Abstract
Pathways of proton entry have been identified in the proton-translocating heme–copper oxidases, but the proton exit pathway is unknown. Here we report experiments with cytochrome bo3 in Escherichia coli cells that may identify the beginning of the exit pathway. Systematic mutations of arginines 438 and 439 (R481 and R482 in the E. coli enzyme), numbering as in cytochrome aa3 from bovine heart mitochondria, which interact with the ring D propionates of the two heme groups, reveal that the D propionate of the oxygen-binding heme is involved in proton pumping; its anionic form must be stabilized in order for proton translocation to occur. This may locate the beginning of the pathway by which pumped protons exit from the enzyme structure.
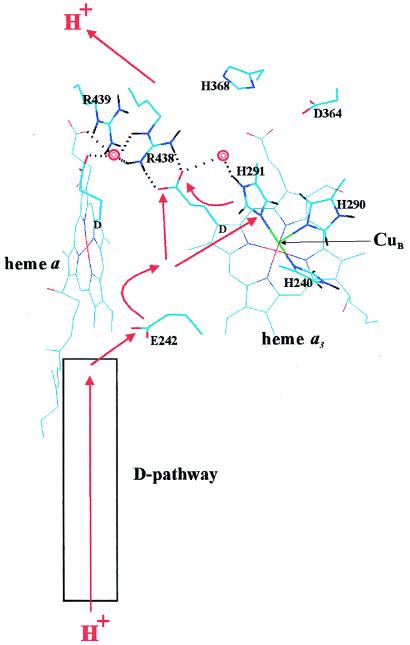
The structurally and functionally related respiratory heme–copper oxidases are redox-linked proton translocators (1–5) that convert free energy from the reduction of O2 to an electrochemical proton gradient across mitochondrial or bacterial membranes, to be subsequently used for ATP synthesis by the membrane-bound H+-ATP synthase (6). The mechanism of proton translocation by this class of enzymes is intensively studied at present, and much of this research has recently been devoted to locating the pathways in the protein structure by which protons are transferred during translocation. Mutagenesis experiments with heme–copper oxidases from Escherichia coli (cytochrome bo3; refs. 7 and 8), Rhodobacter sphaeroides (cytochrome aa3; ref. 9) and Paracoccus denitrificans (cytochrome aa3; ref. 10) have established that protons to be pumped are transferred into the protein via the so-called D-pathway, which has been identified in the crystal structures of cytochrome aa3 from P. denitrificans and bovine heart mitochondria (refs. 11–13; Fig. 1). After this, the protons reach the invariant E242 residue† (14) in a hydrophobic cavity near the middle of the membrane, probably via bound water molecules (14–16). Little is known about proton conduction beyond E242, except that it may require isomerization of the side chain of this residue (15–17). However, because the D-pathway also conducts at least two of the four protons consumed in O2 reduction to water at the binuclear heme–copper center, there must also be connectivity to this site from E242, again possibly via bound water molecules (refs. 15 and 16; see Fig. 1). The pathway of the pumped protons beyond E242 is not known, including the exit path to the opposite side of the membrane. Proton transfer via the D-channel to E242 is believed to be largely passive. Hence, a structural assignment of the beginning of the proton exit path may be valuable for the elucidation of the mechanism of translocation, which must include the structural and functional details of how this process is linked to the chemistry of O2 reduction. It is conceivable that the “molecular machinery” responsible for this linkage should reside between the residue E242 and the beginning of the path by which protons exit the membrane. Experiments reported here may help to define the latter.
Figure 1.
Proton transfer in the heme–copper oxidases. The view is approximately in the membrane plane with the proton input and output sides of the membrane below and above, respectively. The figure is based on the x-ray coordinates of cytochrome aa3 from Paracoccus denitrificans (ref. 13; Protein Data Bank identification code 1AR1), drawn by using the program HyperChem 4.5 (Hypercube, Ontario, Canada). The D rings of hemes a and a3 are marked and the D-propionate groups are highlighted. CuB in the foreground (green), and some key residues are both highlighted and marked. Two crystallographically visible water molecules (13) are shown as red double rings. Dotted lines indicate potential hydrogen bonds discussed in the text. The red arrows outline possible paths of pumped protons across the enzyme’s membrane domain (see text).
MATERIALS AND METHODS
Bacterial growth conditions and purification of histidine-tagged cytochrome bo3 enzymes from E. coli were as described previously (18). Site-directed mutagenesis was performed according to published methods (8) and was confirmed by DNA sequencing (ALFexpress DNA Sequencer, Pharmacia). This confirmation was also done for all mutant cells from fermentor cultivations as well as for the cells used in proton translocation measurements.
Ubiquinol oxidase (cytochrome bo3) activity in the mutant and wild-type bacterial membranes and isolated enzymes were measured as described previously (7). Proton translocation in cells was determined by the oxygen pulse method (7, 8).
RESULTS AND DISCUSSION
A domain on the proton output side of the ring D propionates of the two heme groups (Fig. 1) contains several polar and charged residues and a cluster of bound water molecules, as revealed by the x-ray structures of the cytochrome aa3 enzymes from P. denitrificans and bovine heart mitochondria (12, 13), as well as from calculations based on these structures (16, 19). This domain, which appears well connected protonically to the outside of the membrane, has indeed been implicated in proton exit (11, 13, 19), but functional support for this route is lacking because no mutations in this domain have been found to affect proton translocation specifically (7, 14, 20). Electrostatic calculations based on the crystal structures of the oxidized enzyme suggest that all four heme propionates are stabilized in their anionic state (21). The D-propionate of the oxygen-binding heme (heme a3 or o3) is stabilized by charge interactions with the conserved arginines 438 and 439, as well as by hydrogen bonds from R438, W126 (11, 12), and perhaps from a water molecule between this propionate and the CuB ligand H291 (ref. 13; Fig. 1). The D-propionate of the low-spin heme (heme a in aa3 enzymes; heme b in bo3 enzymes) is also stabilized by charge interactions with R438 and R439 (21), and by hydrogen bonds from the ɛNH group of R439 and the backbone NH of W126 (12, 13).
To explore a possible role in proton translocation of the conserved arginines and the two D-propionates with which they interact, we have systematically mutagenized the two arginines. In agreement with the report by Kawasaki et al. (22), changing the very well conserved R438 (R481 in cytochrome bo3 from E. coli used here) to glutamine decreased enzyme turnover only a little. There was also no effect on proton translocation efficiency, as measured in multiturnover O2-pulse experiments with E. coli cells (Table 1). However, changing R438 to either asparagine or leucine abolished proton translocation, although the turnover decreased only to ≈50% (Table 1). In contrast, mutation of R438 to methionine caused almost complete loss of activity as well as major structural perturbations as indicated by altered optical heme spectra, which was not found with any of the other mutants studied here (not shown; cf. ref. 22).
Table 1.
Range of cytochrome bo3 enzyme activity in wild-type (wt) and mutant E. coli membranes (m) and isolated enzyme (i), and proton translocation efficiency in cells
| Strain (Cyt bo3 numbering) | Enzyme location | Activity, % of wt | H+/e− |
|---|---|---|---|
| Wild type | m, i | 100 | 1.6–2.2 |
| R481Q | m | 70–80 | 1.6–2.0 |
| i | 54–64 | ||
| R481M | i | 2–8 | — |
| R481N | i | 51–65 | 1.0–1.3 |
| R481L | i | 35–45 | 1.0–1.3 |
| R482Q | m | 75–95 | 1.7–2.1 |
| i | 46–60 | ||
| R482N | m | 48–58 | 1.7–2.2 |
| i | 33–39 | ||
| R482L | m | 75–85 | 1.7–2.0 |
| R481Q/R482Q | i | 35–45 | 0.9–1.2 |
Wild-type activity range: 400–750 (m) and 350–560 (i) electrons per sec per bo3. Methods for determining proton translocation and enzyme activity and for mutagenesis are described in refs. 7, 8, and 14. In the quinol oxidase cytochrome bo3 1 H+/e− is released to the outside of the cells due to the oxidation of hydroquinone.
On the other hand, while arginine-439 (R482 in cytochrome bo3) could be changed to glutamine, asparagine, or even leucine without loss of proton translocation (Table 1), changing both arginines to glutamines abolished proton translocation even though either single mutation had no effect. Enzyme turnover decreased to ≈40% in this double mutant—i.e., only slightly more than for each single mutation (Table 1).
Inspection of the structure suggests that the amide group of a glutamine in place of R438 (R438Q) could still maintain hydrogen bonding to the D-propionate of the oxygen-binding heme (heme a3 in cytochrome aa3; heme o3 in cytochrome bo3) without much change in the position of the propionate, but this is not possible with asparagine (R438N) or leucine (R438L). The loss of proton translocation in the latter two mutants suggests that the R438/heme o3 (a3) D-propionate pair is critical for the proton-translocating mechanism. However, because proton translocation efficiency is normal in the R438Q mutant we conclude that this function does not absolutely demand a protonatable or a positively charged side chain in the 438 site. The loss of proton translocation may therefore be ascribed to changes induced in the properties of the D-propionate of heme o3 (a3) with which R438 interacts most strongly.
The anionic form of the D-propionate of heme o3 (a3) may be stabilized by ≈11.0 and 6.5 pKa units because of charge interactions with R438 and R439, respectively (21). Further stabilization by hydrogen bonding from R438 may amount to ≈5 pKa units. In the R438Q, N, and L mutants the stronger charge interaction is lost, but hydrogen bonding uniquely remains in R438Q. Interestingly, it seems that in this case this subtle difference determines whether proton translocation occurs. In the R439 mutants only the weaker charge interaction with the heme o3 (a3) D-propionate is lost and proton translocation remains. However, in the double mutant both charge interactions are abolished and destabilization of this propionate is expected to be considerable. Now proton pumping is lost despite the fact that the hydrogen bond from the 438 locus may be retained.
These predicted effects of the mutations on the heme o3 (a3) D-propionate are summarized in Table 2 and compared with the data on proton translocation. The listed interactions show only the general trend (21), and do not, for example, include possible compensatory effects that might occur in a mutant. At any rate, they suggest that loss of proton translocation correlates with the extent of destabilization of the anionic form of the heme o3 (a3) D-propionate. Such destabilization would favor the uncharged propionic acid state, which may be the cause of the observed decoupling of proton translocation from the chemistry of O2 reduction.
Table 2.
Predicted approximate stabilization of the anionic state of the D-propionate group of heme o3 (a3) by its charge interactions (21) with R438 (R481 in cytochrome bo3) and R439 (R482 in bo3) and its hydrogen bond interaction with R438 (or Q438)
| Strain (Cyt bo3 numbering) | Predicted stabilization, pKa units
|
Proton translocation (see Table 1) | |||
|---|---|---|---|---|---|
| Charge interaction
|
Hydrogen bond | Sum | |||
| With R481 | With R482 | ||||
| Wild type | 11 | 6.5 | 5 | 22.5 | Yes |
| R481Q | 0 | 6.5 | 5 | 11.5 | Yes |
| R481N, L | 0 | 6.5 | 0 | 6.5 | No |
| R482Q, N, L | 11 | 0 | 5 | 16 | Yes |
| R481Q/R482Q | 0 | 0 | 5 | 5 | No |
The simplest rationale is that the D-propionate of heme o3 (a3) normally functions as a proton acceptor in a crucial step of the proton translocation mechanism, which would define the beginning of the exit path of pumped protons. However, our results do not exclude that proton transfer may also involve the neighboring D-propionate of the low-spin heme, which appears to be more strongly stabilized than the D-propionate of heme o3 (a3) by charge interactions with the arginines (21), and may thus not be sufficiently perturbed by the single arginine mutations. In this connection it may be of special interest to note that a water molecule which lies within hydrogen-bonding distance from both propionates of the low-spin heme b (a), is also within hydrogen-bonding distance from the Nɛ and NH2 nitrogens of R438 (ref. 13; Fig. 1). Since the latter is a hydrogen bond donor to the D-propionate of heme o3 (a3), both propionates of the low-spin heme are also at least potentially connected to the pathway of proton exit.
The D-propionate of heme o3 (a3) may accept protons from E242 either directly (11), via bound water molecules (16), or more indirectly via water molecules and the copper ligand H291 (Fig. 1) as proposed in a recent version (19) of the histidine cycle model of proton translocation (23). At any rate, our results suggest that a key problem of the proton translocation mechanism is now to elucidate how proton transfer is accomplished from E242 to the D-propionate of heme o3 (a3) in a way that is coupled to the oxygen reduction chemistry at the binuclear heme–copper center.
Acknowledgments
We are grateful to Robert B. Gennis and Jeff Thomas, who participated in early mutagenesis experiments, to Gerhard Hummer, Aimo Kannt, Joel E. Morgan, and Michael Verkhovsky for insightful discussion, and to Tarja Salojärvi for excellent technical assistance. This work was supported by grants from the Sigrid Jusèlius Foundation, the Academy of Finland, and the University of Helsinki.
Footnotes
The amino acid numbering is for subunit I of cytochrome aa3 from bovine heart mitochondria if not indicated otherwise.
References
- 1.Wikström M. Nature (London) 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 2.Wikström M. Nature (London) 1984;308:558–560. doi: 10.1038/308558a0. [DOI] [PubMed] [Google Scholar]
- 3.Van Verseveld H W, Krab K, Stouthamer A H. Biochim Biophys Acta. 1981;635:525–534. doi: 10.1016/0005-2728(81)90111-0. [DOI] [PubMed] [Google Scholar]
- 4.Solioz M, Carafoli E, Ludwig B. J Biol Chem. 1982;257:1579–1582. [PubMed] [Google Scholar]
- 5.Puustinen A, Finel M, Virkki M, Wikström M. FEBS Lett. 1989;249:163–167. doi: 10.1016/0014-5793(89)80616-7. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 7.Thomas J W, Puustinen A, Alben J O, Gennis R B, Wikström M. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 8.García-Horsman J A, Puustinen A, Gennis R B, Wikström M. Biochemistry. 1995;34:4428–4433. doi: 10.1021/bi00013a035. [DOI] [PubMed] [Google Scholar]
- 9.Fetter J R, Qian J, Shapleigh J, Thomas J W, García-Horsman J A, Schmidt E, Hosler J, Babcock G T, Gennis R B, Ferguson-Miller S. Proc Natl Acad Sci USA. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfitzner U, Odenwald A, Ostermann T, Weingard L, Ludwig B, Richter O-M H. J Bioenerg Biomembr. 1998;30:89–97. doi: 10.1023/a:1020515713103. [DOI] [PubMed] [Google Scholar]
- 11.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 12.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 13.Ostermeier C, Harrenga A, Ermler U, Michel H. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verkhovskaya M L, Garcìa-Horsman A, Puustinen A, Rigaud J-L, Morgan J E, Verkhovsky M I, Wikström M. Proc Natl Acad Sci USA. 1997;94:10128–10131. doi: 10.1073/pnas.94.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riistama S, Hummer G, Puustinen A, Dyer R B, Woodruff W H, Wikström M. FEBS Lett. 1997;414:275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- 16.Hofacker I, Schulten K. Proteins Struct Funct Genet. 1998;30:100–107. [PubMed] [Google Scholar]
- 17.Pomès R, Hummer G, Wikström M. Biochim Biophys Acta. 1998;1365:255–260. [Google Scholar]
- 18.Morgan J E, Verkhovsky M I, Puustinen A, Wikström M. Biochemistry. 1995;34:15633–15637. doi: 10.1021/bi00048a005. [DOI] [PubMed] [Google Scholar]
- 19.Wikström M, Morgan J E, Hummer G, Woodruff W H, Verkhovsky M I. In: Frontiers of Cellular Bioenergetics: Molecular Biology, Biochemistry and Physiopathology. Papa S, Guerrieri F, Tager J M, editors. New York: Plenum; 1999. , in press. [Google Scholar]
- 20.Qian J, Shi W, Pressler M, Hoganson C, Mills D, Babcock G T, Ferguson-Miller S. Biochemistry. 1997;36:2539–2543. doi: 10.1021/bi962721+. [DOI] [PubMed] [Google Scholar]
- 21.Kannt A, Lancaster C R D, Michel H. Biophys J. 1998;74:708–721. doi: 10.1016/S0006-3495(98)73996-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki M, Mogi T, Anraku Y. J Biochem (Tokyo) 1997;122:422–429. doi: 10.1093/oxfordjournals.jbchem.a021770. [DOI] [PubMed] [Google Scholar]
- 23.Morgan J E, Verkhovsky M I, Wikström M. J Bioenerg Biomembr. 1994;26:599–608. doi: 10.1007/BF00831534. [DOI] [PubMed] [Google Scholar]