Abstract
Efficient cellular DNA replication requires the activity of a 5′-3′ exonuclease. These enzymes are able to hydrolyze DNA⋅DNA and RNA⋅DNA substrates exonucleolytically, and they are structure-specific endonucleases. The 5′-3′ exonucleases are conserved in organisms as diverse as bacteriophage and mammals. Crystal structures of three representative enzymes identify two divalent-metal-binding sites typically separated by 8–10 Å. Site-directed mutagenesis was used to investigate the roles of three lysine residues (K83, K196, and K215) situated near two metal-binding sites in bacteriophage T5 5′-3′ exonuclease. Neither K196 nor K215 was essential for either the exo- or the endonuclease activity, but mutation of these residues increased the dissociation constant for the substrate from 5 nM to 200 nM (K196A) and 50 nM (K215A). Biochemical analysis demonstrated that K83 is absolutely required for exonucleolytic activity on single-stranded DNA but is not required for endonucleolytic cleavage of flap structures. Structural analysis of this mutant by x-ray crystallography showed no significant perturbations around the metal-binding sites in the active site. The wild-type protein has different pH optima for endonuclease and exonuclease activities. Taken together, these results suggest that different mechanisms for endo- and exonucleolytic hydrolysis are used by this multifunctional enzyme.
Efficient lagging-strand synthesis in all cells requires the participation of enzymes known as 5′-3′ exonucleases. These proteins remove ribonucleotide primers from the 5′ end of Okazaki fragments prior to ligation with the 3′ end of the upstream DNA fragment and are also involved in DNA repair processes. Many 5′-3′ exonucleases have been sequenced and extensively characterized including human DNase IV, the small fragments of Escherichia coli, Streptococcus pneumoniae, and Thermus aquaticus DNA polymerases as well as those encoded by bacteriophages T4 and T5 (reviewed in refs. 1 and 2). Several of these enzymes are multifunctional, possessing exonucleolytic activity on double-stranded DNA and RNA⋅DNA duplexes (RNase H activity). They also possess structure-specific endonuclease activity allowing them to cleave bifurcations or flap structures containing a free 5′ end (3–5). As these enzymes exhibit both endo- and exonuclease activities on substrates with free 5′ ends, it has been suggested (3) that they should be described as 5′ nucleases to accurately reflect their properties.
Lundquist and Olivera (6) proposed a model to explain nick-translation in which a nicked duplex undergoing strand-displacement synthesis could be processed by DNA polymerase I (Pol I). First, the 3′ end of the nick is extended by the polymerase activity of Pol I concomitant with displacement of a short region corresponding to the 5′-terminated strand at the nick site. The 5′-nuclease domain of Pol I then clips off the short single-stranded flap, effectively moving the nick in the 5′ to 3′ direction. Such a process was elegantly elaborated by Lyamichev and coworkers (3), who showed that prokaryotic 5′ nucleases are able to cleave in such an endonucleolytic manner close to the single-strand/duplex junction in flap substrates. Eukaryotic 5′ nucleases such as FEN1 have structure-specific activity similar to that displayed by eubacterial 5′-3′ exonucleases (3, 4, 7). This activity is thought to involve the enzyme loading or threading on to the free 5′ end of the flap substrate and tracking to the duplex junction, where it then cleaves the displaced strand (8). It has been suggested that the 5′-3′ exonucleolytic and 5′ endonuclease activities of the 5′ nucleases are manifestations of one and the same mechanism and simply reflect the enzymatic processing of different substrate conformations (1). However, the observation that T5 exonuclease is able to degrade single-stranded substrates such as oligo(dC) and oligo(dT) in an exonucleolytic manner contradicts this hypothesis (9).
Crystallographic studies on T5 exonuclease showed that the enzyme contained a hole, suggesting that single-stranded (but not duplex) DNA could pass through the protein and provided the basis for a molecular model (10) for the deduced threading of the substrate’s 5′ end through homologous enzymes (3, 8). The structural (11) and sequence similarities present in this group of enzymes have identified conserved amino acid residues that either form, or are in close proximity to, the binding sites for two divalent metal ions in T. aquaticus Pol I 5′-nuclease domain, T4 RNase H, and T5 exonuclease crystal structures (10, 12–14). As divalent metal ions are required for catalysis, but are not essential for substrate binding (5, 15) it is likely that the metal ions are located in the active site and that at least one actively participates in the chemical cleavage step. Indeed, a two metal ion mechanism analogous to that seen in the 3′-5′ exonuclease activity of the Klenow fragment (16) has been proposed for the 5′ nucleases (14).
Here we report the results of biochemical and mutagenesis studies designed to investigate our model (10) of DNA binding in the prototypic T5 5′ nuclease. This model predicts that certain lysine residues should be involved in DNA binding. We have determined the dissociation constants for wild-type and mutant enzymes and assessed the affect of mutations on both endo- and exonucleolytic function. The results support our model and also suggest that T5 exonuclease endo- and exonuclease activities are distinct and utilize different mechanisms to effect substrate cleavage.
MATERIALS AND METHODS
Determination of Structure-Specific DNA-Cleavage Profiles.
The pseudo-Y substrate was prepared by annealing the 5′-32P-end-labeled FLAP oligonucleotide d(GATGTCAAGCAGTCCTAACTTTGAGGCAGAGTCC), 3 nM, with the BRIDGE oligonucleotide d(GGACTCTGCCTCAAGACGGTAGTCAACGTG), 100 nM, essentially as described (5). In the pseudo-Y substrate residues 21–34 of FLAP are paired with bases 1–14 of BRIDGE, generating a structure which consists of this duplexed region and two single-stranded arms, the 5′ end of FLAP (nucleotides 1–20) and the 3′ end of BRIDGE (nucleotides 15–30). Reaction mixtures also contained 25 mM potassium glycinate (pH 9.3) and 100 mM KCl and were heated to 80°C for 5 min and then left at room temperature for 1 hr. Reaction mixtures were then diluted to contain 4.5 nM unlabeled oligonucleotide and 150 pM labeled oligonucleotide in 25 mM potassium glycinate, pH 9.3/100 mM KCl/10 mM MgCl2/5% (vol/vol) glycerol/1 mM DTT containing acetylated BSA at 0.1 mg/ml. A flap structure was prepared essentially as above by including the oligonucleotide d(CACGTTGACTACCGTC) in the annealing reaction at 4.5 nM. This is complementary to the 3′ end of BRIDGE.
To study the affect of pH on the reactions, different buffers were used: Mes at pH 5.5 and pH 6.0, Hepes at pH 7.0, Tris⋅HCl at pH 8.0, potassium glycinate at pH 9.3, and butylamine at pH 11.3, all at a final concentration of 25 mM, except the butylamine buffers, which were 50 mM. Reaction mixtures contained 7.5 fmol of labeled substrate (pseudo-Y) in a volume of 10 μl, and 2-μl aliquots of the mixture were withdrawn and the reaction was stopped by addition to 18 μl of formamide/EDTA stop mix. This was necessary due to the deleterious affect of the low-pH buffers on the running of the gel. An unbuffered solution was used to dilute enzyme to be added to the reactions buffered by butylamine due to its low buffering capacity. It was found that under these conditions the butylamine could buffer the effect on pH of the added metal ions. Control reaction mixtures, in which enzyme activity was analyzed at a lower pH and compared with the activity that was observed at pH 11.3, also contained enzyme diluted in an unbuffered solution. Reaction products were separated on a 7 M urea/15% polyacrylamide gel, length 50 cm, run in 1× TBE (Tris/borate/EDTA) at 50 W for 1.5 hr, and visualized on a Bio-Rad Molecular Imager phosphoimager. Quantification of the image was carried out by using Molecular Dynamics software.
Site-Directed Mutagenesis.
Oligonucleotide site-directed mutagenesis was carried out on a bacteriophage M13 derivative carrying the T5 D15 exonuclease gene (9) as described previously (5) in conjunction with the following oligonucleotides; K83A 5′-d(CACGATTACCTGCATACTCTGG), K196A 5′-d(TCCCATAATGGCCGCCAGGGAGATAAAC), K215A 5′-d(AATATTATATCCGCGTGCTGCTCCTATTCC), in which anticodon changes are underlined. Dideoxynucleotide sequencing was used to determine that only the desired sequence changes had been introduced. Standard methods for cloning of mutated genes were used (17). Mutated genes were subcloned into the expression vector pJONEX4 as described for the wild-type exonuclease gene (18).
Protein Expression, Purification, and Assays.
Mutant proteins were expressed and purified until free of any detectable contaminating 3′-5′ exonuclease or endonuclease activity as described previously (5, 9, 18). Proteins were assayed for both endonuclease and exonuclease activity. The amount of acid-soluble nucleotide released from high molecular weight DNA was determined with a standard spectrophotometric assay (19). Curves were plotted from the data obtained and estimates of the initial velocity were calculated.
Electrophoretic Mobility-Shift Assay (EMSA).
Pseudo-Y substrates were diluted to contain 4.5 nM BRIDGE oligonucleotide, 150 pM labeled FLAP oligonucleotide in 25 mM potassium glycinate, pH 9.3/100 mM KCl/1 mM EDTA/5% glycerol/1 mM DTT, containing acetylated BSA at 0.1 mg/ml and T5 exonuclease at the concentration stated (enzyme diluted in 25 mM potassium glycinate/50% glycerol/1 mM EDTA/1 mM DTT, containing acetylated BSA at 0.1 mg/ml). The reaction mixtures (10 μl) were incubated on ice for 10 min and analyzed on a nondenaturing 17% acrylamide gel, in 50 mM Tris-Bicine, pH 8.3/1 mM EDTA/1 mM DTT at 4°C for 2 hr at 15 V/cm. The gel was analyzed by using a Molecular Imager (Bio-Rad) and Molecular Dynamics software.
Crystallization and Structure Determination.
Crystals were obtained as described (20), by first setting up drops and equilibrating at 4°C, followed by seeding with native microcrystals at room temperature. Crystals of the K83A mutant approximately 1.0 mm × 1.0 mm × 0.15 mm appeared after about 2 weeks. After the crystals had been cryoprotected in 65% saturated ammonium sulfate/25 mM sodium citrate, pH 6.2/25% sucrose, they were flash frozen for data collection at 100 K on a Siemens area detector and the data were processed with xds (21). The space group of the K83A mutant was determined to be P212121. The molecules were located by molecular replacement with the native structure model (PDB ID code 1exn) by using amore (22) and xplor (Version 3.1, Yale University, New Haven, CT) to verify the solution. Two molecules were found in the asymmetric unit. The positions of the two molecules were rigid body refined to give an initial R factor of 46.6%. Portions of the main chain were restrained by noncrystallographic symmetry (as for the native structure), and several cycles of refinement followed by rebuilding with o were carried out (23). Data collection and refinement statistics for K83A (PDB ID code 1xo1) are shown in Table 1.
Table 1.
Crystallographic data for the K83A mutant protein
| Space group | P212121 |
| Cell dimensions, Å | a = 66.89, b = 76.81, c = 120.10 |
| Resolution range, Å | 25.0–2.5 |
| Reflections (unique) | 158,564 (21,915) |
| Completeness % | 99.6 |
| Rsym[(|Iobs − Iav|)/I], % | 8.2 |
| Refinement resolution range, Å | 6.0–2.5 |
| Protein atoms | 4,404 |
| Water molecules | 316 |
| Reflections (test set) | 19,728 (1,941) |
| R factor (Rfree), % | 22.6 (30.9) |
| Average B factor, Å2 | 25.4 |
| rms deviation in bond lengths, Å | 0.006 |
| rms deviation in bond angles, ° | 1.35 |
| rms deviation between molecules in asymmetric unit, Å | 0.284* |
Residues 20–34, 42–83, and 104–290.
RESULTS
Differential pH Sensitivity of Nuclease Reaction.
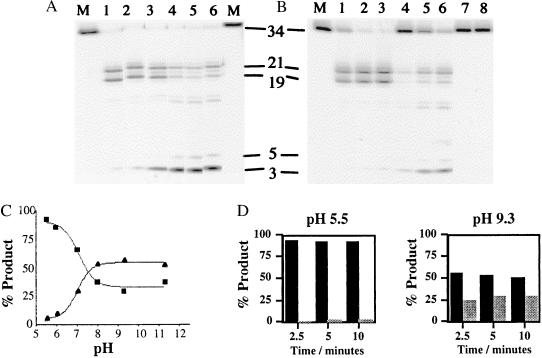
Analysis of the hydrolysis products of a pseudo-Y substrate revealed that endonucleolytic cleavage was enhanced at low pH, whereas exonuclease activity was markedly inhibited. At pH 9.3 the full-length FLAP oligonucleotide was rapidly converted to tri- and pentanucleotides with concomitant production of the endonucleolytically derived product (21- and 19-mers). Under the conditions used in the assay most of the initial pseudo-Y substrate was converted to smaller products after 10 min. However, at low pH the rate of disappearance of the 34-mer starting material was similar to that observed at higher pH, but the major products were the endonucleolytically derived 19- and 21-mers. Quantitative analysis of the phosphoimager data showed that exonucleolytically derived product accounted for 5% of the products at completion at pH 5.5, whereas they made up 80% of the products at pH 9.3. The relative contribution of exonuclease and endonuclease activity at differing pH is readily appreciated by inspection of Fig. 1.
Figure 1.
The product distribution of T5 exonuclease reaction varies with pH. (A) Activity was assayed on the pseudo-Y substrate, with 10 mM MgCl2 cofactor. Lanes marked M show the result of reactions in mixtures that contained no enzyme, and lanes 1–6 contained 4 nM wild-type enzyme in buffers at pH 5.5, 6.0, 7.0, 8.0, 9.3, and 11.3, respectively. The sizes (nucleotides) of the products are as indicated. (B) Time courses of wild-type 5′-3′ exonuclease at pH extremes. Wild-type exonuclease, at a concentration of 0.8 nM, was incubated with the labeled pseudo-Y substrate at 37°C for 2.5, 5, or 10 min at pH 5.5 (lanes 1–3) or pH 9.3 (lanes 4–6) in the presence of 10 mM MgCl2. The products of the reaction were separated on a 7 M urea/15% acrylamide gel. Untreated substrate, lane M, and substrates incubated at 37°C for 10 min at each pH in the absence of enzyme (lanes 7 and 8) are shown. (C) Phosphoimager data from A showing the percentage of product plotted against pH for both exonucleolytic (▴) and endonucleolytic (■) cleavage. (D) Graphical representation of the time course at pH 5.5 and 9.3 from data obtained from B. Endonucleolytic product is shown by dark bars, exonucleolytic product, by light bars. Differences between C and D in the absolute levels of exo- and endonuclease activity reflect the concentration of the enzyme used. In C all of the original substrate has been degraded.
Properties of Mutant Exonucleases.
Purified protein carrying the K215A mutation was found to retain a level of activity close to that of the wild type (specific activity of 45 units and 55 units, respectively; 1 unit is 1 nmol of released nucleotides per min per μg of protein in the standard spectrophotometric assay at 37°C). The K196A protein showed 1/10 the specific activity (4.5 units), and enzyme with the K83A mutation was found to be almost inactive in this exonuclease assay (0.3 unit, Fig. 2).
Figure 2.
Exonuclease activities of wild-type and mutant exonucleases. Exonucleolytic digestion of high molecular weight DNA was assayed spectrophotometrically. Results of three separate assays were plotted (except for K83A mutant; two assays), and fitted to a rectangular hyperbolic function. The r2 values (goodness of fit) ranged from 0.91 (K83A) to 0.99 (K215A). Different amounts of enzyme were added to each standard 600-μl assay: 1 μg of K196A, 0.8 μg of K215A, 0.5 μg of wild type, and 5 μg of K83A. Initial velocities for each reaction were determined. The specific activities calculated were as follows: wild type, 55 units; K215A, 45 units; K196A, 4.5 units; and K83A, 0.3 unit.
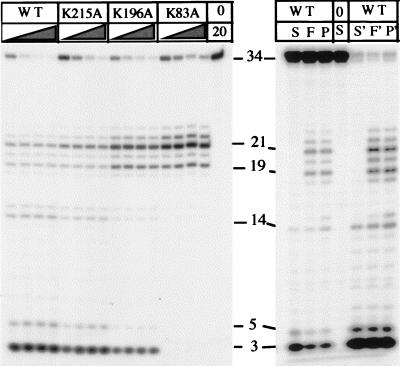
The enzymes were assayed for their ability to cleave a pseudo-Y structure substrate (Fig. 3). The wild type and K215A mutant produced similar patterns of cleavage product—i.e., exonucleolytic cleavage of the single-stranded portion of the substrate releasing 3-mers and 5-mers, and endonucleolytic cleavage of the hinge region of the substrate, releasing oligonucleotide 19 and 21 nucleotides long. The ratio of endo- to exonucleolytic cleavage by the K215A mutant was similar to that shown by the wild-type enzyme. The K196A mutant was less efficient than the wild-type enzyme. Ten times as much mutant as wild-type enzyme was required to effect a similar extent of substrate degradation. Exo- and endonucleolytic cleavage occurred, but the extent of 5′-3′ exonucleolytic cleavage was decreased compared with endonuclease cleavage. The K83A mutant showed no evidence of exonucleolytic cleavage. Only structure-specific endonucleolytic cleavage, resulting in the production of 21- and 19-mers, was observed. Wild-type enzyme released 3-mers and 5-mers exonucleolytically from single-stranded substrate (FLAP oligonucleotide alone) in accordance with previously published results (9). The products of flap structure cleavage (mimicking a strand-displaced substrate) were similar to those obtained with pseudo-Y substrate.
Figure 3.
Degradation of pseudo-Y substrate by T5 5′-3′ exonuclease lysine mutants. (Left) Enzymes were incubated with the substrate at 37°C for 2.5, 5, 10, or 20 min in the presence of 10 mM MgCl2, and reaction products were separated by denaturing PAGE. The concentration of the K83A and K196A mutants (40 nM) was 10 times higher in the reaction than that of the K215A mutant and the wild-type enzyme (4 nM). A control reaction lacking enzyme was incubated for 20 min (lane 20). (Right) Wild-type enzyme (20 nM) was incubated with pseudo-Y substrate (P), single-stranded oligonucleotide (S), or a full flap structure (F). Reactions were sampled at 2 and 30 min (first and last three lanes, respectively).
Analysis of DNA-Binding Properties.
Binding of nucleases to pseudo-Y substrates was assayed by electrophoretic mobility shift in the absence of divalent metal cofactor (Fig. 4). An approximation to the dissociation constant (Kd) for the binding reaction was calculated as the protein concentration at which half the substrate remained unbound (24). Wild-type enzyme had a Kd of approximately 5 nM in this assay. The dissociation constants of all three lysine mutants were increased compared with wild-type enzyme, with K215A having the least affect (Kd = 50 nM). The K196A and K83A substitutions greatly weakened substrate binding (Kd ≈ 200 nM).
Figure 4.
Substrate binding by the T5 5′-3′ exonuclease lysine mutants. (A) Pseudo-Y substrate was incubated with the enzymes on ice, and the enzyme–substrate complex was separated from the unbound substrate by electrophoresis on a nondenaturing 17% acrylamide gel. The substrate concentration was constant in each experiment, and the enzyme concentration was varied as shown. (B) Data from the gel retardation experiments were plotted as percentage of free substrate against the enzyme concentration on a logarithmic scale. At least 30 values were plotted for each enzyme. The means of the values were plotted (though all points were used in the data analysis). Error bars represent maximum and minimum data values for each protein concentration. The enzyme concentration required to bind half the substrate was determined graphically. Dissociation constants were 5, 48, 194, and 220 nM for the wild type, K215A, K196A, and K83A, respectively.
Mutant Structure.
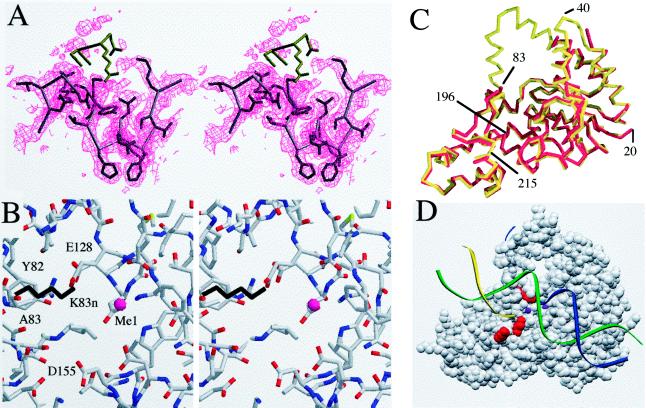
The K83A protein crystallized under conditions similar to those required for the native protein (20). The mutant crystallized in a different space group from the native protein, but the overall structural features of both proteins are very similar. The exceptions lie mainly in regions that are now found to be disordered in the mutant protein. Whereas all residues in the native molecule between 19 and 290 were visible in the electron density map, the K83A mutant has two disordered regions, residues 35–41 and 84–103 (Fig. 5). The electron density map of this region is shown in Fig. 5A and includes the top of the helical arch observed in the native structure. Other regions that differ slightly include the 200–223 finger and the region from 233 to 247. These regions were found to be slightly flexible in the native crystals (11).
Figure 5.
(A) Electron density map of the region around the mutated residue K83. The density for this residue is defined in a 2Fo − Fc map contoured at 1σ. The positions of the next few residues of the native molecule are colored in brown, and these residues are disordered. The next residue is G84; this position is expected to show flexibility. The helical arch does not show evidence of order in the electron density map. The first well defined residue in helix 5 is F104. The absence of density preceding this residue suggests that the helix is unwound and exists in an undefined conformation. (B) A stereo view of the metal-binding site MeI as determined in the native molecule. The K83 side chain from the native molecule is shown in black. The positions of the metal’s ligands are largely unchanged compared with the native (data not shown). The only amino group in close proximity to MeI is that of K83 (distance between NH2 and MeI = 4.2 Å). (C) A Cα trace of the native (red) and K83A mutant structures (yellow) superimposed. Disordered residues in the mutant include 35–41 and 84–103. The regions around the acidic residues composing the metal-binding site are very similar. (D) A space-filling model of the protein, showing the three residues mutated in this study in red (top to bottom: K83, K196, and K215) in relation to the proposed threading mechanism. DNA is shown as colored strands and the metal sites observed are shown as purple spheres.
DISCUSSION
Phage T5 5′-3′ exonuclease, the product of gene D15, is required for efficient viral DNA replication and is a member of a large group of homologous 5′-3′ exonucleases (25). These include the N-terminal domains of many eubacterial Pol I-like proteins, other phage-encoded 5′-3′ exonucleases, and flap endonuclease I (FEN-1) (4). The prokaryotic enzymes share primary sequence motifs that include acidic residues shown to be involved in cofactor binding by recent crystal structures of T4 RNase H (13), T5 exonuclease (10), and Taq DNA polymerase (14). Mutagenesis studies have also implicated some of these residues in cofactor binding (25–28). So far the mechanism of the reactions catalyzed by this group of enzymes remains to be fully established.
pH Profile.
Previous studies on the enzyme had identified a pH optimum of 9.3 (29) by using assays designed to detect acid-soluble nucleotides and small polynucleotides (up to 10-mers or so) released from high molecular weight duplex DNA substrates. However, it has since been shown that the 5′ nucleases also display structure-specific endonuclease activity on pseudo-Y and strand-displaced (flap) substrates (3, 4, 10). Processing of pseudo-Y substrates by T5 exonuclease was pH sensitive. The relative levels of endo- and exonucleolytic cleavage varied markedly. High pH favored the exonuclease reaction as previously reported, whereas the release of short oligonucleotides (<6-mers) was inhibited at lower pH. However, the overall rate of substrate loss was similar at low pH. Instead, the rate of endonucleolytic cleavage had increased, with most product present in the form of 19- and 21-mers. The time courses of the reactions at pH 5.5 and 9.3 were investigated. Analysis of the data confirmed that the overall rates of loss of starting material were similar at both pH extremes, but that the profile of cleavage product distribution was different. This finding suggests that the two activities might either proceed by chemically distinct hydrolytic mechanisms or perhaps have separate pH-sensitive binding sites. Conserved residues in the active site include K83 and Y82. The counterpart of the latter residue in T4 RNase H was mutated to phenylalanine without significant loss of catalysis. Thus, the role of the conserved tyrosine is obscure, but the phenolic hydroxyl is not essential for catalytic activity (25).
Mutational Analysis.
We reported the crystal structure of native T5 exonuclease (10) and proposed a physical model for the substrate threading mechanisms put forward for homologous systems (3, 30). In this model, three lysine residues are predicted to contact closely a bound DNA substrate. K83 lies in the helical arch region and is perfectly conserved throughout all the prokaryotic 5′-3′ exonuclease sequences so far determined (12). This residue is sited close to metal site I. K196 is less well conserved and lies between the two metal ion binding sites. K215 is more widely conserved than K196 but has no obvious counterpart in the T4 and T7 exonucleases (12). The K215A mutation had the least impact on either the pattern of hydrolysis or substrate affinity. Both were reduced from wild-type levels, but at high substrate concentrations K215A retained nearly 80% of wild-type exonuclease activity. This mutation caused a decrease in the ability of the protein to bind a pseudo-Y substrate: the Kd increased approximately 10-fold (from 5 nM to 50 nM). As the product spectrum was indistinguishable from that obtained with wild-type protein, it appears that this residue plays a substantial role in DNA binding but is not involved in catalysis.
Replacement of either residue K83 or K196 with alanine had more dramatic effects. Both the K196A and K83A mutations showed similar, though greatly reduced, substrate affinity (Kd ≈ 200 nM). The K196A protein hydrolyzed substrate both endo- and exonucleolytically, whereas the K83A protein had lost the ability to function as an exonuclease. That K83A retains endonuclease activity is further strong evidence that endo- and exonuclease activities are separate and distinct phenomena.
Structural Studies.
The K83A protein was crystallized and the structure was solved to determine whether a difference in the structure compared with that of the wild-type protein had caused the observed change in nucleolytic activities. The structure of K83A was found to be very similar to wild-type protein except that the helical arch visible in the latter was disordered in the mutant protein. It is interesting to note that this region is disordered in homologues of the T5 5′ nuclease—i.e., T4 RNase H (13) and the 5′-nuclease domain of Taq DNA polymerase (14). The helical arch was well defined in the native crystal form (P43) of T5 5′ nuclease partly due to the crystal contacts with neighboring molecules. In the mutant crystal form the back of helix 4 of the arch can contact a symmetry-related molecule, but the front of the arch is exposed to solvent. We believe this disorder is an indicator of flexibility inherent in this class of 5′ nucleases and may serve a functional role (11).
The only other differences found in the mutant crystal structure were residues 200–223 and 233–247. These are remote from the active site and also differed between the two molecules in the asymmetric unit of the native crystals. Thus with the overall structure remaining the same as in the native molecule and given that the metal-binding site remains intact, we conclude that loss of exonuclease function in the mutant protein is due to the loss of the K83 ɛ-amino group.
Possible Roles of K83.
The reduced binding affinity of K83A and the minor structural perturbations observed in the crystal structure suggest that this lysine residue is more important for substrate binding than is K215. The increase in Kd for the K83A mutant is similar to that observed for the K196A enzyme, yet this protein did display detectable endo- and exonuclease activities. It could be argued that the two nuclease activities do not share a common substrate binding site and that K83 fulfills an essential role only for substrate binding in the exonucleolytic reaction. However, given the overall positive surface potential in and around the active site (10), one would expect to be able to detect some traces of K83A exonuclease activity, as was observed with K196A. Even high concentrations (30-fold higher than that used in Fig. 3) of K83A failed to show exonucleolytic cleavage (data not shown). Thus, we suggest that K83 may play a role as a general base in exonucleolytic cleavage.
The pH profile of the exonucleolytic reaction catalyzed by the wild-type T5 enzyme suggests a decrease in activity below pH 7.5. This behavior implies that a residue with a pKa of close to 7 is important for this reaction. Histidine is the obvious candidate residue in terms of a suitable pKa; however, no histidine residues are conserved in the 5′-3′ exonuclease family. Furthermore, inspection of all three published 5′ nuclease crystal structures fails to identify a suitable candidate histidine.
Could loss of exonuclease activity be explained by protonation of a residue leading to reduction in substrate binding? The possibility that the terminal phosphate group of the 5′-phosphorylated substrates used in this study (pKa of approximately 6.5) (31) are important for the reaction was considered. At low pH the di-anionic form would be protonated, reducing the charge and thus the potential to interact with a positively charged residue such as K83 or an arginine. Though this model explains the fall-off in exonuclease activity at acidic pH, it predicts that unphosphorylated oligonucleotides would be less strongly bound than the phosphorylated counterpart. This was not the case for T5 exonuclease (data not shown) or for the homologous T7 gene 6 5′-3′ exonuclease, which also shows no discrimination between 5′ phosphate or hydroxyl groups (32). The acid-sensitive residue could be K83, given that both low pH and replacement of K83 with alanine inhibit wild-type exonucleolytic activity. With exonucleolytic activity falling below pH 8, it could be that this lysine acts as a general base in the deprotonated state. Though such a low pKa is unusual for a lysine residue, it is not unknown; an essential lysine residue in yeast glutathione reductase has a pKa of 7.3 (33). The distance between the ɛ-N of K83 and metal site I is 4.2 Å, and the distance between the ɛ-N of K83 and metal site II is 8.2 Å. Thus, we suggest the K83 amino group’s role is to act as a proton acceptor in the mechanism as follows: in the absence of substrate, the ɛ-amino group of K83 is in close proximity to the positive charge carried by metal site I and is also influenced by the positively charged guanidinium group of R86 (also a conserved residue). This micro-environment surrounding K83 would tend to destabilize the protonated form, thus lowering its pKa. Upon DNA binding, the phosphodiester backbone would shield K83 from the influence of the metal sites. K83 could then abstract a proton from water (or assist the bound metal ion in so doing), generating a hydroxide ion to effect cleavage. Upon or during release of substrate, the protonated K83 would become reexposed to the destabilizing micro-environment, allowing loss of a proton (transferred to the leaving 3′-alkoxide ion), thus regenerating the catalyst. Metal site I, in close proximity to K83, has been implicated as having a catalytic function in T4 RNase H, whereas similar alterations to metal site II failed to abolish activity in the latter enzyme (25).
An alternative mechanism has been proposed (14), provoked by the observation of two divalent-metal-binding sites situated in the active site of the Taq 5′ nuclease. One metal ion could promote formation of a hydroxide ion while the second stabilizes the pentacoordinate transition state, as has been shown for the 3′-5′ exonuclease activity of Pol I (33). However, this mechanism does not explain the apparent requirement of K83 and is further undermined by the results of the T4 RNase H and FEN-1 mutagenesis studies, which imply that only one metal site is required for catalysis (25, 26) and one is involved in substrate binding. Furthermore, the metal sites in T5 exonuclease are too far apart to participate in the two-metal mechanism as postulated (8.1 Å in T5 exonuclease versus 3.9 Å in the 3′-5′ exonuclease of Pol I). Although the exact mechanism(s) of T5 exonuclease-catalyzed hydrolysis remains to be determined, this study provides strong evidence that endo- and exonucleolytic hydrolyses proceed by distinctly different mechanisms. Given the extensive similarities between known prokaryotic 5′ nucleases (11, 12), it is likely that they employ mechanisms similar to those used by T5 exonuclease.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council for a studentship to support S.J.G. J.R.S. is supported by The Wellcome Trust (052123).
ABBREVIATION
- Pol I
DNA polymerase I
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1xo1).
References
- 1.Lieber M R. BioEssays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 2.Ceska T A, Sayers J R. Trends Biochem Sci. 1998;23:331–336. doi: 10.1016/s0968-0004(98)01259-6. [DOI] [PubMed] [Google Scholar]
- 3.Lyamichev V, Brow M A D, Dahlberg J E. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 4.Harrington J J, Lieber M R. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garforth S J, Sayers J R. Nucleic Acids Res. 1997;25:3801–3807. doi: 10.1093/nar/25.19.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lundquist R C, Olivera B M. Cell. 1982;31:53–60. doi: 10.1016/0092-8674(82)90404-4. [DOI] [PubMed] [Google Scholar]
- 7.Murante R S, Huang L, Turchi J J, Bambara R A. J Biol Chem. 1994;269:1191–1196. [PubMed] [Google Scholar]
- 8.Murante R S, Rust L, Bambara R A. J Biol Chem. 1995;270:30377–30383. doi: 10.1074/jbc.270.51.30377. [DOI] [PubMed] [Google Scholar]
- 9.Sayers J R, Eckstein F. J Biol Chem. 1990;265:18311–18317. [PubMed] [Google Scholar]
- 10.Ceska T A, Sayers J R, Stier G, Suck D. Nature (London) 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 11.Artymiuk P J, Ceska T A, Suck D, Sayers J R. Nucleic Acids Res. 1997;25:4224–4229. doi: 10.1093/nar/25.21.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutman P D, Minton K W. Nucleic Acids Res. 1993;21:4406–4407. doi: 10.1093/nar/21.18.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueser T C, Nossal N G, Hyde C C. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Eom S H, Wang J M, Lee D S, Suh S W, Steitz T A. Nature (London) 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 15.Harrington J J, Lieber M R. J Biol Chem. 1995;270:4503–4508. doi: 10.1074/jbc.270.9.4503. [DOI] [PubMed] [Google Scholar]
- 16.Beese L S, Steitz T A. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Sayers J R, Eckstein F. Nucleic Acids Res. 1991;19:4127–4132. doi: 10.1093/nar/19.15.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser M J. Methods Enzymol. 1980;65:255–263. doi: 10.1016/s0076-6879(80)65035-6. [DOI] [PubMed] [Google Scholar]
- 20.Ceska T A, Sayers J R, Eckstein F, Suck D. J Mol Biol. 1993;233:179–182. doi: 10.1006/jmbi.1993.1496. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 22.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 23.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Carey J. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 25.Bhagwat M, Meara D, Nossal N G. J Biol Chem. 1997;272:28531–28538. doi: 10.1074/jbc.272.45.28531. [DOI] [PubMed] [Google Scholar]
- 26.Shen B, Nolan J P, Sklar L A, Park M S. Nucleic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizrahi V, Huberts P. Nucleic Acids Res. 1996;24:4845–4852. doi: 10.1093/nar/24.24.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Derbyshire V, Ng K, Sun X C, Grindley N D F, Joyce C M. J Mol Biol. 1997;268:284–302. doi: 10.1006/jmbi.1997.0967. [DOI] [PubMed] [Google Scholar]
- 29.Paul A V, Lehman I R. J Biol Chem. 1966;241:3441–3451. [PubMed] [Google Scholar]
- 30.Murante R S, Rumbaugh J A, Barnes C J, Norton J R, Bambara R A. J Biol Chem. 1996;271:25888–25897. doi: 10.1074/jbc.271.42.25888. [DOI] [PubMed] [Google Scholar]
- 31.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 32.Kerr C, Sadowski P D. J Biol Chem. 1972;247:311–318. [PubMed] [Google Scholar]
- 33.Pandey A, Iyengar L, Katiyar S S. J Enzyme Inhib. 1997;12:143–154. doi: 10.3109/14756369709035815. [DOI] [PubMed] [Google Scholar]