Abstract
Purpose
To facilitate the molecular characterization of naturally occurring cataracts in dogs by providing the radiation hybrid location of 21 cataract-associated genes along with their closely associated polymorphic markers. These can be used for segregation testing of the candidate genes in canine cataract pedigrees.
Methods
Twenty-one genes with known mutations causing hereditary cataracts in man and/or mouse were selected and mapped to canine chromosomes using a canine:hamster radiation hybrid RH5000 panel. Each cataract gene ortholog was mapped in relation to over 3,000 markers including microsatellites, ESTs, genes, and BAC clones. The resulting independently determined RH-map locations were compared with the corresponding gene locations from the draft sequence of the canine genome.
Results
Twenty-one cataract orthologs were mapped to canine chromosomes. The genetic locations and nearest polymorphic markers were determined for 20 of these orthologs. In addition, the resulting cataract gene locations, as determined experimentally by this study, were compared with those determined by the canine genome project. All genes mapped within or near chromosomal locations with previously established homology to the corresponding human gene locations based on canine:human chromosomal synteny.
Conclusions
The location of selected cataract gene orthologs in the dog, along with their nearest polymorphic markers, serves as a resource for association and linkage testing in canine pedigrees segregating inherited cataracts. The recent development of canine genomic resources make canine models a practical and valuable resource for the study of human hereditary cataracts. Canine models can serve as large animal models intermediate between mouse and man for both gene discovery and the development of novel cataract therapies.
As the leading cause of worldwide blindness and low vision, cataracts affect millions of individuals, and billions of dollars are spent annually on cataract medical and surgical expenses [1]. The etiology of cataracts varies; they can be hereditary, traumatic, or secondary to ocular inflammatory or systemic diseases, or as a sequelae to taking certain pharmaceuticals/medications. The onset of cataract also varies. Congenital cataracts occur in over two out of 10,000 live births [2] and are the leading cause of treatable visual impairment in children worldwide [3,4]. Approximately 1/3 of all congenital cataracts are inherited [5]. Age-related cataracts, however, are the most common type of cataract. Approximately 26.6 million Americans older than 40 years have cataract or have had cataract surgery, and this number is projected to increase to 39.6 million by the year 2020 [6]. The risk factors associated with the development of age-related cataract include ultraviolet light exposure [7], cigarette smoking [8], alcohol consumption [9], and nutrition [10]. In addition, the role of genetics in age-related cataract has been reviewed and suggests an interaction between genetic background and environmental risk factors [11].
Most of our knowledge of cataractogenesis comes from genetic studies of hereditary cataracts in man and mice. Mutations causing nonsyndromic inherited cataract have been identified in at least 15 genes in humans [12], and at least 18 genes in the mouse (Mouse Genome Informatics). Here, we propose the development of the dog as a complementary large animal model for molecular studies of cataracts to facilitate gene discovery and the development of novel and alternative cataract therapies. There are over 300 breeds of dog, and of these, 20 breeds are known to have hereditary cataracts while another 125 breeds are suspected of having hereditary cataract [13]. It is likely that some hereditary cataracts in dogs will be caused by mutations in genes already identified in humans or mice. However, to date no mutations causing cataract have been identified in any canine pedigrees. Because of the recent progress in canine genomics [14–19], and the success in identifying canine models for retinal diseases [20–22], it is likely that similar progress will be possible with molecular studies of canine cataracts.
To begin the molecular identification of cataract phenotypes in the dog we have selected 21 genes known to cause cataracts in man and/or mouse. These include structural proteins (6), crystallin genes (6), and transcription factors (9). We have determined the canine chromosomal locations of each gene using a radiation hybrid (RH) 5000 rad panel [23] and compared our experimentally derived chromosomal locations for each gene with that of the draft sequence of the canine genome. RH-mapping and genome sequencing are complementary procedures which identify gene locations, order, and distance, along with regions of conserved synteny across species. Combining data from RH-maps with that from genomic sequences maximizes the information obtained from both methodologies [24] and helps to resolve discrepancies that might arise with the draft sequence information currently available. The identification of cataract gene locations along with their nearest polymorphic markers provides an integrated resource essential for segregation analysis of cataract phenotypes in canine cataract pedigrees. This resource will aid in the development of the dog as a complementary large animal model for the molecular study of cataracts and will help to advance our knowledge of naturally occurring cataracts in mammalian species. This may one day contribute to the development of new therapies to prevent, delay, or even reverse cataracts.
METHODS
Selection of cataract gene orthologs
Twenty-one gene orthologs were selected as cataract candidate genes in dogs based on their ability to cause cataracts in man and/or mouse. These candidate genes include structural proteins: beaded filament structural protein 2 (BFSP2), connexin 46 (CX46), connexin 50 (CX50), lens intrinsic membrane protein 2 (LIM2), major intrinsic protein of lens fiber (MIP), and secreted protein acidic cysteine-rich (SPARC); crystallins: βB2-crystallin (CRYBB2), γA-crystallin (CRYGA), γB-crystallin (CRYGB), γC-crystallin (CRYGC), γD-crystallin (CRYGD), and γS-crystallin (CRYGS); transcription factors: CEH10 homeodomain-containing homolog (CHX10), homolog of Drosophila eyes absent 1 (EYA1), forkhead Box E3 (FOXE3), heat shock transcription factor 4 (HSF4), avian musculoaponeurotic fibrosa-rcoma oncogene homolog (MAF), paired-box 6 transcription factor (PAX6), paired-like homeodomain transcription factor 3 (PITX3), homolog of Drosophila sine oculis homeobox (SIX5), and sex-determining region-Y (SRY)-box 1 (SOX1).
Canine orthologous sequences
Human gene cDNAs obtained from the National Center for Biotechnology Information (NCBI) database were analyzed against drafts of the canine genome to obtain orthologous sequences. The corresponding accession numbers for the human cDNA sequences are listed in Table 1. Human gene cDNAs for BFSP2, CX46, CX50, MIP, PAX6, and PITX3 were analyzed against the original 1.5X (poodle) canine genome sequence through collaboration with The Institute for Genomic Research (TIGR). Human gene cDNAs for CHX10, CRYBB2, CRYGA, CRYGB, EYA1, FOXE3, HSF4, MAF, SIX5, SOX1, and SPARC were analyzed against the publicly available 1.5X (poodle) canine genome using the BLAST function, and cDNAs for CRYGC, CRYGD, CRYGS, and LIM2 were analyzed against the 7.5X (boxer) canine whole genome shotgun sequence using the BLAT function. Primers were designed from putative intronic or untranslated regions of canine orthologs of each cataract candidate gene using Amplify software (Bill Engels, University of Wisconsin, Madison, WI). Primers were designed to amplify canine-specific sequences suitable for RH mapping and are summarized in Table 1. The average length of primers was 25 nucleotides with an average melting temperature of 56.2 °C, and the average PCR product size was 363 bp. All PCRs were performed on an MJ Research Tetrad 2 thermocycler (Global Medical Instrumentation Inc., Ramsey, MN). To ensure canine specificity, initial PCRs were carried out at 25 μl volume on control canine, hamster, and canine:hamster (1:5 ratio) DNA. PCR conditions were as follows: 2 min 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 56 °C, 30 s at 72 °C, and a final extension of 5 min at 72 °C. The PCR product sizes ranged from 144–463 bp. PCRs were optimized by adjusting the annealing temperature where needed.
Table 1.
PCR primer sequences used to place cataract orthologs on canine chromosomes using an RH5000 panel
| Gene | GenBank accession number | Primer sequences | Tm (°C) | Product size (bp) |
|---|---|---|---|---|
| Structural protein genes | ||||
| BFSP2 | NM_003571 | F: CCTCTTCATAGTTCCTTGAC | 50.7 | 419 |
| R: CCCTACAGTTCTATGCTTCCAG | 55.8 | |||
| CX46 | NM_021954 | F: CCAGAGAATGCTAATTTGTCCC | 54.9 | 365 |
| R: CAGCACATGCACTTACACAGG | 57.6 | |||
| CX50 | NM_005267 | F: GGGATAACCAGAGGGTAGCAC | 57.8 | 386 |
| R: GGATCTCGAAGTTCTCCTACC | 54.9 | |||
| LIM2 | NM_030657 | F: GGCAACAAGTGCTACCTGC | 56.9 | 353 |
| R: CCCAGTTTCCAACCTAGTTC | 53.0 | |||
| MIP | NM_012064 | F: GGGCACGAGTACGAGCACTG | 61.0 | 308 |
| R: GCCTGCACGCTTCGCAAGC | 63.5 | |||
| SPARC | NM_003118 | F: CTGATCATGTGGCCTTGAGC | 56.2 | 343 |
| R: GGCCTAATCTGATCTGCTAAG | 52.5 | |||
| Crystallin genes | ||||
| CRYBB2 | NM_000496 | F: GGAGAGCAGTTTGTGTTCGAG | 55.9 | 356 |
| R: GTGCAATGTGGCAACCCTTAG | 56.9 | |||
| CRYGA | NM_014617 | F: CCTACGAAGGAATCCCTTTTG | 53.2 | 349 |
| R: GAGTGGAGCTCATTGAGCCG | 58.1 | |||
| CRYGB | NM_005210 | F: GCTCCTGGCATCCAGTGGG | 61.0 | 404 |
| R: CTGCTCCTGCTGCTCTACTC | 57.3 | |||
| CRYGC | NM_020989 | F: CTCTAACCCAGACTCTGAACC | 54.3 | 373 |
| R: GGACCCAGCAGCCCTCCAG | 63.3 | |||
| CRYGD | NM_006891 | F: GACAGCGGCTGCTGGATGC | 62.1 | 381 |
| R: CCAGGACAGGACCTATTGCTG | 57.4 | |||
| CRYGS | NM_017541 | F: CAGCTCATCCCAAGACTGAATG | 55.7 | 390 |
| R: GACACCATCCAGCACCTTACTG | 57.7 | |||
| Transcription factors | ||||
| CHX10 | NM_182894 | F: CGTAAGTTAAGTACCAGAGGG | 52.1 | 390 |
| R: CTAAACCTGTGATTTCTGTGCC | 54.0 | |||
| EYA1 | NM_172060 | F: CTACTGCCCAAACTCATCAC | 53.2 | 386 |
| R: CCCACAGAAAGAATGTAGATG | 50.7 | |||
| FOXE3 | NM_012186 | F: GCAAGTCACTGCAGGGACTG | 58.6 | 342 |
| R: GTTCTTTAGTGGTGCTGGAGG | 55.5 | |||
| HSF4 | NM_001538 | F: GATTCCTGAGCTCTACCATAG | 51.7 | 463 |
| R: GCCAGGGTCTGGTTGAAGC | 58.9 | |||
| MAF | NM_005360 | F: GTTAACAGCACGGAGGTTACAC | 55.9 | 340 |
| R: CCCTTCCACTGCATCTGAGTC | 57.6 | |||
| PAX6 | NM_001604 | F: GCCACATCTTCAGTACAAAG | 49.6 | 300 |
| R: TAGTTCAGGCATTGACTGATG | 50.3 | |||
| PITX3 | NM_005029 | F: CCTCCCTGAACAGGGTGATAG | 57.5 | 304 |
| R: CCGCACCATTGCACACCACG | 63.0 | |||
| SIX5 | NM_175875 | F: GACCCGCAGCTTCTCAAGC | 59.0 | 371 |
| R: CCTTCCTACTGCAAAGTGAGC | 55.8 | |||
| SOX1 | NM_005986 | F: CTCAAGAATGATATCCACTGCTTC | 53.5 | 299 |
| R: GTACATTTCAGAGTCAATGTGGC | 54.1 | |||
The column labeled “Tm” indicates primer melting temperature. PCR product size and corresponding NCBI reference sequences are shown.
RH5000 panel
Canine cataract candidate genes were positioned on a canine radiation hybrid map using an RH5000 panel consisting of 118 cell lines made by fusing 5,000 rad-irradiated canine fibroblast cells with TK-HTK3 hamster cells [23]. Information on the RH5000 panel may be obtained from the Canine Genetics Research Projects web site. The RH5000 panel has a retention frequency of 22% and a resolution limit of 600 kb [17]. The PCR reactions on the RH5000 panel were performed with 15 μl volumes, using the same PCR conditions already described, and run in duplicate or triplicate. PCR products were analyzed on 2% agarose gels containing 0.05% ethidium bromide at 50–65 volts. Gel bands were visualized using a UV transilluminator, and images were taken of each gel. Gels were then scored for presence, absence, or ambiguity of the band of interest in each of the 118 cell lines.
Radiation hybrid map construction
Radiation hybrid data collected from the 21 cataract gene orthologs was merged with previous data from 3,270 markers [17], and analyzed using Multimap® software [25]. Two-point linkage analysis was performed for each canine ortholog to determine its nearest markers and chromosomal location. A subset of the linked markers was selected and used as a framework map based on their map locations as previously provided [17]. The Multimap® multipoint mapping algorithm was then used to place the cataract gene orthologs on canine chromosomes in relation to the markers within the framework map.
The May 2005 7.5X canine draft sequence predicted locations of each cataract ortholog were compared with the corresponding locations of each PCR amplicon used for RH-mapping. The resulting genome locations for each ortholog and corresponding sequence-tagged site (STS) are listed in Table 2.
Table 2.
Twenty-one cataract genes mapped onto the canine genome
| Gene | Chromosome | Closest markers | LOD | θ | STS location | Canine ortholog location |
|---|---|---|---|---|---|---|
| BFSP2 | CFA23 | REN156G20 | 22.448 | 0.086 | 33427673–33428089 | 33388029–33450927 |
| REN210D03 | 17.827 | 0.169 | ||||
| CX46 | CFA25 | EST18C6 | 9.688 | 0.276 | 20988791–20989147 | 20986634–20987933 |
| BAC_372_A23 | 9.617 | 0.261 | ||||
| CX50 | CFA17 | CFOR12C10 | 20.47 | 0.056 | 61506499–61506884 | 61498923–61505209 |
| PEZ8 | 19.834 | 0.058 | ||||
| LIM2 | CFA1 | KLK2 | 18.201 | 0.035 | 108473188–108473537 | 108472780–08479290 |
| REN06N11 | 17.642 | 0.066 | ||||
| MIP | CFA10 | FH2537 | 25.039 | 0.09 | 3684287–3684594 | 3684852–3689232 |
| ATP5B | 21.428 | 0.132 | ||||
| SPARC | CFA4 | FH4018 | 14.097 | 0.048 | 60879945–60880285 | 60873070–60887224 |
| BAC_374_E21 | 12.897 | 0.093 | ||||
| CRYBB2 | CFA26 | CRYBB1 | 25.249 | 0.043 | 22335395–22335748 | 22331283–22338858 |
| REN131L06 | 24.11 | 0.064 | ||||
| CRYGA | CFA37 | AHT133 | 20.482 | 0.092 | 19453604–19453950 | 19453388–19455353 |
| REN105M20 | 20.482 | 0.092 | ||||
| CRYGB | CFA37 | BAC_381_J22 | 15.426 | 0.123 | 20124535–20124930 | 19440507–19442393 |
| FH2708 | 13.738 | 0.157 | ||||
| CRYGC | CFA37 | BAC_381_J22 | 24.954 | 0 | 19431676–19432046 | 19431504–19433284 |
| EST22D1 | 23.468 | 0.026 | ||||
| CRYGD | CFA37 | CRYGB | 15.027 | 0.045 | 19425779–19426157 | 19424588–19426264 |
| BAC_374_E1 | 8.03 | 0.354 | ||||
| CRYGS | CFA34 | BAC_374_K23 | 27.708 | 0.021 | 22165843–22166230 | 22165569–22166681 |
| REN266K05 | 27.283 | 0.022 | ||||
| CHX10 | CFA8 | EST10D4 | 23.799 | 0.025 | 50510969–50511356 | 50492693–50511026 |
| EST27E3 | 21.735 | 0.051 | ||||
| EYA1 | CFA29 | STS246A16 | 19.01 | 0.119 | 23177126–23177509 | 23176722–23342854 |
| C29_188 | 18.473 | 0.138 | ||||
| FOXE3 | CFA15 | FH3888 | 23.211 | 0.046 | 16294587–16294926 | 16295972–16298254 |
| FH3886 | 22.806 | 0.047 | ||||
| HSF4 | CFA5 | AHTH201REN | 13.127 | 0.052 | 85205320–85205780 | 85204186–85208678 |
| REN122J03 | 11.927 | 0.1 | ||||
| MAF | CFA5 | FH3450 | 9.517 | 0.244 | 74976764–74977099 | 74970434–74976948 |
| FH3113 | 9.427 | 0.224 | ||||
| PAX6 | CFA18 | WT1 | 17.245 | 0.148 | 38687699–38688008 | 38671041–38693640 |
| FH3824 | 17.15 | 0.148 | ||||
| PITX3 | CFA28 | EST3H1 | 19.137 | 0.083 | 17763487–17763792 | 17761261–17772191 |
| COL17A1 | 18.606 | 0.085 | ||||
| SIX5 | CFA1 | BAC_375_N13 | 21.594 | 0 | 112730879–112731247 | 112726865–12731063 |
| FH2598 | 21.594 | 0 | ||||
| SOX1 | CFA22 | EST3G2 | 26.711 | 0.022 | 62864830–62865127 | 62861615–62864949 |
| BAC_376_O1 | 24.774 | 0.045 |
PCR primers (Table 1) from 21 cataract genes were used for radiation hybrid-mapping using an RH5000 panel. Multimap® two-point linkage analysis was used to identify the chromosome location and two closest markers [17], along with the corresponding LOD score, and θ value (distance). To confirm these locations, PCR amplicons were analyzed against the 7.5X canine draft sequence to obtain sequence-tagged sites (STS) locations, which were compared with the corresponding canine ortholog locations within the draft sequence.
RESULTS & DISCUSSION
Two-point linkage analysis
Twenty-one cataract gene orthologs were positioned on canine chromosomes using an RH5000 panel of 118 cell lines. Orthologs were located on 16 of the 39 canine chromosomes with more than one ortholog represented on three chromosomes (CFA1, 5, and 37; Table 2). The nearest chromosomal markers were determined for each cataract gene ortholog using Multimap® two-point linkage analysis [25]. Logarithm of odds (LOD) scores for the two closest linked markers ranged from 8.0 (CRYGD:BAC_374_E1) to 27.7 (CRYGS:BAC_374_K23), with an average LOD score of 18.9 (Table 2). Theta (θ), a measure of the frequency of breakage between two markers, varies from 0 (no breakage between markers) to 1 (complete breakage between markers), and represents the distance between markers. Theta ranged from 0 (CRYGC:BAC_381_J22, SIX5:BAC_375_N13, and SIX5:FH2598) to 0.354 (CRYGD:BAC_374_E1) with an average θ of 0.095 for the two closest linked markers (Table 2).
Two-point linkage analysis placed the γ-crystallin genes (CRYGA, CRYGB, CRYGC, and CRYGD) on CFA37. There are estimated to be seven γ-crystallin genes in humans; these include CRYGA, CRYGB, CRYGC, CRYGD, and CRYGS, and two pseudogenes, CRYGE and CRYGF. There is sequence homology between CRYGA, CRYGB, CRYGC, CRYGD, CRYGE, and CRYGF and these genes form a cluster at HSA 2q33-35, and in mouse they form a cluster on chromosome 1 (MMU1). In humans, CRYGS maps to 3q25-qter, and in mouse to MMU16. Similarly, canine CRYGS was not part of the γ-crystallin cluster on CFA37, but mapped to CFA34 near BAC_374_K23 and marker REN266K05. Mutations causing cataracts have previously been identified in murine Cryga [26,27], Crygb [26], Crygc [27], Crygd [27,28], and Crygs [29,30] as well as human CRYGC [31], CRYGD [31], and CRYGS [32]. As a result, there is a high likelihood that the γ-crystallins will also be involved in canine cataracts.
LOD scores for the two closest linked markers to γ-crystallin genes A, B, C, and D ranged from 8.0 (CRYGD:BAC_374_E1) to 24.95 (CRYGC:BAC_381_J22) and θ ranged from 0 (CRYGC:BAC_381_J22) to 0.354 (CRYGD:BAC_374_E1; Table 2). The two closest linked markers to CRYGD were CRYGB from a previous study [17] (LOD=15.027, θ=0.045) and BAC_374_E1 (LOD=8.03, θ=0.354). Although in the previous study, the CRYGB marker was “unlinked” to any other marker, the corresponding 171 bp amplicon was located on CFA37 (19,425,908–19,426,078). Also, in the previous study, markers were incorporated into a 1,500 marker map using Multimap® pairwise calculation at a threshold of LOD>8 [15]. The CRYGB marker in that study may have linked to other markers in the data set at a lower threshold. Decreased marker density in the vicinity of a gene’s map location can also result in unlinked gene markers. However, marker density in the vicinity of CRYGB should have been sufficient in the previous study since they mapped the same markers, which linked to the γ-crystallins in this study.
The next closest linked marker to CRYGD (BAC_374_E1: LOD=8.03, θ=0.354) was also on CFA37. The decrease in LOD score from 15.027 (CRYGD:CRYGB) to 8.03 supports linkage but the increase in θ from 0.045 (CRYGD:CRYGB) to 0.354 indicates an increase in the distance between these two markers. All of the markers that linked to CRYGD with LOD>4 were located on CFA37 (data not shown). In addition, the CRYGD PCR amplicon was located on CFA37 (19,425,779–19,426,157) within the predicted location for CRYGD based on reference sequence NM006891 (19,424,588–19,426,264). Thus, based on two-point linkage analysis results and the location of the CRYGD PCR amplicon, we place CRYGD on CFA37.
αA- and αB-crystallin genes were previously mapped to CFA31 (CRYAA) and CFA5 (CRYAB) [33], and βA1- and βB1-crystallin to CFA9 (CRYBA1) [17] and CFA26 (CRYBB1) [17,33]. Here we placed CRYBB2 on CFA26 near CRYBB1 and REN131L06. Similarly, CRYBB1 and CRYBB2 both map to 22q11.2–12.2 in man, and chromosome 5 in mouse (MMU5). MIP was mapped to CFA10 near FH2537 and ATP5B, in agreement with previously reported data [33]. Both LIM2 and SIX5 mapped to CFA1, and HSF4 and MAF to CFA5. All other orthologs (excluding the γ-crystallins) mapped to individual chromosomes as summarized in Table 2.
Sequence-tagged sites
Sequence-tagged sites (STS) for each cataract ortholog were obtained by analyzing the corresponding PCR amplicons against the May 2005 7.5X canine draft sequence. With the exception of CRYGB, all of the resulting STS were located inside or within 1,300 bp of the predicted gene location based on human reference sequences (Table 2). The CRYGB STS (20,124,535–20,124,930) was not within the predicted location for canine CRYGB (19,440,507–19,442,393) based on the human reference sequence (NM_005210). Primers used to amplify CRYGB were designed from the 1.5X (poodle) whole genome shotgun reference sequence AACN010184836. This sequence was selected from 1.5X canine sequences having homology to human CRYGB mRNA (NM_005210). When analyzed against the 7.5X (boxer) canine genomic sequence, AACN010184836 had sequence homology to four unique sites on CFA37 (19,417,160–19,418,711; 19,426,006–19,426,485; 19,432,821–19,455,395; 20,124,527–20,126,527). These included the human reference sequence (NM_005210) CRYGB predicted site (19,440,507–19,442,393). However, the actual AACN010184836 sequence was located on CFA37 from 20,124,527–20,126,527, and the 396 bp portion of this sequence which corresponded to the CRYGB amplicon only had homology to CFA37 at 20,124,535–20,124,930 (which is the CRYGB amplicon location). Therefore, even though the CRYGB primers were designed from the AACN010184836 sequence which had homology to human CRYGB, this sequence and the PCR amplicon derived from it were >682 kb downstream from the expected CRYGB site.
Furthermore, there are four CRYGB predicted sites on CFA37 based on human protein homology (19,415,025–19,433,068; 19,424,652–19,455,135; 19,440,575–19,442,393; and 20,126,034–20,128,506). Human protein sequence homology is determined by analyzing human protein sequences against the human genome to find exonic sequences corresponding to amino acid sequences. The amino acid sequences are then analyzed against the canine genomic sequence using tBLASTn (protein query vs. translated database) to find predicted canine exons. Three of the four CRYGB predicted sites based on protein homology overlap and cover a 40 kb region (19,415,026–19,455,135). This region is 670 kb upstream of the fourth predicted CRYGB site (20,126,033–20,128,569). The CRYGB STS amplified here was located 1,104 bp upstream to this fourth site. Because there are multiple predicted sites for CRYGB on CFA37 in the dog genome, and these sites share sequence homology, it is difficult to design primers specific for any single CRYGB predicted site. The multiple sites of CRYGB sequence homology make the actual location of the canine CRYGB gene unclear. As a result, our CRYGB STS should not be used to predict the exact location of canine CRYGB.
Radiation hybrid-mapping
Markers linked to cataract orthologs, as determined by two-point linkage analysis, were used to create a framework map with previously established marker positions [17]. This framework map was then used to determine the map locations of the cataract orthologs (Figure 1). The orthologs were positioned between the two closest markers using the multipoint mapping algorithm of Multimap® software. In two cases, the cataract orthologs mapped to the same exact position as a BAC end marker in the framework map (θ=0). For example, SIX5 mapped to CFA1 at the same location as BAC_375_N13, and CRYGC mapped to CFA37 at the same location as BAC_381_J22. Since BACs represent large (155 kb) DNA fragments [34], the mapping of these two genes to the same location as the two BACs suggests that these genes may be located within the BACS.
Figure 1.
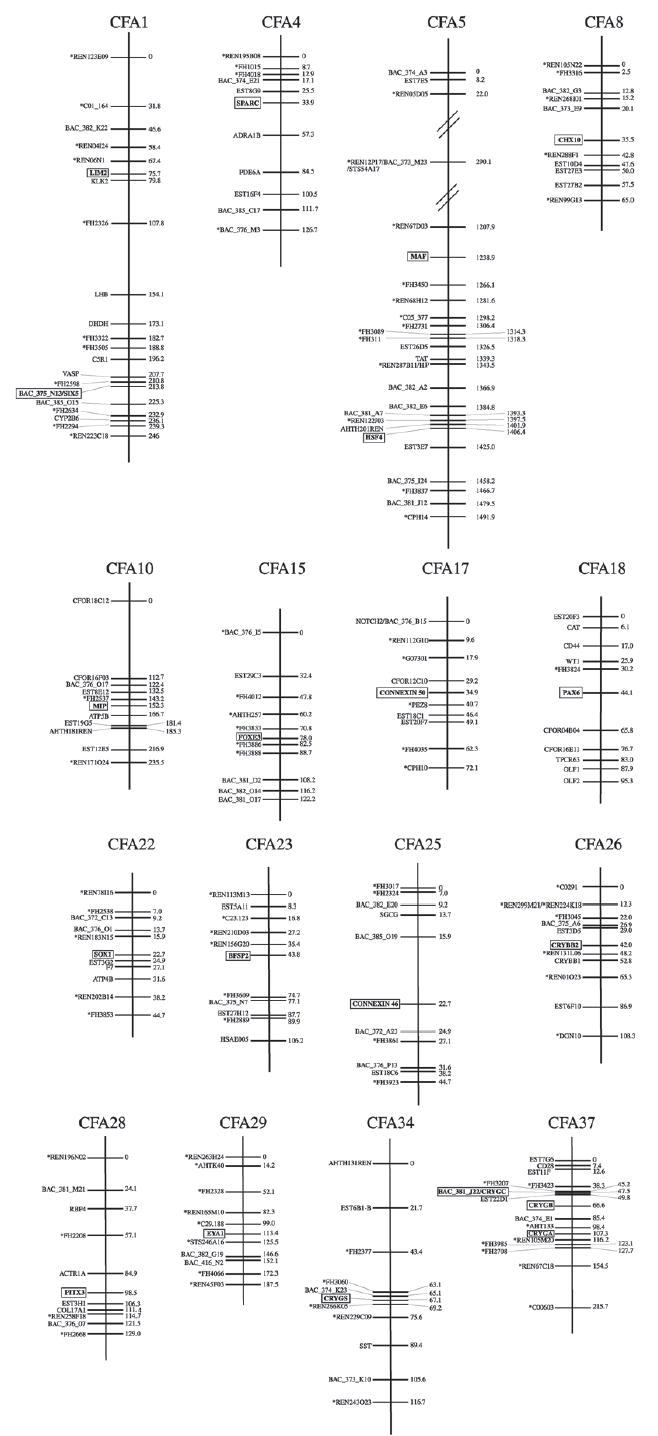
Radiation hybrid maps locating 20 cataract genes to canine chromosomes. In each Canis familiaris (CFA) chromosome representation, unboxed markers on left represent framework markers [17]. Markers with an asterisk are polymorphic. Boxed markers are location of canine orthologs of cataract associated genes. Numbers on right are distances in centiRays from the top framework marker.
The RH maps generated by the Multimap® multipoint mapping algorithm were generally consistent with the results of two-point linkage analysis, and the orthologs mapped on top of, between, or near the closest linked markers as determined by two-point linkage. An exception was CRYGD which was placed on CFA37 based on two-point linkage results and the location of the CRYGD STS (Table 2) but could not be accurately localized within the framework map. The framework map used to localize the γ-crystallin genes on CFA37 included the γ-crystallin gene vectors (CRYGA, CRYGB, CRYGC, CRYGD) along with framework markers whose locations were previously determined [17]. As a result, the framework map did not include the unlinked CRYGB marker from the previous study [17], which was the closest linked marker to CRYGD based on two-point linkage analysis. Instead, the closest linked marker to CRYGD in the framework map was BAC_374_E1 which linked to CRYGD with LOD=8.03, and θ=0.354. All of the other markers in the framework map which linked to CRYGD had a LOD score <8.03 and a θ value >0.354. These θ values indicated an increased frequency of breakage (increased distance) between CRYGD and the markers on the framework map and decreased the likelihood of accurately localizing CRYGD on the map. As a result, although we placed CRYGD on CFA37 based on two-point linkage analysis and the location of the CRYGD amplicon, we were unable to accurately localize this gene on the framework map using the Multimap® multipoint mapping algorithm due to the decreasing LOD scores and increasing θ values of the markers linked to CRYGD. The lack of markers linked to CRYGD with high LOD scores and low θ values may indicate low marker density in the vicinity of CRYGD. Even though all of the γ-crystallins orthologs (CRYGA, CRYGB, CRYGC, CRYGD) were located on CFA37 and were mapped against each other, this did not resolve the problem of marker density in the vicinity of CRYGD. To resolve this problem CRYGD should be mapped on the new RH9000 panel [24].
γA-, γB-, and γC-crystallin genes mapped to CFA37 as expected based on results of two-point-linkage analysis. The 5′->3′ RH-map order on CFA37 (CRYGC, CRYGB, CRYGA; Figure 1) matched the order of the predicted gene locations within the 7.5X draft sequence (Table 2, Column G). However, this RH-map order did not correspond with the order of the STS locations within the 7.5X draft sequence (CRYGC, CRYGA, CRYGB; Table 2, Column F). Two-point linkage analysis linked the CRYGB ortholog to multiple markers on CFA37. The two closest linked markers were BAC_381_J22 (LOD=15.426, θ=0.123), and FH2708 (LOD=13.738, θ=0.157). The Multimap® multipoint mapping algorithm placed the CRYGB ortholog on CFA37 between EST22D1 and BAC_374_E1, which are also between BAC_381_J22 and FH2708. Thus, two-point linkage analysis and radiation hybrid mapping results are consistent and place CRYGB on CFA37 in the same region, however this location does not correspond with the CRYGB STS location.
RH maps are physical maps based on the statistical analysis of the pattern of marker retention in a panel of RH-cell lines. As a result, the order and location of markers within RH-maps do not always correspond exactly with the order and location of markers within genomic sequences. Marker order and position on the canine RH5000 map [17] for example, are not equivalent with their order and position in the 7.5X canine genomic sequence in all instances. Thus, it is not surprising that the CRYGB marker in this study mapped to a location on the RH-map that does not exactly correspond to the physical location of this marker within the canine genome. Nevertheless, RH-maps are valuable in localizing markers and genes to specific regions and providing statistically close markers for association and linkage testing.
Furthermore, the discrepancy between the order of CRYGA, CRYGB, and CRYGC on the RH-map and their STS in the draft sequence may also reflect the limited power for fine resolution of the RH5000 panel, which has a resolution limit of 600 kb. The distance between CRYGA and CRYGB STS is about 670 kb, and the distance between CRYGB and CRYGC STS is about 692 kb. These distances are close to the resolution limit for the RH5000 panel. Further, the distance between CRYGA and CRYGC STS is only 21,558 bp and is outside the resolution limit for the panel. Therefore, the inconsistency between the RH-map order and the STS order for CRYGA, CRYGB, and CRYGC may be due to the resolution limit of the RH5000 panel. The new RH9000 panel has a resolution of 200 kb [24] and can be used to resolve mapping issues for the γ-crystallin genes in the dog.
The canine chromosomal locations of the 21 cataract orthologs were consistent with human and canine established chromosomal synteny [35–37]. All of the cataract orthologs were mapped to canine chromosomal locations which were syntenic with the corresponding human gene locations, except for four orthologs (SPARC, HSF4, CX46, and PITX3) that mapped to locations directly adjacent to the corresponding syntenic region of the human gene location. Although CRYGD was not localized on the RH-map, the results of the two-point linkage analysis did place this gene on CFA37 in a region with homology to HSA2q33–35 where the gene is located in humans.
To facilitate the molecular characterization of naturally occurring cataracts in the dog we have provided the RH location of 20 cataract-associated genes along with their associated polymorphic markers (Figure 1). Primers for selected polymorphic markers near cataract-associated genes can be obtained from the RH5000 website and serve as a resource for association and linkage testing in canine pedigrees segregating inherited cataracts. Although CRYGD is not represented in Figure 1, two-point linkage analysis did link CRYGD to BAC_374_E1 which localizes it on CFA37 to some degree. Also, the CRYGD STS location (19,425,779–19,426,157) can be used to identify proximal markers in the canine genome sequence. Though the RH5000 panel was used to map cataract-associated genes in this study, these results have not been incorporated into the RH5000 map. Nevertheless, the RH5000 map can be used to locate the markers referred to in this study (Table 2 and Figure 1). In addition, while the RH5000 and RH9000 maps will not be merged, a subset of 545 BACs has been mapped on both RH-panels and is part of both maps allowing navigation between the RH5000 and RH9000 maps.
Animal models provide a tremendous tool in the study of cataract, and molecular studies in mice have been instrumental in advancing our knowledge of the mechanisms of cataract formation. The dog has recently emerged as a promising model organism for gene discovery and genomic studies, and is a complementary species to the mouse for molecular genetic studies. There are over 300 known breeds of dogs, and most function as genetic isolates due to restricted mating. As a result, there is a decrease in genetic diversity within breeds and an increase in diversity between breeds such that genetic diseases within one breed tend to be caused by the same mutations (founder effect). This facilitates the mapping of disease loci. In addition, developments in canine genomic resources that include integrated RH maps [14–17,24], the availability of the canine genomic sequence [18,19], and comparative chromosome maps of the human and dog genomes which identify homologous segments and conserved synteny between canine and human karyotypes [35–37] make it practical to develop and use canine models.
Although mutations causing human cataracts have been identified in 15 genes there are many other genes with mutations yet to be identified. The study of naturally occurring cataracts in the dog could reveal novel genes or mutations not yet identified in either man or the mouse. The dog represents a valuable yet largely untapped resource to study the molecular genetics of cataracts. This study takes some initial steps toward developing the tools needed to begin studies on the canine cataract model. We have identified the chromosomal locations of 20 canine orthologs of cataract-associated genes and their nearest polymorphic markers (Figure 1) that can be used for association and linkage testing in cataract pedigrees. This resource, along with the recent advances in canine genomics, will help facilitate the development of such models for the study of naturally occurring inherited cataracts. Once developed, canine cataract models should help to identify new cataract genes and disease-causing mutations, assist in the development of new therapies, and provide an intermediate large animal model between mouse and human for preclinical trials of new therapies.
Acknowledgments
We would like to thank Vicki Meyers-Wallen, Anna Kukekova, James Kijas, Orly Goldstein, Keith Watamura, and John McElwee for their support and interest in this work. Funding was provided by the National Institutes of Health (grant F32-EY13677 and RO1 EY15173), Cornell University Graduate Research Assistantship, the Morris Animal Foundation/Seeing Eye, Inc., the Van Sloan Fund for Canine Genetic Research, and the Research to Prevent Blindness Foundation.
References
- 1.Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–51. [PMC free article] [PubMed] [Google Scholar]
- 2.Wirth MG, Russell-Eggitt IM, Craig JE, Elder JE, Mackey DA. Aetiology of congenital and paediatric cataract in an Australian population. Br J Ophthalmol. 2002;86:782–6. doi: 10.1136/bjo.86.7.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans CA. 1995 Presidential Address. Public health: vision and reality Am J Public Health. 1996;86:476–9. doi: 10.2105/ajph.86.4.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans J, Rooney C, Ashwood F, Dattani N, Wormald R. Blindness and partial sight in England and Wales: April 1990-March 1991. Health Trends. 1996;28:5–12. [Google Scholar]
- 5.He W, Li S. Congenital cataracts: gene mapping. Hum Genet. 2000;106:1–13. doi: 10.1007/s004390051002. [DOI] [PubMed] [Google Scholar]
- 6.Congdon N, Vingerling JR, Klein BE, West S, Friedman DS, Kempen J, O’Colmain B, Wu SY, Taylor HR Eye Diseases Prevalence Research Group. Prevalence of cataract and pseudophakia/aphakia among adults in the United States. Arch Ophthalmol. 2004;122:487–94. doi: 10.1001/archopht.122.4.487. [DOI] [PubMed] [Google Scholar]
- 7.McCarty CA, Taylor HR. A review of the epidemiologic evidence linking ultraviolet radiation and cataracts. Dev Ophthalmol. 2002;35:21–31. doi: 10.1159/000060807. [DOI] [PubMed] [Google Scholar]
- 8.Klein BE, Klein R, Lee KE, Meuer SM. Socioeconomic and lifestyle factors and the 10-year incidence of age-related cataracts. Am J Ophthalmol. 2003;136:506–12. doi: 10.1016/s0002-9394(03)00290-3. [DOI] [PubMed] [Google Scholar]
- 9.Morris MS, Jacques PF, Hankinson SE, Chylack LT, Jr, Willett WC, Taylor A. Moderate alcoholic beverage intake and early nuclear and cortical lens opacities. Ophthalmic Epidemiol. 2004;11:53–65. doi: 10.1076/opep.11.1.53.26439. [DOI] [PubMed] [Google Scholar]
- 10.Mares-Perlman JA, Lyle BJ, Klein R, Fisher AI, Brady WE, VandenLangenberg GM, Trabulsi JN, Palta M. Vitamin supplement use and incident cataracts in a population-based study. Arch Ophthalmol. 2000;118:1556–63. doi: 10.1001/archopht.118.11.1556. [DOI] [PubMed] [Google Scholar]
- 11.Hejtmancik JF, Kantorow M. Molecular genetics of age-related cataract. Exp Eye Res. 2004;79:3–9. doi: 10.1016/j.exer.2004.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–15. doi: 10.1016/j.survophthal.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Gelatt KN, Wallace MR, Andrew SE, MacKay EO, Samuelson DA. Cataracts in the Bichon Frise. Vet Ophthalmol. 2003;6:3–9. doi: 10.1046/j.1463-5224.2003.00258.x. [DOI] [PubMed] [Google Scholar]
- 14.Priat C, Hitte C, Vignaux F, Renier C, Jiang Z, Jouquand S, Cheron A, Andre C, Galibert F. A whole-genome radiation hybrid map of the dog genome. Genomics. 1998;54:361–78. doi: 10.1006/geno.1998.5602. [DOI] [PubMed] [Google Scholar]
- 15.Breen M, Jouquand S, Renier C, Mellersh CS, Hitte C, Holmes NG, Cheron A, Suter N, Vignaux F, Bristow AE, Priat C, McCann E, Andre C, Boundy S, Gitsham P, Thomas R, Bridge WL, Spriggs HF, Ryder EJ, Curson A, Sampson J, Ostrander EA, Binns MM, Galibert F. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–95. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, Sabacan L, Scott A, Evanno G, Parker HG, Kirkness EF, Hudson R, Guyon R, Mahairas GG, Gelfenbeyn B, Fraser CM, Andre C, Galibert F, Ostrander EA. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:65. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyon R, Lorentzen TD, Hitte C, Kim L, Cadieu E, Parker HG, Quignon P, Lowe JK, Renier C, Gelfenbeyn B, Vignaux F, DeFrance HB, Gloux S, Mahairas GG, Andre C, Galibert F, Ostrander EA. A 1-Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci U S A. 2003;100:5296–301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM, Venter JC. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- 19.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wil-son A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 20.Kijas JW, Cideciyan AV, Aleman TS, Pianta MJ, Pearce-Kelling SE, Miller BJ, Jacobson SG, Aguirre GD, Acland GM. Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 2002;99:6328–33. doi: 10.1073/pnas.082714499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidjanin DJ, Lowe JK, McElwee JL, Milne BS, Phippen TM, Sargan DR, Aguirre GD, Acland GM, Ostrander EA. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum Mol Genet. 2002;11:1823–33. doi: 10.1093/hmg/11.16.1823. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Acland GM, Wu WX, Johnson JL, Pearce-Kelling S, Tulloch B, Vervoort R, Wright AF, Aguirre GD. Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum Mol Genet. 2002;11:993–1003. doi: 10.1093/hmg/11.9.993. [DOI] [PubMed] [Google Scholar]
- 23.Vignaux F, Hitte C, Priat C, Chuat JC, Andre C, Galibert F. Construction and optimization of a dog whole-genome radiation hybrid panel. Mamm Genome. 1999;10:888–94. doi: 10.1007/s003359901109. [DOI] [PubMed] [Google Scholar]
- 24.Hitte C, Madeoy J, Kirkness EF, Priat C, Lorentzen TD, Senger F, Thomas D, Derrien T, Ramirez C, Scott C, Evanno G, Pullar B, Cadieu E, Oza V, Lourgant K, Jaffe DB, Tacher S, Dreano S, Berkova N, Andre C, Deloukas P, Fraser C, Lindblad-Toh K, Ostrander EA, Galibert F. Facilitating genome navigation: survey sequencing and dense radiation-hybrid gene mapping. Nat Rev Genet. 2005;6:643–8. doi: 10.1038/nrg1658. [DOI] [PubMed] [Google Scholar]
- 25.Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994;6:384–90. doi: 10.1038/ng0494-384. Erratum in: Nat Genet 1994; 7:215. [DOI] [PubMed] [Google Scholar]
- 26.Klopp N, Favor J, Loster J, Lutz RB, Neuhauser-Klaus A, Prescott A, Pretsch W, Quinlan RA, Sandilands A, Vrensen GF, Graw J. Three murine cataract mutants (Cat2) are defective in different gamma-crystallin genes. Genomics. 1998;52:152–8. doi: 10.1006/geno.1998.5417. [DOI] [PubMed] [Google Scholar]
- 27.Graw J, Neuhauser-Klaus A, Klopp N, Selby PB, Loster J, Favor J. Genetic and allelic heterogeneity of Cryg mutations in eight distinct forms of dominant cataract in the mouse. Invest Ophthalmol Vis Sci. 2004;45:1202–13. doi: 10.1167/iovs.03-0811. [DOI] [PubMed] [Google Scholar]
- 28.Smith RS, Hawes NL, Chang B, Roderick TH, Akeson EC, Heckenlively JR, Gong X, Wang X, Davisson MT. Lop12, a mutation in mouse Crygd causing lens opacity similar to human Coppock cataract. Genomics. 2000;63:314–20. doi: 10.1006/geno.1999.6054. [DOI] [PubMed] [Google Scholar]
- 29.Sinha D, Wyatt MK, Sarra R, Jaworski C, Slingsby C, Thaung C, Pannell L, Robison WG, Favor J, Lyon M, Wistow G. A temperature-sensitive mutation of Crygs in the murine Opj cataract. J Biol Chem. 2001;276:9308–15. doi: 10.1074/jbc.M010583200. [DOI] [PubMed] [Google Scholar]
- 30.Bu L, Yan S, Jin M, Jin Y, Yu C, Xiao S, Xie Q, Hu L, Xie Y, Solitang Y, Liu J, Zhao G, Kong X. The gamma S-crystallin gene is mutated in autosomal recessive cataract in mouse. Genomics. 2002;80:38–44. doi: 10.1006/geno.2002.6803. [DOI] [PubMed] [Google Scholar]
- 31.Heon E, Priston M, Schorderet DF, Billingsley GD, Girard PO, Lubsen N, Munier FL. The gamma-crystallins and human cataracts: a puzzle made clearer. Am J Hum Genet. 1999;65:1261–7. doi: 10.1086/302619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H, Ma Z, Li Y, Liu B, Li Z, Ding X, Gao Y, Ma W, Tang X, Li X, Shen Y. Gamma-S crystallin gene (CRYGS) mutation causes dominant progressive cortical cataract in humans. J Med Genet. 2005;42:706–10. doi: 10.1136/jmg.2004.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker HG, Yuhua X, Mellersh CS, Khan S, Shibuya H, Johnson GS, Ostrander EA. Meiotic linkage mapping of 52 genes onto the canine map does not identify significant levels of microrearrangement. Mamm Genome. 2001;12:713–8. doi: 10.1007/s00335-001-2057-3. [DOI] [PubMed] [Google Scholar]
- 34.Li R, Mignot E, Faraco J, Kadotani H, Cantanese J, Zhao B, Lin X, Hinton L, Ostrander EA, Patterson DF, de Jong PJ. Construction and characterization of an eightfold redundant dog genomic bacterial artificial chromosome library. Genomics. 1999;58:9–17. doi: 10.1006/geno.1999.5772. [DOI] [PubMed] [Google Scholar]
- 35.Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999;61:145–55. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- 36.Yang F, O’Brien PC, Milne BS, Graphodatsky AS, Solanky N, Trifonov V, Rens W, Sargan D, Ferguson-Smith MA. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- 37.Sargan DR, Yang F, Squire M, Milne BS, O’Brien PC, Ferguson-Smith MA. Use of flow-sorted canine chromosomes in the assignment of canine linkage, radiation hybrid, and syntenic groups to chromosomes: refinement and verification of the comparative chromosome map for dog and human. Genomics. 2000;69:182–95. doi: 10.1006/geno.2000.6334. [DOI] [PubMed] [Google Scholar]


