarity and dynamic properties are conferred by the hydrolysis of ATP on actin filaments. Release of inorganic phosphate (Pi) from filaments after ATP hydrolysis promotes depolymerization. We identify a yeast actin mutation, Val-159 to Asn, which uncouples Pi release from the conformational change that results in filament destabilization. Three-dimensional reconstructions of electron micrographs reveal a conformational difference between ADP-Pi filaments and ADP filaments and show that ADP V159N filaments resemble ADP-Pi wild-type filaments. Crystal structures of mammalian β-actin in which the nucleotide binding cleft is in the “open” and “closed” states can be used to model actin filaments in the ADP and ADP-Pi conformations, respectively. We propose that these two conformations of G-actin may be related to two functional states of F-actin.
Regulation of actin filament dynamics is essential for cytokinesis, cell motility, and control of cell shape and polarity. The ATP cycle of actin is fundamental to this regulation. Actin polymerizes preferentially from ATP-bound actin, and this bound ATP is hydrolyzed after assembly. The subsequent release of inorganic phosphate (Pi) destabilizes the filament and promotes actin filament disassembly (1, 2). The addition of excess Pi causes actin to behave as an equilibrium polymer, with the same critical concentration at both ends of the filament (3, 4). Beryllium fluoride and aluminum fluoride also can stabilize actin filaments by binding to the Pi site and mimicking ADP-Pi or ATP filaments (5, 6).
The different assembly properties of ADP versus ATP or ADP-Pi actin suggests that there is a conformational change in actin after Pi release. Much has been learned about the nucleotide hydrolysis-dependent conformational changes that occur in G proteins and motor proteins (for review see ref. 7). However, the crystal structures of rabbit muscle actin in the ADP- and ATP-bound states show only minor structural differences (8). Actin has two major domains separated by a nucleotide binding cleft. The smaller domain can be further divided into subdomains 1 and 2, and the larger domain consists of subdomains 3 and 4. Proteolysis studies and fluorescence resonance energy transfer assays reveal shifts in the position of subdomain 2 in ATP vs. ADP monomeric actin (9–11). Consistent with this finding, three-dimensional reconstructions of electron micrographs of actin filaments revealed a disordering of subdomain 2 in ADP compared with ADP-BeF actin filaments (12, 13).
To study the mechanism by which actin uses the energy of ATP hydrolysis to control filament dynamics, we introduced mutations into the ATP binding pocket of the yeast actin gene. Here we discuss the study of actin in which Val-159 is mutated to Asn (V159N). The amide group of V159 makes a hydrogen bond with the γ-phosphate oxygen O1C in ATP-bound actin and makes no nucleotide contacts in ADP-bound actin (8). This residue therefore is in a good position to act as a “sensor” for the presence of ATP or ADP-Pi vs. ADP and to participate in Pi release. Expression of this mutant actin in yeast results in a decrease in the rate of actin filament turnover in vivo (14). In this study, we examine the biochemical and structural properties of this mutant actin to determine the mechanism by which the V159N mutation stabilizes actin filaments.
MATERIALS AND METHODS
Construction of Mutations.
The mutation was constructed as described (15) and confirmed in the final yeast strain by sequencing a PCR product from the region containing the mutation.
Actin Purification.
Actin was purified from yeast expressing act1–159 or wild-type actin by DNaseI affinity chromatography, frozen in liquid N2, and stored at −80°C (16). Before all experiments, the actin was subjected to an assembly and disassembly cycle as follows. Actin was polymerized by addition of 0.1 M KCl, 0.5 mM ATP, and 2 mM MgCl2. KCl was added to a final concentration of 0.3 M, and the filamentous actin was sedimented, resuspended in G buffer (10 mM Tris, pH 7.5/0.2 mM CaCl2/0.5 mM ATP/1 mM DTT), and dialyzed for 16 hr at 4°C against two changes of G buffer. The G-actin then was clarified by centrifugation at 90,000 rpm for 30 min in a TLA100 rotor at 4°C, and actin concentration was measured by absorbance at 290 nm (e = 26, 600 M−1) and densitometry of SDS/PAGE gel bands. Actin cycled in this manner had no cofilin by Western blotting (molar ratio less than 1:100 cofilin/actin).
Measuring Polymerization and Depolymerization Dynamics.
The critical concentration was measured by pelleting filamentous actin from steady-state reactions containing different concentrations of actin. Supernatants and pellets were analyzed by SDS/PAGE, and actin was quantified by densitometry using an IS-2000 digital imaging system (Alpha Innotech, San Leandro, CA). Polymerization and depolymerization were monitored by light scattering at 400 nm using a Hitachi F-4010 fluorescence spectrophotometer, in a quartz cuvette thermostatted at 25°C. Gelsolin-capped filaments were assembled by mixing 6 μM actin (1:5 pyrene rabbit muscle actin/yeast actin) with 10 nM human gelsolin. Latrunculin A was added to 50 μM, and disassembly was monitored by recording pyrene fluorescence (excitation 365 nm, emission 407 nm). Gelsolin-capped filaments were observed by adding 0.2 units of rhodamine phalloidin (Molecular Probes R-415) to 10 μl of actin filaments. The filaments were diluted 100-fold in F buffer (10 mM Tris, pH 7.5/2 mM MgCl2/0.2 mM CaCl2/1 mM ATP/0.1 M KCl) with 100 mM DTT and observed immediately by fluorescence microscopy. Filament lengths were measured by using image pro plus software (Phase 3 Imaging, Glen Mills, PA).
Pi Release.
Pi release was measured as described by Melki et al. (17) by using the EnzChek kit (Molecular Probes). Actin used in these experiments was cycled as described above, except that that final clearing spin was 2 min at 90,000 rpm instead of 30 min. Twenty-five micrometers of actin, 0.2 mM MESG (2-amino-mercapto-7 methylpurine ribonucleoside), and 0.1 units of purine nucleotide phosphorylase (PNP) with or without 25 μM cofilin in G buffer (total volume of 95 μl) were incubated for 5 min at 25°C. Polymerization was initiated by addition of 5 μl of 20× polymerization salts (2 M KCl, 40 mM, MgCl2, 10 mM ATP), and the absorbance at 360 nm was monitored by using a Spectramax 340 spectrophotometer (Molecular Devices). Data were collected and analyzed by using softmaxRpro software. Pi release was normalized by subtracting a trace of the identical reaction missing the PNP.
Preparation of Samples for Electron Microscopy.
Frozen G-actin (wild type and V159N) was thawed quickly at room temperature and then passed through a polymerization-depolymerization cycle. Samples of wild-type and V159N actin filaments were prepared for electron microscopy as described (18). For BeF4−-wild-type filaments, G-actin (0.25 mg/ml) in G buffer was polymerized for 3 hr after addition of 50 mM KCl and 2 mM MgCl2 at room temperature. F-actin was spun down in a TLX-tabletop ultra-centrifuge. The pellet was resuspended in a 50 mM KCl, 2 mM MgCl2, 1 mM ADP solution, and incubated in this solution for 1–2 hr, before spinning down the F-actin again. The pellet was resuspended in a BeF4− solution (50 mM KCl/2 mM MgCl2/1 mM ADP/5 mM NaF/100 μM BeSO4/10 mM Tris⋅HCL, pH 7.6), and incubated overnight at 4°C before preparation for electron microscopy.
Three-Dimensional Reconstructions.
The reconstructions of yeast F-actin represent averages of 24 filaments (wild type), 49 filaments (ADP-BeF4−), and 37 filaments (V159N mutant), with each filament containing approximately 75 subunits for the wild type and 100 subunits for the others. Twelve layer lines (l = 0, n = 0; l = 1, n = 2; l = 2, n = 4; l = 3, n = 6; l = 4, n = −5; l = 5, n = −3; l = 6, n = −1; l = 7, n = 1; l = 8, n = 3; l = 12, n = −2; l = 13, n = 0; l = 14, n = 2) were extracted automatically from each filament transform by using an algorithm that found the layer line position by maximizing the intensity and minimizing the intensity-weighted near-far phase residual. By dividing each data set into two halves, and using a 45° phase-residual criterion in the comparison between the two independent data sets, the resolution of each reconstruction was determined to be 23.5 Å (wild type), 20.4 Å (ADP-BeF2−), and 22.2 Å (V159N mutant). The surfaces shown for all reconstructions (real actin filaments and atomic models) correspond to 100% of the expected molecular volume of actin, assuming a partial specific volume for protein of 0.75 cm3/gr.
RESULTS
V159N Actin Depolymerizes Slowly and Has a Low Critical Concentration.
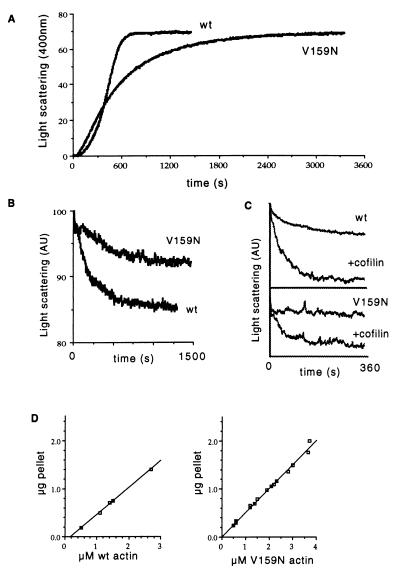
We purified actin from yeast expressing V159N (act1–159) as their sole source of actin (14) and compared polymerization and depolymerization kinetics to those of yeast actin purified from an isogenic wild-type strain. The mutant actin nucleates assembly slightly faster than wild-type actin, but subsequent assembly occurs at a slower rate (Fig. 1A). The rate of depolymerization upon dilution of filamentous actin is reduced 3-fold in the V159N mutant compared with wild type (Fig. 1B). The dilution induced depolymerization of wild-type yeast actin is accelerated by the addition of cofilin (19). When cofilin is added to the V159N mutant actin under the same conditions, the rate of depolymerization is increased, but not to the same extent as with wild-type actin (Fig. 1C). We measured the critical concentration, the concentration at which net polymerization equals net depolymerization, by pelleting actin filaments from steady-state reactions at various actin concentrations. The critical concentration of V159N was quite low (less than 20 nM) compared with wild-type yeast actin (160 nM) (Fig. 1D). The critical concentration measured for wild-type yeast actin is consistent with previously published reports (20). The low critical concentration suggests that this mutation results in an overall stabilization of actin filaments.
Figure 1.
Polymerization and depolymerization of V159N actin were measured by light scattering at 400 nm. (A) Polymerization of 5 μM wild-type or V159N actin was initiated by the addition of 0.1 M KCl, 2 mM MgCl2, and 0.5 mM ATP. (B) Depolymerization was measured by diluting 5 μM F-actin to 0.5 μM in the polymerization buffer. The half time of depolymerization for wild-type actin is 114 s (n = 4, SD = 18), and for V159N actin is 355 s (n = 5, SD = 155). (C) Cofilin-accelerated depolymerization was observed by diluting 5 μM F-actin to 0.5 μM in polymerization buffer containing 0.5 μM yeast cofilin. (D) The critical concentration was measured by pelleting filamentous actin from steady-state reactions containing different concentrations of actin. Supernatants and pellets were analyzed by SDS/PAGE, and actin was quantified by densitometry. The critical concentration of V159N was measured three times, and the values ranged from 0 to 20 nM. The critical concentration of wild-type yeast actin is 160 nM.
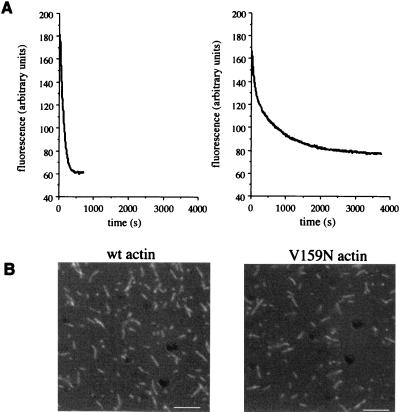
Depolymerization upon dilution of filamentous actin depends on the on and off rates of actin monomers at both the barbed (fast growing) and pointed (slow growing) ends of the filaments. To observe depolymerization specifically from the pointed end, we assembled filaments capped by gelsolin at their barbed ends and monitored depolymerization in the presence of latrunculin A, a drug that prevents reassembly of actin monomers (21) (Fig. 2A). Under these conditions, depolymerization can be observed in the absence of polymerization. In this experiment, the rate of depolymerization is reduced approximately 2.5-fold for V159N compared with wild-type actin. An apparent decrease in the depolymerization rate also could result from a low number of filament ends because of an increase in the average filament length. We measured the lengths of filaments used for this experiment (Fig. 2B) and found that the average lengths of the gelsolin-capped filaments for wild-type and V159N actin are 1.4 μm and 1.1 μm, respectively (n = 300 for each), consistent with a slower rate of polymerization of V159N actin. Because the V159N filaments are not longer than wild-type filaments, the apparent slow depolymerization is not the result of an increase in average filament length. Therefore, the primary reason for slow depolymerization appears to be a slow off-rate of monomers from the pointed end.
Figure 2.
Depolymerization of gelsolin-capped actin filaments. (A) Latrunculin A (50 μM) was added to gelsolin-capped filaments, and depolymerization was monitored with pyrene fluorescence. The half time of depolymerization is 96 s for wild-type (wt) filaments and 230 s for V159N filaments. (B) To compare length distributions, actin filaments used for the depolymerization assay were stained with rhodamine phalloidin and observed by fluorescence microscopy. (Bar = 5 μm.)
The Rate of Pi Release from V159N Actin Filaments During Assembly Is Normal.
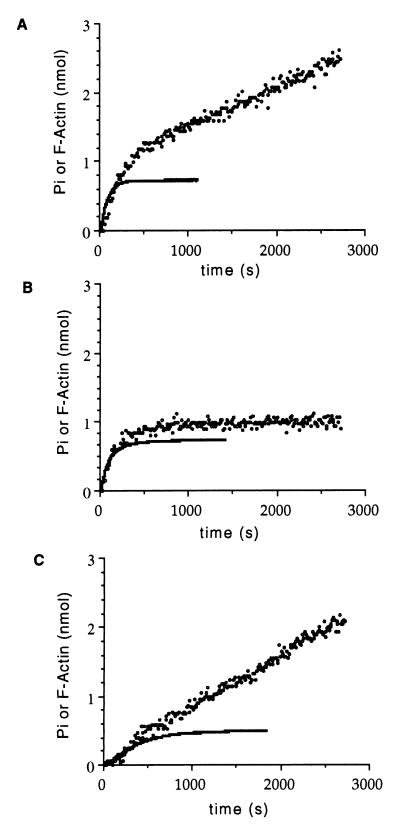
We next determined whether the slow depolymerization rate of V159N filaments was the result of a decrease in the rate of Pi release. We measured Pi release under conditions in which the initial rates of assembly are similar between mutant and wild-type actin. As shown in Fig. 3 A and B, the initial rates of Pi release are similar in the mutant and wild-type actins (0.0048 s−1 for wild type and 0.0042 s−1 for V159N actin). Once steady-state assembly levels have been achieved, wild-type actin continues to release Pi (5.8 × 10−4⋅s−1) as actin monomers depolymerize, exchange ADP for ATP, and repolymerize. However, Pi release from V159N actin filaments is dramatically reduced (3.8 × 10−5⋅s−1) once steady-state assembly levels have been achieved and 1 mol of Pi has been released per mol of actin polymerized. We postulated that slow steady-state Pi release results from slow depolymerization of V159N actin filaments. Consistent with this hypothesis, addition of cofilin, which stimulates depolymerization, greatly increases the rate of Pi release for V159N actin filaments at steady state (Fig. 3C). Because Pi release is normal during assembly of V159N actin, these filaments should be composed primarily of ADP actin. We therefore propose that V159N actin does not undergo the conformational change that normally accompanies Pi release and allows rapid depolymerization.
Figure 3.
Pi release during actin filament assembly. Polymerization of actin (25 μM) was monitored by light scattering at 400 nm, and Pi release measured with the EnzChek phosphate assay kit (Molecular Probes). The amount of total polymerized actin then was measured by pelleting the final polymerization mix and measuring the protein concentration of the pellet by using an SDS gel containing actin standards. (A) Wild-type yeast actin. (B) V159N actin. (C) V159N actin plus 25 μM cofilin. The solid lines represent actin assembly and the circles are free Pi.
V159N Actin Filaments Resemble BeF3−-ADP Actin Filaments.
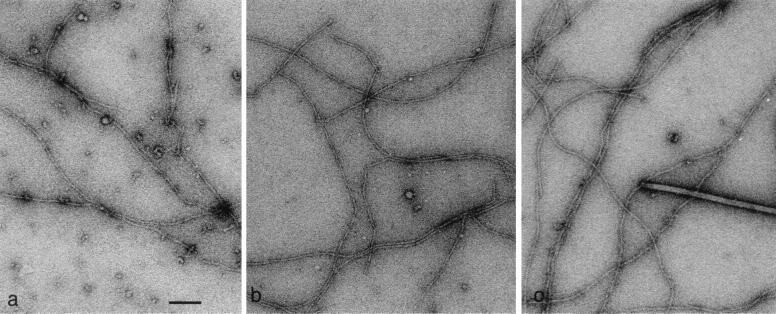
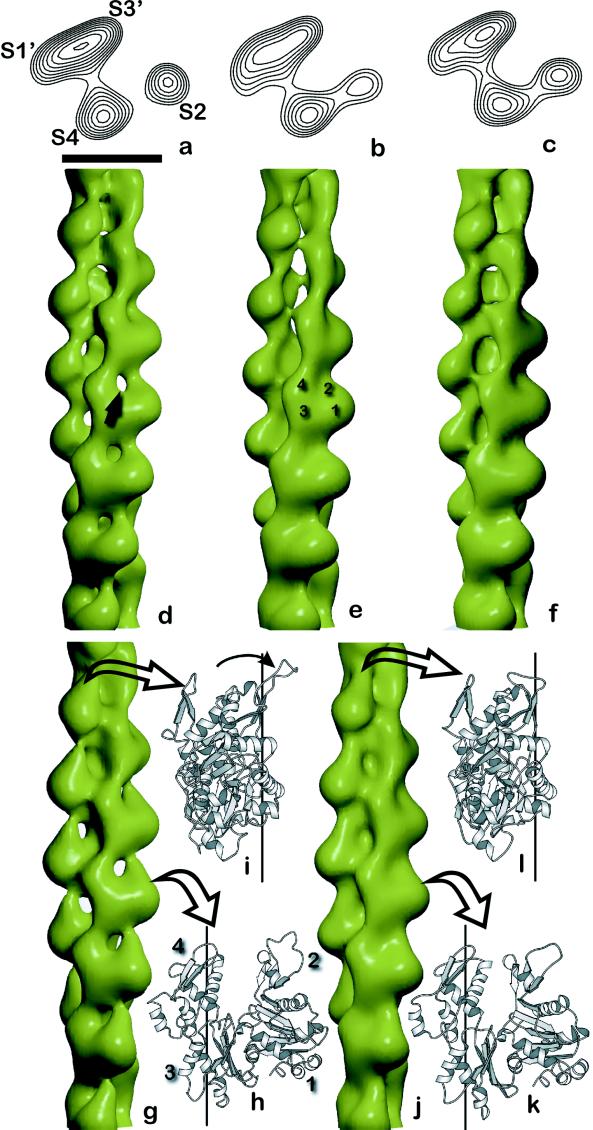
We directly tested this hypothesis by using electron microscopy and three-dimensional reconstructions of actin filaments. Fig. 4 shows electron micrographs of yeast wild-type, V159N, and BeF3−-ADP actin filaments. Wild-type actin consists primarily of ADP actin subunits, whereas BeF3−-ADP acts as a stable analog for the ATP or ADP-Pi state (5). As shown in Fig. 5 a–f, the structure of BeF3−-ADP actin filaments closely resembles that of V159N filaments, even though the V159N filaments have released their Pi. This finding strongly suggests that the V159N actin filaments do not undergo a conformational change after Pi release and suggests that this conformational change is required for the ability to rapidly depolymerize.
Figure 4.
Electron micrographs of yeast wild-type F-actin (a), ADP-BeF4− filaments (b), and V159N filaments (c). Tobacco mosaic virus particles with a helical pitch of 23 Å were used as a size standard (c). (Bar in a = 1,000 Å.)
Figure 5.
Three-dimensional electron microscopy reconstructions and actin filament models. Reconstructions of yeast actin filaments (a–f) compared with models generated from β-actin (g–l). The wild-type yeast actin, shown as a surface view (d) and in a single cross section (a), has an open nucleotide-binding cleft (arrow, d). The open cleft results in a lack of density between subdomains 2 (S2) and 4 (S4), as seen in cross section (a). Subdomains 1 (S1′) and 3 (S3′) from a subunit on the opposite strand also are labeled in a, and four subdomains from a single actin subunit are labeled in e. When the wild-type F-ADP filaments are incubated with BeF4− (b and e), the nucleotide-binding cleft closes. The V159N mutant actin (c and f) also shows a closure of the nucleotide-binding cleft. The opening of the nucleotide-binding cleft also can be seen in a crystal structure of nonmuscle β-actin (27) (h and i). The G-actin subunit from this crystal structure (h) has been oriented to the Lorenz et al. model for F-actin (26), using a least-squares alignment of Cα atoms in subdomains 1, 3, and 4 (3.3-Å rms deviation). The filament axis after such an alignment is shown by the vertical line (h, front view; i, side view), and the open arrows in g indicate the corresponding views of the subunits in a low-resolution filament surface generated from the atomic model. The corresponding “closed” state of β-actin (26) after alignment to the Lorenz et al. model (3.1-Å rms deviation) is shown in k and l, with the low-resolution surface generated from this subunit in j. The main difference between these two forms is the rotation of subdomain 2 by about 15°, shown by the arrow in i. (The scale bar in a is 50 Å and applies to a–c.)
The primary structural difference between wild-type ADP yeast actin filaments and V159N or BeF3−-ADP filaments is that the nucleotide-binding cleft is open in ADP wild-type filaments. Yeast actin filaments with an S14A mutation also have a closed cleft similar to that of V159N actin (18) and appear to have slow dynamics as determined by a decrease in ATP hydrolysis during assembly (20). An additional difference between the ADP wild-type and BeF3−-ADP wild-type filaments is that the connectivity between the two long-pitch strands appears to be significantly reduced in the BeF3−-ADP state. This finding is consistent with previous observations suggesting that the strength of the long-pitch bonds is significantly greater than the bonds across the strands in the ADP-Pi state (22).
Because each of the reconstructions represents an average of a rather large data set, we have been able to do several tests to exclude the possibility that the observed opening and closing of the nucleotide binding cleft is the result of an artifact. A larger average of 42 wild-type filaments was generated (containing 84 independent near-far data sets), and it was nearly indistinguishable from the average of 24 filaments shown in Fig. 5. Subsets of the “best” filaments (based on phase residuals against the average) have been independently averaged together for the wild-type and V159N filaments, and these are nearly identical to the averages shown. The BeF2−ADP filaments may not be entirely homogeneous, as some subsets do show an opening of the cleft. This result is likely explained by the incomplete saturation of both BeF binding sites (6). Most importantly, statistical difference maps (23) between the V159N and wild-type filaments show a highly significant peak (4.3 σ) at the position of the open nucleotide-binding cleft, demonstrating that this feature does not arise from noise or fluctuations in the averages. We also know that as the resolution of the reconstruction decreases, the cleft will appear to close. However, the resolutions of the reconstructions that show the closed cleft (V159N and BeF2−-ADP filaments) are actually higher (22.2 Å and 20.4 Å, respectively) than the wild-type (23.5 Å) reconstruction that reveals the open nucleotide binding cleft. These data are published as supplemental material to this article on the PNAS web site (www.pnas.org).
The Two Conformations of Mammalian Cytoplasmic β-Actin Can Be Modeled into Filaments that Resemble ADP and V159N Filaments.
Additional insight into the two different conformations of the nucleotide-binding cleft comes from x-ray crystallography. A single high-resolution model for the F-actin filament has been generated by refining the structure of a crystallographically determined skeletal muscle G-actin subunit against x-ray fiber diffraction data (24, 25). However, a nonmuscle β G-actin subunit has been found in two different conformations: with the nucleotide-binding cleft in a closed conformation (26) and in an open conformation (27). These two structures differ primarily by a rotation of subdomain 2. By starting with the Lorenz et al. model of F-actin (24), we have generated two additional models by replacing the filament protomer with β G-actin in an “open” state (Fig. 5 g–i) and in a “closed” state (Fig. 5 j–l). The open-state model (Fig. 5g) provides a very good fit to the yeast wild-type filaments (Fig. 5d), whereas the closed-state model (Fig. 5j) provides a very good fit to both the wild-type ADP-Pi state (Fig. 5e) and the V159N mutant (Fig. 5f). A difference map between the two low-resolution atomic models is very similar to the difference map between the wild-type and V159N filaments, suggesting that the differences we observe can be explained by the differences between the two atomic models (see supplemental material on www.pnas.org). Our biochemical and structural data suggest that these two states represent actin conformations that favor polymerization (closed) and depolymerization (open) and demonstrate that the highly conserved residue, Val-159, is important for inter-conversion between these two states.
The crystallographic models in Fig. 5 g and j provide additional tests of the electron microscopy data. The layer lines from the two models were used as references, and most of the V159N and BeF3−-ADP actin filaments yielded lower-phase residuals against the closed model, whereas most of the wild-type filaments yielded lower-phase residuals against the open model.
DISCUSSION
Yeast actin with the V159N mutation nucleates assembly faster than wild-type yeast actin, but then assembles with slightly slower kinetics. V159N actin also depolymerizes much more slowly than wild-type actin and has a lower critical concentration. The rate-limiting step for nucleation of actin filaments is the formation of an actin trimer (28, 29). The slow depolymerization rate of V159N actin suggests that nucleation may be faster because of a slower dissociation of dimers or trimers of actin; however, further studies will be required to test this hypothesis.
Val-159 is in the nucleotide binding cleft and makes a hydrogen bond to the γ-phosphate of the bound ATP. However, the V159N mutation does not affect the rate of Pi release during filament assembly. This finding suggests that neither ATP hydrolysis nor Pi release are affected by this mutation. Structural studies of V159N actin filaments reveal that they more closely resemble BeF2−ADP yeast actin filaments than ADP yeast actin filaments, suggesting that they fail to undergo the normal conformational change that accompanies Pi release. In support of this conclusion, we observe that BeF2 does not further stabilize V159N actin filaments under conditions in which it stabilizes wild-type yeast actin filaments (data not shown). The observed conformational change consists primarily of a closure of the nucleotide binding cleft caused by a shift of subdomain 2 after Pi release.
Yeast actin is more similar to vertebrate cytoplasmic actins than to skeletal muscle actins in both sequence and function (30, 31). We therefore propose that this mechanism of regulating dynamics may be used by other dynamic cytoplasmic actins. In addition, other studies have implicated shifts in subdomain 2 as the conformational shift between ADP and ADP-Pi actin in skeletal muscle actin (9, 10, 12).
Actin shares structural similarity with hexokinase, the hsp70 family of proteins (32–34) and, presumably, the actin-related proteins (35). The catalytic mechanism of hexokinase depends on an opening and closing of the ligand binding cleft (36), and we now have shown that control of actin polymerization and depolymerization depends on a related mechanism. A recent paper has suggested that the differences between the two β-actin/profilin crystal structures are inconsistent with the current model for F-actin (37). However, our demonstration that two different crystal structures of G-actin can be oriented into the current model of F-actin (24) in agreement with electron microscopy reconstructions of two different states of F-actin lends strong support to the current model and provides additional insights into actin dynamics.
Supplementary Material
Acknowledgments
We thank Paul Janmey for the gift of human platelet gelsolin, Phil Crews for latrunculin A, and Pekka Lappalainen for providing purified yeast cofilin. We thank M.-F. Carlier for advice on enzyme-linked phosphate release assays, and Bruce Goode, Avi Rodal, and Jamie Cope for comments on the manuscript. This work was supported by National Institutes of Health Grants to E.H.E. (AR42023) and D.G.D. (GM 42759) and a fellowship to L.D.B. and an American Cancer Society grant to D.G.D.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Carlier M F, Pantaloni D. Biochemistry. 1986;25:7789–7792. doi: 10.1021/bi00372a001. [DOI] [PubMed] [Google Scholar]
- 2.Carlier M F. Adv Biophys. 1990;26:51–73. doi: 10.1016/0065-227x(90)90007-g. [DOI] [PubMed] [Google Scholar]
- 3.Rickard J E, Sheterline P. J Mol Biol. 1986;191:273–280. doi: 10.1016/0022-2836(86)90264-0. [DOI] [PubMed] [Google Scholar]
- 4.Wanger M, Wegner A. Biochim Biophys Acta. 1987;914:105–113. doi: 10.1016/0167-4838(87)90052-5. [DOI] [PubMed] [Google Scholar]
- 5.Combeau C, Carlier M F. J Biol Chem. 1988;263:17429–17436. [PubMed] [Google Scholar]
- 6.Allen P G, Laham L E, Way M, Janmey P A. J Biol Chem. 1996;271:4665–4670. doi: 10.1074/jbc.271.9.4665. [DOI] [PubMed] [Google Scholar]
- 7.Vale R D. J Cell Biol. 1996;135:291–302. doi: 10.1083/jcb.135.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabsch W, Mannherz H G, Suck D, Pai E F, Holmes K C. Nature (London) 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 9.Moraczewska J, Strzelecka-Golaszewska H, Moens P D, dos Remedios C G. Biochem J. 1996;317:605–611. doi: 10.1042/bj3170605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strzelecka-Golaszewska H, Moraczewska J, Khaitlina S Y, Mossakowska M. Eur J Biochem. 1993;211:731–742. doi: 10.1111/j.1432-1033.1993.tb17603.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim E, Motoki M, Seguro K, Muhlrad A, Reisler E. Biophys J. 1995;69:2024–2032. doi: 10.1016/S0006-3495(95)80072-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orlova A, Egelman E H. J Mol Biol. 1992;227:1043–1053. doi: 10.1016/0022-2836(92)90520-t. [DOI] [PubMed] [Google Scholar]
- 13.Orlova A, Egelman E H. J Mol Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 14.Belmont L D, Drubin D G. J Cell Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wertman K F, Drubin D G, Botstein D. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melki R, Fievez S, Carlier M F. Biochemistry. 1996;35:12038–12045. doi: 10.1021/bi961325o. [DOI] [PubMed] [Google Scholar]
- 18.Orlova A, Chen X, Rubenstein P A, Egelman E H. J Mol Biol. 1997;271:235–243. doi: 10.1006/jmbi.1997.1163. [DOI] [PubMed] [Google Scholar]
- 19.Lappalainen P, Drubin D G. Nature (London) 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Rubenstein P A. J Biol Chem. 1995;270:11406–11414. doi: 10.1074/jbc.270.19.11406. [DOI] [PubMed] [Google Scholar]
- 21.Coue M, Brenner S L, Spector I, Korn E D. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- 22.Egelman E H, Orlova A. Curr Opin Struct Biol. 1995;5:172–180. doi: 10.1016/0959-440x(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 23.Egelman E H, Yu X. Science. 1989;245:404–407. doi: 10.1126/science.2667137. [DOI] [PubMed] [Google Scholar]
- 24.Lorenz M, Popp D, Holmes K C. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 25.Holmes K C, Popp D, Gebhard W, Kabsch W. Nature (London) 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 26.Schutt C E, Myslik J C, Rozycki M D, Goonesekere N C, Lindberg U. Nature (London) 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 27.Chik J K, Lindberg U, Schutt C E. J Mol Biol. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- 28.Pollard T. Curr Opin Cell Biol. 1990;2:33–40. doi: 10.1016/s0955-0674(05)80028-6. [DOI] [PubMed] [Google Scholar]
- 29.Mullins R D, Heuser J A, Pollard T D. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheterline P, Clayton J, Sparrow J. Actin. London: Academic; 1995. [PubMed] [Google Scholar]
- 31.Karlsson R, Aspenstràm P, Bystràm A S. Mol Cell Biol. 1991;11:213–217. doi: 10.1128/mcb.11.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flaherty K M, DeLuca-Flaherty C, McKay D B. Nature (London) 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- 33.Flaherty K M, McKay D B, Kabsch W, Holmes K C. Proc Natl Acad Sci USA. 1991;88:5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurley J. Annu Rev Biophys Biomol Struct. 1996;25:137–162. doi: 10.1146/annurev.bb.25.060196.001033. [DOI] [PubMed] [Google Scholar]
- 35.Mullins R D, Kelleher J F, Pollard T D. Trends Cell Biol. 1996;6:208–212. doi: 10.1016/0962-8924(96)20017-0. [DOI] [PubMed] [Google Scholar]
- 36.Steitz T A, Shoham M, Bennett W S., Jr Philos Trans R Soc London B. 1981;293:43–52. doi: 10.1098/rstb.1981.0058. [DOI] [PubMed] [Google Scholar]
- 37.Page R, Lindberg U, Schutt C E. J Mol Biol. 1998;280:463–474. doi: 10.1006/jmbi.1998.1879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.