Two articles in this issue of the JCI (1, 2) describe haploinsufficiency of the transcription factor Nkx2-1 (also known as TTF1) as the cause of a heretofore unrecognized human syndrome characterized by neurological, thyroid, and pulmonary dysfunction. Definition of the disorder and elucidation of its cause is a real tribute to the astute clinicians who looked beyond unusual laboratory values and carefully considered the multisystem manifestations in very young patients. Affected infants had elevated levels of thyroid-stimulating hormone, and indeed some were identified by neonatal screening protocols; but this was not typical hypothyroidism, since serum thyroxine (T4) levels were normal in several infants. CNS findings were prominent but pleiotropic, including microcephaly, basal ganglia malformations, choreoathetosis, ataxia, muscular hypotonia, and developmental delays. Pulmonary problems further complicated the presentation, and symptoms ranged from neonatal, sometimes lethal respiratory distress syndromes to severe recurrent infections. While management of each individual phenotype undoubtedly consumed considerable medical attention, recognition of the composite clinical triad, CNS, thyroid, and pulmonary disease, provided the critical foothold that ultimately solved this medical mystery.
Comparable multisystem deficits had been described in homozygous Nkx2-1–null mice (3), which prompted genetic analyses of the human homologue NKX2.1 (also referred to as TTF1, T1TF1, and T/EBP) in these patients. Definition of mutations in NKX2.1 elucidated the molecular basis for this novel clinical triad and further indicated transcription factor haploinsufficiency as the basis for yet another human syndrome.
The diversity of transcription factor haploinsufficiency disorders
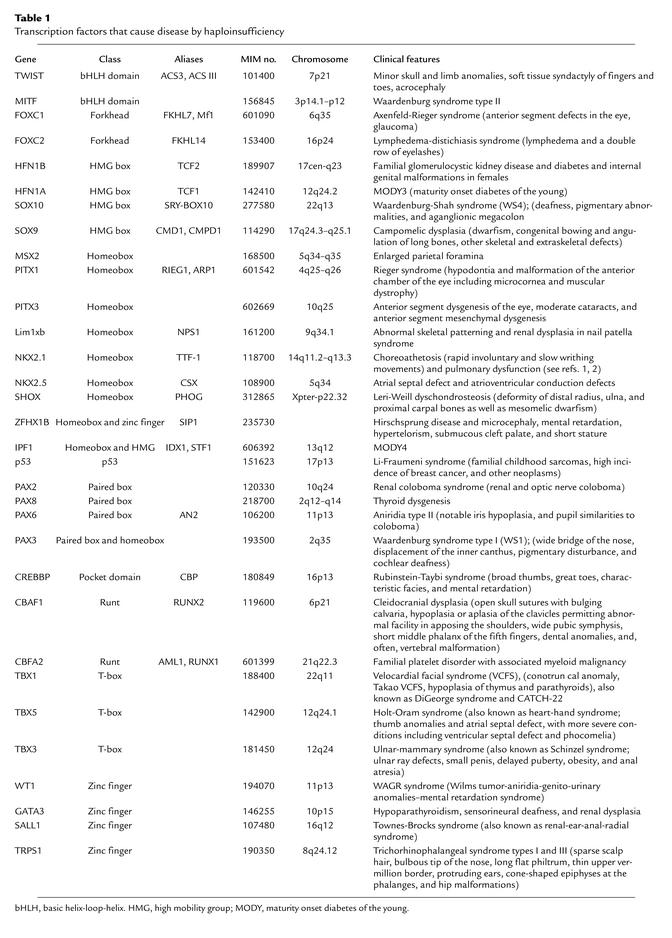
Molecular genetic studies of multiple complicated clinical phenotypes have demonstrated that a surprising number of these maladies are due to mutations in genes encoding transcription factors (see Table 1 and ref. 4). The usual manifestations of Holt-Oram syndrome (hand malformations of the embryonic radial rays and heart defects including atrial septal defect, ventricular septal defect, and complex malformations; MIM no. 142900), lymphedema-distichiasis syndrome (double-rowed eyelashes; MIM no. 153400), and Waardenburg syndrome (lateral displacement of the inner canthus of each eye, pigmentary disturbance including frontal white blaze of hair, heterochromia iridis, white eyelashes, leukoderma, and cochlear deafness; MIM no. 193500) are now defined as the consequences of dominant mutations in TBX5, FOXC2, and PAX3, respectively. The fact that these genes regulate the gene expression in organs with disparate functions and distinct embryologic origins helps account for the remarkable constellation of phenotypes produced by disease mutations.
Table 1.
Transcription factors that cause disease by haploinsufficiency
The mechanism by which such transcription factor defects cause disease is often haploinsufficiency. Whether by deletion, insertion, or a 5′ point mutation that prematurely terminates translation, human mutations often inactivate one allele, producing haploinsufficiency, or a reduction by half in the protein levels of the encoded transcription factor. As is evident from a review of the plethora of clinical manifestations associated with genetic syndromes, half-normal levels of many transcription factors are simply not enough.
Geneticists have recognized for many years that interpreting the consequences of a gene mutation can be important for understanding the mechanisms for disease. Dominantly inherited mutations that produce clinical disorders are generally understood to affect protein levels (haploinsufficiency occurs more commonly than overexpression) or to alter protein function, producing a gain or loss of function. Differentiating between haploinsufficiency and poison peptides can be difficult when only human subjects are available for study. Mutation type can provide a clue, but often not more than that. Even identification of a deletion as the cause of disease may not be sufficient to indicate haploinsufficiency as the mechanism. If a deletion removes only the carboxyl end of a protein, the mutant truncated peptide may function as a poison polypeptide. Similarly, some nonsense mutations can cause disease by creating an inactive peptide, while others cause disease by creating a mutant mRNA that is rapidly degraded by nonsense-mediated decay (5). Only the finding of a disease-causing mutation that prevents transcription, abolishes mRNA expression posttranscriptionally, or encodes extremely small (and likely unstable) peptide fragments is sufficient evidence to warrant the conclusion of haploinsufficiency as the mechanism for disease.
The most compelling evidence often comes from genetically engineered animals in which one allele is fully inactivated. The article by Pohlenz et al. in this issue of the JCI (2) describes the phenotype of a heterozygous mutant mouse bearing a null Nkx2-1 allele; the mice exhibit components of the human phenotype, thereby demonstrating that reduced levels of Nkx2-1 are sufficient to cause disease. Comparable data for other human syndromes (Table 1) also confirm the importance of transcription factor haploinsufficiency as a mechanism for disease.
Because transcription factors regulate the expression of one or more downstream target genes, the finding that a mutation that reduces by 50% the level of these factors has any clinical effect is surprising for several reasons. First, studies of prokaryotic genes have suggested that gene transcription can be an all-or-none response. Second, neither cell-based assays nor in vitro analyses of mammalian transcription factors usually indicate dramatic effects in response to twofold changes in levels of a transcription factor. Third, of the myriad genetically engineered mice produced with haploinsufficiency of a given transcription factor (i.e., heterozygous knockout mice), most heterozygous mice are reported to have no demonstrable phenotype. As recognized by Pohlenz et al. (2) and others (e.g., see ref. 6), the apparent absence of a phenotype might reflect nothing more than incomplete study, but it is also possible that reduced levels of certain factors (gata4, Irx4, fos, and myc among them) are truly inconsequential, at least in some genetic backgrounds.
A comparison of the more than 30 different human syndromes caused by transcription factor haploinsufficiency (Table 1) raises many interesting questions. Why are half-normal levels of many transcription factors insufficient? Do altered levels of any transcription factor always cause disease, or does haploinsufficiency of only certain gene families cause disease? What biology underpins the seemingly unrelated tissues that exhibit phenotypes in transcription factor haploinsufficiency? What accounts for clinical variation amongst patients with mutations that produce haploinsufficiency? Clear answers to these questions are lacking, but the ideas below may offer models with which to address them.
Effects of half-normal expression of certain transcription factors
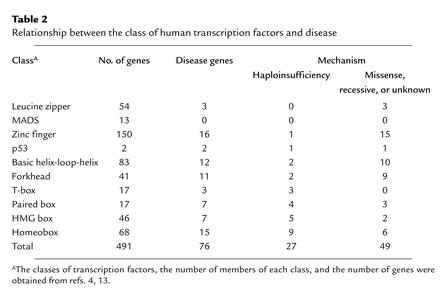
Transcription factors play a critical role in enhancing or inhibiting a target gene expression by binding to the promoter sequences, modulating the binding of RNA polymerase, and thus regulating gene transcription. More than 500 human transcription factors are encoded in the human genome (4), and these can be subdivided into classes or families based on the structure of their DNA-binding domains (ref. 7; Table 2). Several features of the different classes of transcription factors may indicate why haploinsufficiency of some, but not others, causes disease.
Table 2.
Relationship between the class of human transcription factors and disease
Evolution has introduced redundancy into many gene families that encode transcription factors (for a review see ref. 8, and for examples see refs. 9, 10). Duplication of many homeobox genes, for example, has resulted in gene clusters that are scattered throughout the genome. Experiments in mice indicate that inactivation of only one gene in a homeobox gene cluster often has no effect, while deletion of an entire cluster produces demonstrable phenotypes. Overlapping regulation of downstream target genes by multiple transcription factors provides a simple mechanism to account for the absence of phenotype, despite transcription factor haploinsufficiency.
Some transcription factors direct excess expression of target gene products. Reduced levels of proteins that are abundantly expressed or proteins with redundant functions may not have cellular consequences. For example, heterozygous mice bearing a defective α-tropomyosin allele have a 50% reduction in the levels of cardiac α-tropomyosin mRNA, but no apparent effect on myocyte function or heart physiology (11). In other cases, exquisite precision in gene expression may be required for viability. If early embryonic lethality occurs as a result of reduced expression of a particular protein, haploinsufficiency of transcription factors regulating expression of such genes would escape detection.
Transcription factors regulate gene expression by binding the target gene promoter and turn the target gene “on” or “off”; in a particular cell, target gene expression is an all-or-none phenomenon. Transcription factor haploinsufficiency alters the likelihood that a promoter is activated, and this likelihood is influenced by promoter sequence–transcription factor interactions. While some transcription factors may interact at only one site, better definitions of target sequences and genome databases indicate that many transcription factors probably interact at multiple sites on any one promoter. Initiation of gene transcription may require occupancy of several, if not all, binding sites. But since the number of binding sites within a cell is very small compared with the number of transcription factor molecules, the likelihood for binding site occupancy should be proportional to the concentration of transcription factor. Assuming n binding sites in a promoter and no interactions between sites, the likelihood that all sites will be occupied is directly proportional to the transcription factor concentration to the nth power. Thus, the more promoter binding sites for a transcription factor, the more sensitive gene expression will be to transcription factor concentration. In contrast, promoters that contain only a single transcription factor–target site should be relatively insensitive to transcription factor concentration. Therefore, given a mutation that causes haploinsufficiency, a twofold change in the concentration of a transcription factor might lead to either minimal or substantial effects on gene expression.
In a simple analysis, assuming no cooperativity in transcription factor binding, a promoter with three DNA-binding sites for a single transcription factor would be eight times less active in an individual with a mutation causing haploinsufficiency than in an individual with two functional transcription factor alleles. Presumably, transcriptional regulation that is more severely affected by transcription factor haploinsufficiency produces greater phenotypic expression. Clearly, the consequences of transcription factor haploinsufficiency are further complicated by cooperative interactions between transcriptional regulators, including homotypic interactions and heterotypic interactions with other DNA- or protein-binding factors. Since promoter sequences provide a platform for a large number of direct or indirect protein interactions, the consequences of reduced levels of even one transcription factor could be considerable. By extension of the model above, the more independent interactions each factor has with regulatory elements (proteins and sequences), the more sensitive gene expression may be to transcription factor concentration.
Transcription factor gene families and haploinsufficiency
Given the limited data about levels of individual transcription factors and of gene family members that are required for appropriate cell biology and organ physiology, continued analysis of the clinical consequences of transcription factor mutations in humans clearly has merit. A molecular cause has been defined in only a fraction of the 3644 inherited clinical syndromes defined by OMIM (4). If many of these syndromes arise from transcription factor gene mutations, considerable information may be gleaned from delineating disease-causing mutations and defining mechanisms.
Some generalities have already emerged from the molecular causes of human syndromes in Table 1. Surprisingly, haploinsufficiency of members of several gene families appears more likely to produce disease than do other mechanisms (Table 2). Deficiencies in three distinct T-box genes and nine homeobox genes have been defined as the cause of human disorders. Whereas only 1 of 15 mutations reported in zinc finger transcription factors and 2 of 12 defects in factors with basic helix-loop-helix motifs cause disease by haploinsufficiency, mutations in these transcription factors produce disease via other mechanisms.
Effects of transcription factor haploinsufficiency on disparate tissues
Mammalian genetics studies have demonstrated efficiencies in gene transcription; functionally unrelated genes are sometimes regulated by the same factor. While logic might suggest that genes expressed in the same spatial or temporal pattern could share transcriptional regulators, it is surprising that even these identifiers do not clearly demarcate transcription factor targets. Often these molecules regulate genes in different organs at distinct developmental stages. For example, Tbx5 cooperates with the homeobox protein Nkx2-5 to regulate cardiac expression of connexin40 and atrial natriuretic factor (6, 12). But since human TBX5 mutations cause hand and heart malformations, this factor must also regulate genes involved in forelimb development. Notably, cardiac development is virtually complete by the time limb development commences, and TBX5 partner proteins appear to be unique, since Nkx2-5 expression has not been found in the forelimbs.
Presumably, this efficient utilization of transcription factors reflects duplication of particularly useful protein-DNA and protein-protein interactions during evolution to define fundamental principles of organogenesis. For example, the establishment of axis (i.e., left-right, anterior-posterior) might be an underlying function for TBX5 expression in both the hand and the heart. While a definition of target genes may provide a molecular basis for the diversity of phenotypes resulting from transcription factor mutations, the converse is also true; careful analyses of clinical manifestations of these mutations should provide clues about the matrices necessary for engineering a tissue and integrating organ systems.
Clinical variation among patients with transcription factor haploinsufficiency
Although a variety of mutations can inactivate one gene allele, one might predict that identical phenotypes would be produced by each defect that causes transcription factor haploinsufficiency. The results are otherwise. Even family members with precisely the same genetic cause of haploinsufficiency do not have identical clinical features. For example, deletions of TBX1 on chromosome 22, which causes velocardial facial syndrome (CATCH-22 syndrome; MIM no. 188400), are recognized to produce considerable intrafamilial variation in the range and severity of dysmorphic features and cardiac malformations.
Two explanations to account for variable phenotype are differences in environment and differences in modifier genes. Since transcription factor mutations that cause congenital syndromes mediate their effects during in utero development, assessing the effects of environmental differences can be difficult. Identical twins provide a unique opportunity to address this issue in humans.
In contrast, there are multiple models that provide genetic explanations for variation in phenotypes of individuals with mutations causing transcription factor haploinsufficiency. Modifying background genes are likely to be particularly relevant to this class of mutations. First, polymorphisms within or flanking DNA target sequences may alter the transcriptional activity of factors derived from the normal allele. Second, the multiple interactions between transcription factors and other proteins provide great opportunity for polymorphisms within coregulators to influence transcriptional activity of target genes. Finally, the molecular mechanisms that regulate transcription of transcription factor genes may also affect protein levels.
Particularly clear evidence for these and other genetic modifiers of transcription factor mutations comes from the analyses of genetically engineered mutations in different inbred mouse strains. Deletion of Tbx5 in SvEv mice causes a myriad of cardiac malformations, including embryonic lethality, whereas the same mutation introduced onto a Black Swiss background produces viable animals with only atrial septal defects (6).
Clinical management of transcription factor haploinsufficiency disorders
Early and accurate diagnosis is an important benefit from the discovery that a human syndrome results from a transcription factor mutation. Because these syndromes are rare and often present complex and seemingly unexplainable multisystem manifestations, gene-based diagnosis can provide definitive information that helps both families and physicians. While recognition of a simple protein deficiency as the cause of human disease often prompts thoughts of replacement strategies, the jump from gene discovery to gene therapy will likely be long and slow. While it is appealing to consider tackling diseases with small, sometimes widely expressed molecules, the plethora of malformations resulting from a two-fold reduction in transcription factor levels suggests considerable stringency in the appropriate dose of these proteins. Surprisingly little is known about the consequences of either overexpression or temporal misexpression of most transcription factors. Continued discovery of critical parameters affecting transcription factor biology not only should provide important information regarding the molecular basis for human disease but may also provide clues about organ morphology and physiology that are as relevant to tissue engineering as they are to development.
Footnotes
See the related articles beginning on pages 469 and 475.
References
- 1.Krude H, et al. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J Clin Invest. 2002;109:475–480. DOI:10.1172/JCI200214341. doi: 10.1172/JCI14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pohlenz J, et al. Partial deficiency of Thyroid transcription factor 1 produces predominantly neurological defects in humans and mice. J Clin Invest. 2002;109:469–473. DOI:10.1172/JCI200214192. doi: 10.1172/JCI14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimura S, et al. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 4.McKusick, V.A. 2002. OMIM™: Online Mend-elian Inheritance in Man. http://www.ncbi.nlm.nih.gov/Omim/.
- 5.Byers PH. Killing the messenger: new insights into nonsense-mediated mRNA decay. J Clin Invest. 2002;109:3–6. DOI:10.1172/JCI200214841. doi: 10.1172/JCI14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruneau BG, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 7.Wingender E, et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 9.Kirby ML, Hunt P, Wallis K, Thorogood P. Abnormal patterning of the aortic arch arteries does not evoke cardiac malformations. Dev Dyn. 1997;208:34–47. doi: 10.1002/(SICI)1097-0177(199701)208:1<34::AID-AJA4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchard EM, et al. Targeted ablation of the murine alpha-tropomyosin gene. Circ Res. 1997;81:1005–1010. doi: 10.1161/01.res.81.6.1005. [DOI] [PubMed] [Google Scholar]
- 12.Hiroi Y, et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- 13.SWISS-PROT protein knowledgebase. Swiss Institute of Bioinformatics and European Bioinformatics Institute. http://us.expasy.org/sprot/.