Abstract
Lens opacity 11 (lop11) is an autosomal recessive mouse cataract mutation that arose spontaneously in the RIIIS/J strain. At 3 weeks of age mice exhibit total cataracts with vacuoles. The lop11 locus was mapped to mouse chromosome 8. Analysis of the mouse genome for the lop11 critical region identified Hsf4 as a candidate gene. Molecular evaluation of Hsf4 revealed an early transposable element (ETn) in intron 9 inserted 61 bp upstream of the intron/exon junction. The same mutation was also identified in a previously mapped cataract mutant, ldis1. The ETn insertion altered splicing and expression of the Hsf4 gene, resulting in the truncated Hsf4 protein. In humans, mutations in HSF4 have been associated with both autosomal dominant and recessive cataracts. The lop11 mouse is an excellent resource for evaluating the role of Hsf4 in transparency of the lens.
Keywords: Cataracts, Mouse, Mapping, Locus, Gene, Mutation, Transposon, Insertion, Hsf4, Expression
Cataracts are the most frequent cause of treatable blindness worldwide [1], with the majority of cataracts occurring in the elderly [2]. Congenital cataracts, though significantly less common than age-related cataracts, are the leading cause of treatable childhood blindness [3]. The incidence of congenital cataracts has been estimated to vary from 0.6 to 6 per 10,000 live births [4], of which about half are hereditary [5]. The identification of genes harboring mutations responsible for hereditary congenital cataracts facilitates a better understanding of the initial molecular events in cataractogenesis, including novel insights into the mechanisms responsible for the development and function of the lens. Mutations in at least 19 genes have been identified as associated with human hereditary congenital cataracts [4].
Mice are excellent animal models for gene discovery and evaluation of molecular processes that lead to the development of cataracts. More then 60 mouse cataract models have been recovered either from large-scale mutagenesis projects or as a result of spontaneous mutations identified in breeding colonies [6]. Controlled breeding and large litter size have allowed for efficient mapping and cloning of mutations in a number of cataract-associated genes [6]. In addition, the mouse cataract models provide excellent tissue resources for detailed evaluation of the phenotype, as well as the differential expression of candidate genes by age, tissue, or disease status. Mouse cataract models are of great benefit in understanding molecular pathways relevant for the transparency of the lens.
In both humans and mice, the majority of mutations responsible for congenital cataracts show autosomal dominant inheritance. However, significant progress has been made in families with autosomal recessive congenital cataracts for which mutations in five genes, LIM2 [7], CRYAA [8], HSF4 [9,10], GCNT2 [11], and CRYBB3 [12], have been identified. In mouse models, mutations in only two genes, Cryaa [13] and Crygs [14], have been identified as associated with recessive congenital cataracts. Therefore, more insight is needed to understand the genes and molecular/cellular processes that lead to recessive congenital cataracts.
The focus of this study is on a mouse model of recessive congenital hereditary cataracts that arose spontaneously in the RIIIS/J strain. The cataract locus was termed lens opacity 11 (lop11). Here we present mapping and cloning of the lop11 locus. Further analysis identified the insertion of an early transposable element in intron 9 of the Hsf4 gene that altered splicing and expression of the Hsf4 gene and resulted in truncated Hsf4 protein. The same Hsf4 transposon insertion identified in lop11 was also identified in the ldis1 mouse cataract locus that had previously been mapped to mouse chromosome 8 [15]. The Hsf4 gene belongs to a family of highly conserved heat shock transcription factors. Mutations in HSF4 have been identified in families with both autosomal dominant [16] and recessive congenital cataracts [9,10]. However, it remains unknown how mutations in HSF4 lead to cataractogenesis. The lop11 mice offer a valuable resource for evaluation of the role of Hsf4 in the lens and molecular mechanisms that lead to cataract development in both humans and mice.
Results
The slit lamp examination of the cataract-affected adult RIIIS/J mouse, termed lens opacity 11 (lop11), showed diffuse cortical and nuclear opacification with white vacuoles irregularly patterned throughout the lens. All portions of the lens were affected. The fundus could not be seen due to the lens opacity. Lids, cornea, and iris were all unremarkable and media was clear. The cataract phenotype in lop11 mice segregated as an autosomal recessive trait. These data were confirmed by (lop11/lop11 × CAST/E)F1 × lop11/lop11 backcross. Clinical examination of the affected F2 lop11/lop11 mice showed cataracts identical to those of the parental lop11/lop11 phenotype. The F2 lop11/lop11 histological examination was consistent with the clinical observations that the lens structure showed vacuoles and extensive disorganization (Fig. 1A). In contrast, F2 lop11/+ showed the wild-type phenotype (Fig. 1B), consistent with the autosomal recessive mode of inheritance.
Fig. 1.
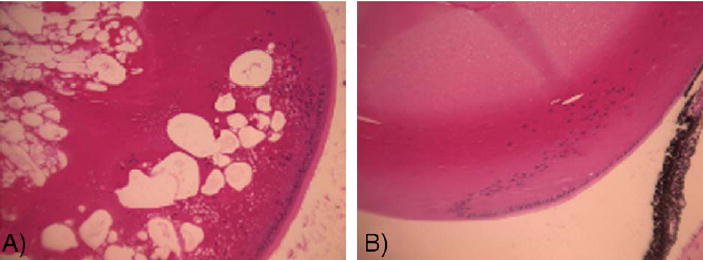
Histological sections of P21 mouse lenses. (A) lop11/lop11 shows the presence of vacuoles throughout the lens. (B) lop11/+ shows no pathological changes.
Genome-wide segregation analysis identified linkage between the lop11 locus and microsatellite markers on chromosome 8. The lop11 locus was mapped further between D8Mit110 and D8Mit313; no recombinants were identified with marker D8Mit198 (Fig. 2). Evaluation of the mouse genome map (http://genome.ucsc.edu/cgi-bin/hgGateway) of the lop11 critical region identified heat shock transcription factor 4 (NM_011939). To evaluate Hsf4 as a candidate gene, we generated RT-PCR products from whole eye mRNA from postnatal day 21 (P21) lop11/lop11 and wild-type C57BL/6 mice. RT-PCR from lop11/lop11 mRNA with primers specific to Hsf4 exons 1 through 6 generated cDNA products identical to the product from C57BL/6 (NM_011939) (Fig. 3A) and from exons 3 through exon 9 (data not shown). The identities of PCR products were confirmed by sequencing. However, RT-PCR with primers specific for Hsf4 exons 10 through 13 did not yield any cDNA products in lop11/lop11. In contrast, mRNA from C57BL/6 produced cDNAs of the expected size (Fig. 3A). Northern blot analysis of the Hsf4 expression in wild-type C57BL/6 and lop11/lop11 mouse eyes was consistent with the RT-PCR data. Hybridization with a 3′ end Hsf4 cDNA failed to detect the Hsf4 transcript in P21 and P1 lop11/lop11 mouse eyes (data not shown). However, hybridization of the same Northern blot with the 5′ end Hsf4 cDNA identified in lop11/lop11 an Hsf4 transcript of smaller molecular weight more abundantly expressed compared to the wild-type Hsf4 expressed in C57BL/6. This higher level of expression of the Hsf4 transcript was shown to be present in both P21 and P1 lop11/lop11 eye tissues (Fig. 3B).
Fig. 2.
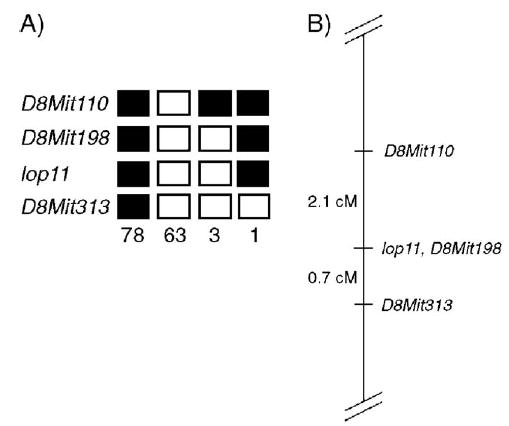
Chromosomal mapping of the lop11 locus. (A) Each column represents the haplotype identified in the backcross progeny: (▪) CAST/E allele, (□) RIIIS/J allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. (B) Linkage map of the lop11 locus; the numbers on the left represent the genetic distances in centimorgans (cM).
Fig. 3.
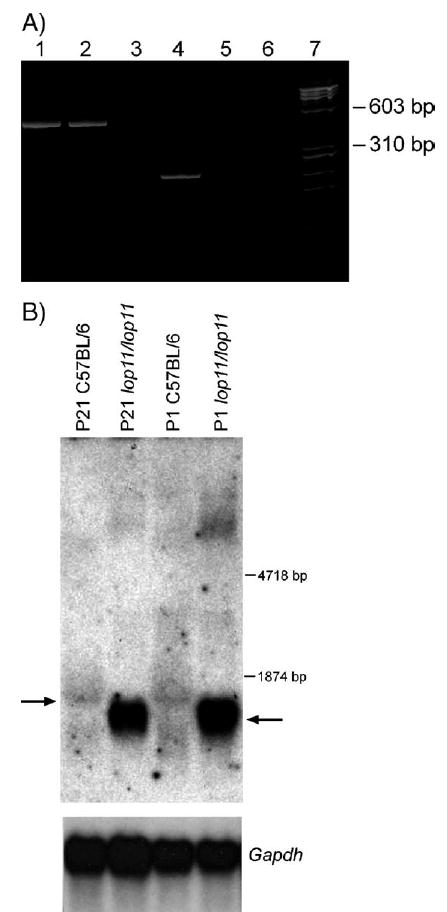
RT-PCR and Northern blot analysis. (A) Acrylamide gel electrophoresis of RT-PCR products derived from amplification of the Hsf4 gene from mouse eye mRNA. An RT-PCR product encompassing exons 1–6 is present in C57BL/6 and lop11/lop11 (lanes 1 and 2). An RT-PCR product encompassing exons 10–13 is present in C57BL/6, but absent from lop11/lop11 (lanes 4 and 5). Lanes 3 and 6 show H2O as a negative control; lane 7 is the φχ-174 RF DNA HaeIII molecular weight marker. (B) Northern analysis from P21 and P1 C57/BL6 and lop11/lop11 whole eye mRNA hybridized with a 5′ end Hsf4 probe (exons 1–6). Arrow to the left points to the wild-type Hsf4 transcript. Arrow to the right points to the lop11–Hsf4 transcript of the smaller molecular weight and higher level of expression. Hybridization of the same blot with Gapdh shows even loading between samples.
Genomic differences in the Hsf4 region between lop11/+ and lop11/lop11 were identified via Southern blotting (Fig. 4A). To determine if these differences were due to polymorphisms between RIIIS/J and CAST/E strains or genomic differences responsible for the cataract phenotype, we initiated sequencing of the Hsf4 genomic region. Our analysis did not detect any sequence difference between lop11/lop11 and C57BL/6 in any of the 13 exons, 650 bp upstream of the start codon, or any of the introns, except the 3′ end of intron 9. Initially, the conventional PCR did not yield a product from the 3′ end of intron 9 for lop11/lop11. However, long-range PCR in lop11/lop11 generated a PCR product about 5.6 kb larger than in C57BL/6 (Fig. 4B) and CAST/Ei (data not shown). Sequencing of the 5.9-kb PCR product from lop11/lop11 revealed an insertion of 5542 bp positioned −61 bp upstream of the 5′ intron/exon junction of exon 10. The Blast analysis of the 5542-bp inserted sequence revealed it to be identical to an early transposable element (ETn) (Accession No. Y17106) (Fig. 4C). The ETn insertion cosegregated with all affected F2 lop11/lop11 progeny. All the unaffected lop11/+ were heterozygous for the ETn insertion. Neither C57BL/6 nor CAST/Ei strains contained the ETn insertion in intron 9 of Hsf4. To determine if ETn was responsible for aberrant splicing of Hsf4, RT-PCR was performed with primers specific for Hsf4 and the ETn sequence (Accession No. Y17106). A chimeric transcript showing aberrant splicing between Hsf4 exon 9 and the pseudo-exon starting at 21 bp of the ETn sequence Accession No. Y17106) was identified (Fig. 4D). The sequence analysis of the Hsf4–ET transcript identified an open reading frame of 1059 bp following a premature stop codon.
Fig. 4.
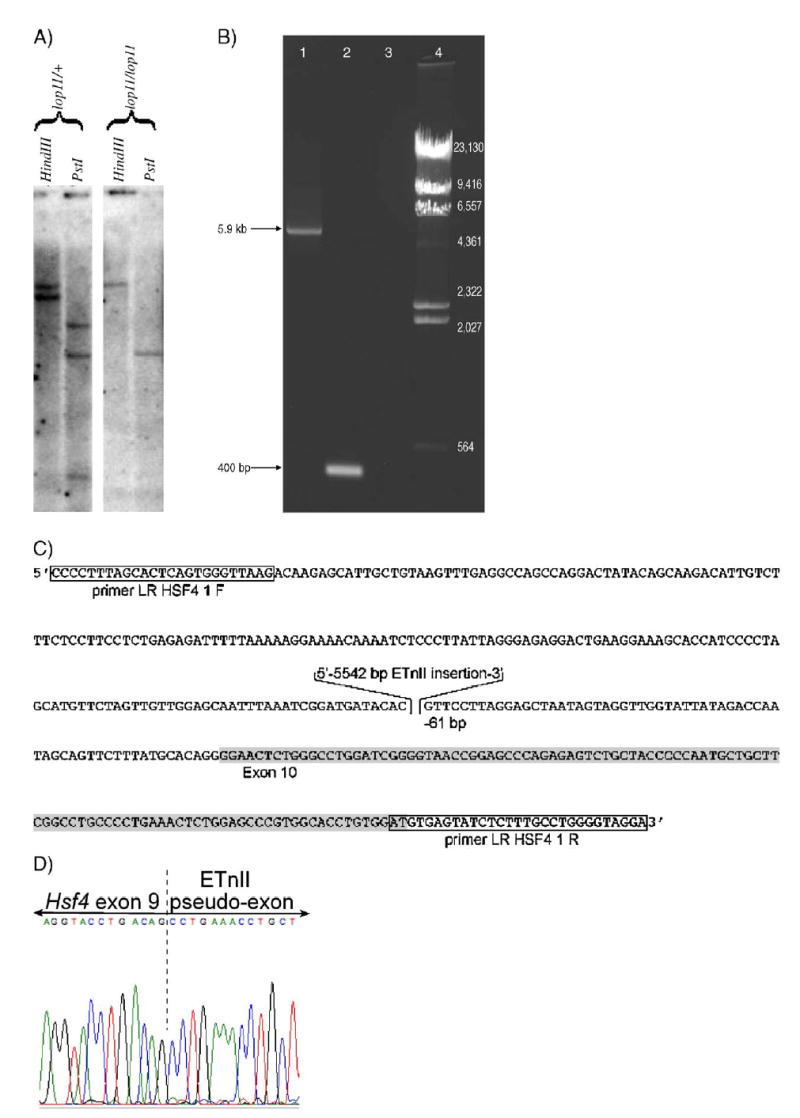
The analysis of the lop11 allele. (A) Southern blot analysis of the Hsf4 region. HindIII- and PstI-digested DNA from lop11/+ and lop11/lop11 hybridized with the 3′ end Hsf4 cDNA probe (exons 10–13). (B) LR-PCR spanning Hsf4 intron 9 showing the ETn insertion: (lane 1) lop11/lop11, (lane 2) C57BL/6, (lane 3) H2O, (lane 4) λ HindIII marker. (C) Structure of the lop11 allele. Partial genomic sequence of the chimeric Hsf4–ET genomic DNA from lop11 mice. Boxed sequences represent the primers used to amplify across the insertion in (B). Highlighted in gray is exon 10. The 5542-bp inserted sequence is the perfect match of the ETnII transposable element (Accession No. Y17106) placed in the 5′ → 3′ orientation −61 bp downstream of the 5′ intron/exon junction of exon 10. (D) Fluorescent sequence traces from the chimeric Hsf4–ET cDNA showing alternative splicing between Hsf4 exon 9 and the pseudo-exon from the ETn insertion. The dashed line separates the 3′ end of Hsf4 exon 9 and the 5′ of the pseudo-exon from the ETnII element (Accession No. Y17106).
To determine if the ETn insertion had any consequences on the Hsf4 protein in lenses of lop11/lop11, we performed Western blots. The Hsf4 protein was identified as an Hsf4-immunoreactive band of about 55 kDa in protein extracts from lenses of C57BL/6 (Fig. 5) that comigrated with a band from Jurkat cell lysates (data not shown) that served as a positive control. However, the 55-kDa Hsf4 was absent in protein extracts from lop11/lop11, but an Hsf4-immunoreactive band of about 40 kDa was identified that was not present in C57BL/6 (Fig. 5). In contrast to the observation on the Northern blot, on the Western blot the 40-kDa Hsf4 mutant band did not appear to be expressed at a higher level compared to the 55-kDa Hsf4 protein.
Fig. 5.
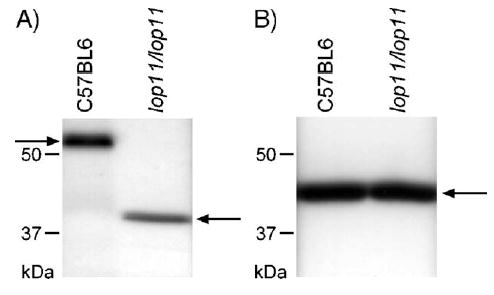
Western blot analysis of protein from P21 mouse lenses from C57BL/6 and lop11/lop11. (A) Western blot using antibody specific to Hsf4; the arrow on the left points to the wild-type Hsf4 protein of about 55 kDa and the arrow on the right points to truncated Hsf4 of about 40 kDa. (B) Western blot using antibody specific for β-actin; the arrow points to the 43-kDa β-actin band.
Following mapping of the lop11 locus, a search of MGI (http://www.informatics.jax.org) identified that another recessive mouse cataract locus, ldis1, maps to chromosome 8 [15] close to lop11. The ldis1 locus was also identified in the RIIIS/J strain. Comparison of the clinical phenotype between lop11 and ldis1 revealed similar cataracts in both strains. Based on the same strain origin and phenotypical and genetic similarities, we hypothesized that ldis1 may carry the same mutation as lop11. Long-range PCR and sequence analysis of ldis1/ldis1 identified the same 5542-bp ETn (Accession No. Y17106) sequence in intron 9 inserted −61 bp upstream of the 5′ intron/exon junction of exon 10 as already identified in lop11.
Discussion
In this study we have shown that the Hsf4 gene in lop11 mice carries the insertion of an early transposable element in intron 9. Following mapping of the lop11 locus, a search of MGI (http://www.informatics.jax.org) identified another recessive mouse cataract mutant termed “lens disrupter 1” (ldis1). This ldis1 locus was in the RIIIS/J strain that was originally obtained from The Jackson Laboratory and subsequently diagnosed for cataracts [15]. Independent of findings on the ldis1 strain, the lop11 strain was identified during systematic screening of mutant and inbred strains for eye defects at The Jackson Laboratory. Sequencing of Hsf4 intron 9 identified the same ETn insertion in ldis1 mice that was found for lop11. Based on the same RIIIS/J strain origin, and similar clinical, genetic, and molecular data, we believe that lop11 and ldis1 are a single mutation that arose spontaneously only once in RIIIS/J. Although we did not set up allelic crosses between ldis1 and lop11 that would unequivocally prove that these two lines are mutations in the same gene, we conclude that cataracts in both lop11 and ldis1 are due to the same Hsf4–ETn insertion.
ETn elements are a family of repetitive sequences transcribed during early mouse embryogenesis, primarily in undifferentiated cells of the inner cell mass and embryonic ectoderm [21]. They range in length from 4.4 to 7.1 kb and lack significant open reading frames [22]. ETn elements have long terminal repeat (LTR) sequences and have been classified into two groups (ETnI and ETnII), which differ only in the 3′ half of the LTR and in the 5′ end of the internal region [22]. The mutagenic power of ETn elements has been established with at least 14 mouse mutations that are due to ETn insertions [22], resulting in various phenotypes, including cataracts. An ETn insertion in the Mip gene in the CatFr mouse resulted in congenital cataracts [23]. The 5542-bp ETn element (Accession No. Y17106) identified in this study as inserted into Hsf4 intron 9 belongs to an ETII family of early transposable elements. The same ETnII element (Accession No. Y17106) was responsible for mutagenesis in SELH/Bc mice in the Tyr gene resulting in the albino phenotype [24].
Our ability to detect Hsf4 mRNA fragments from exons 1 through 9 indicated that the lop11–Hsf4 locus remained transcriptionally active despite the presence of the transposon. However, the full-length wild-type Hsf4 mRNA molecule could not be detected by RT-PCR or Northern blot in lop11. We identified a product of alternative splicing between Hsf4 exon 9 and a pseudo-exon in the transposon sequence resulting in premature termination. The open reading frame of the chimeric Hsf4–ET transcripts predicts the chimeric Hsf4–ETn protein to be composed of 353 amino acids (aa), with 330 aa originating from the Hsf4 gene (exons 1 through 9) and 23 aa from the transposon pseudo-exon. The predicted molecular weight of this chimeric protein would be 38.69 kDa (http://www.sciencegate-way.org/tools/proteinmw.htm). Western blots identified an Hsf4-immunoreactive band of about 40 kDa in lop11, further supporting these findings. We could not detect the wild-type 55-kDa Hsf4 protein as present in lop11 lenses; thus we conclude that the ETn insertion is responsible for the cataract phenotype in the lop11 mouse.
It has been shown that the functional Hsf4 protein is essential for maintenance of lens transparency. Knockout studies demonstrate that Hsf4 expression in the developing lens is required for correct lens development [25,26]. In families with autosomal dominant lamellar and Marner cataracts four different missense mutations have been identified within the HSF4 DNA binding domain [16]. In addition, a splice mutation causing skipping of exon 12 was identified in a family with autosomal recessive cataracts [9]. Recently, two additional mutations, R175P and a frameshift mutation (595_599delGGGCCC), were reported in families with autosomal recessive cataracts [10]. These studies support the critical role of the functional Hsf4 protein for the physiology of the lens.
The role of Hsf4 in the lens has been proposed as a transcriptional regulator of genes necessary for proper lens development. In rat lenses Hsf4 was shown to have a specific interaction with αB-crystallin [27]. Expression profiles previously reported from adult ldis1 mouse whole eyes showed down regulation of γ-crystallins, transforming growth factor, fibroblast growth factor, the bone morphogenic proteins, and the activins [15]. Expression profiles from Hsf4 knockout studies also showed that Hsf4 is an activator of γcrystallin genes and regulates expression of growth factor genes essential for cell growth and differentiation [26]. Our findings point to another possible role for Hsf4. The truncated lop11–Hsf4 transcript identified in lop11/lop11 lenses was more abundant than the wild-type Hsf4 detected in C57BL/6 lenses. Given that the wild-type Hsf4 is absent in lop11, it is possible that the functional Hsf4 protein may participate in negative self-regulation of expression. However, we cannot exclude a possibility that the ETn integration increases the stability of the truncated lop11–Hsf4 transcript or that the ETn integration up regulates expression of the lop11–Hsf4 transcript resulting in the more abundant truncated Hsf4 transcript. Regardless of the mechanism responsible for the more abundant chimeric Hsf–ET transcript, we did not observe higher levels of the mutant Hsf4 protein (Fig. 5). These findings suggest that the mutant Hsf4 protein likely undergoes a degradation process.
In the lop11 mouse the absence of the wild-type Hsf4 protein is consistent with the cataract phenotype. At this point it is unclear if cataracts in lop11 are due only to the absence of functional Hsf4 protein and its transcriptional regulation of essential lens genes such as crystallins and growth factors. Alternatively, cataracts may be due to an inability of the lens to degrade the truncated Hsf4 protein fully, ultimately resulting in cellular cytotoxicity and compromised processing of other lens proteins. Further studies are needed to elucidate the role of the ETn insertion in the Hsf4 gene as it relates to the onset of cataracts in lop11 mice.
It should be noted that Hsf4 has been reported to exist as at least two isoforms, Hsf4a and Hsf4b, due to two alternative splice sites in exons 8 and 9 [28,29]. Two distinct roles have been proposed for the two Hsf4 isoforms: Hsf4a as a transcription suppressor and Hsf4b, which contains 30 additional amino acids, as a transcription activator. In human and mouse lenses only Hsf4b transcript has been identified [25,26]. Our study also identified only the Hsf4b transcript as present in the lens. The Hsf4–ETn insertion identified in lop11 probably affects splicing of both Hsf4a and Hsf4b; we evaluated consequences of the ETn insertion only for the Hsf4b transcript and protein in the lens. However, we have not evaluated the expression of Hsf4b in brain, lung, liver, and skeletal muscle [29] or the expression of Hsf4a in brain, heart, skeletal muscle, and pancreas [28]. The effects of the ETn insertion on expression of Hsf4a and Hsf4b in other tissues, and if the ETn insertion may be responsible for phenotypes other then cataracts reported for the RIIIS/J strain (http://www.informatics.jax.org), are beyond the scope of this study.
Materials and methods
lop11 identification
Inbred mouse strains were systematically screened for any eye defects with a slit lamp and indirect ophthalmoscope as previously described [17]. The inbred strain RIIIS/J showed complete cataracts and was further evaluated.
Linkage mapping
The lop11/lop11 mice were outcrossed to CAST/Ei, and F1 mice were backcrossed to lop11/lop11 to generate 145 backcross progeny. The progeny were evaluated at 3 weeks of age with a slit lamp following mydriasis with 1% atropine, and phenotypes were recorded. The animals were euthanized and tissues were collected. For linkage analysis, genomic DNA was isolated from spleens and typed with polymorphic markers as described previously [18]. Briefly, the initial genome-wide scan was performed using DNA from 25 F2 backcross progeny and 51 polymorphic microsatellites. The markers were selected about 30 cM apart across mouse autosomes [18]. Once linkage to chromosome 8 was established, additional markers from chromosome 8 were selected and all 145 F2 backcross progeny were typed.
Histology
Whole eyes were fixed in 4% paraformaldehyde in phosphate-buffered saline for 24 h, dehydrated for 20 min through increasing concentrations (50, 75, and 95%) of ethanol, paraffin embedded, serially sectioned (5 μm), and stained with hematoxylin and eosin.
Hsf4 exon scanning
Genomic PCR was carried out in 25-μl volumes containing 100 ng genomic DNA, a 0.2 mM concentration of each primer, a 0.315 mM concentration of each dNTP, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.625 U Taq polymerase (Invitrogen, Carlsbad, CA, USA). Reactions were carried out as follows: 95°C (3 min); 30 cycles of 95°C (30 s), annealing temperature as indicated in Table 1 (30 s), 72°C (30 s); final extension 72°C (7 min). Primer sequences are summarized in Table 1. PCR products were electrophoresed on 6% acrylamide gels, stained with ethidium bromide, photographed, and purified with Microcon centrifugal filter devices (Millipore, Billerica, MA, USA). The resultant PCR products were sequenced directly with the AmpliTaq FS sequencing kit (Applied Biosystems, Foster City, CA, USA), and sequencing analysis was done with the ABI 310 genetic analyzer. Comparative sequence analysis was done with DNAStar software (Madison, WI, USA).
Table 1.
PCR primers used to amplify genomic and cDNA segments of Hsf4
| Target | Primer name | Primer sequence (5′–3′) | Annealing temperature (°C) |
|---|---|---|---|
| HSF4 exon 1 | HSF4 1 F | CACTTTCGCGGCTTTGAC | 60 |
| HSF4 1.1 R | TCTCCCCATCTTTGCTTCAC | ||
| HSF4 intron 1 | HSF4 Intron 1 F | CCTCAACTCCTTCAGTGGTC | 60 |
| HSF4 Intron 1 R | TCACTTACGAGGAAGCTGGT | ||
| HSF4 exons 2–4 | HSF4 2 F | TGGACCCCCACAGTGAGTAT | 65 |
| HSF4 2 R | GAGGCTCCAACCAAAGAGC | ||
| HSF4 exons 5–6 | HSF4 3.1 F | GTATCCCCTTGCTGGCAAC | 63 |
| HSF4 3 R | CTGGAGGGTCTTTGGATGTG | ||
| HSF4 intron 6 | HSF4 Intron 6 F | GTGGTGTTGACACCTGCAT | 60 |
| HSF4 Intron 6 R | CATTGAATTTGGCAGATGCT | ||
| HSF4 exon 7 | HSF4 4.1 F | TTCCACTCCAGCATGTGACT | 62 |
| HSF4 4.1 R | CTAGGTCTGGGTGGTCGAGT | ||
| HSF4 intron 7 | HSF4 Intron 7 F | CCTCCAGGACCCCTACTTTA | 60 |
| HSF4 Intron 7 R | CCTGTGTCCTTCAGGAGATG | ||
| HSF4 exon 8 | HSF4 5 F | CCCAGCACAGATCATTTTCA | 60 |
| HSF4 5 R | TGCACCAGAGAAAAGCTTCA | ||
| HSF4 intron 8 | HSF4 Intron 8 F | CTGAAGGACACAGGCTTTCT | 60 |
| HSF4 Intron 8 R | CTGTAGCTCCCTTTCCCTTC | ||
| HSF4 exon 9 | HSF4 6 F | AAAGAAGAGCCGGCCAGT | 62 |
| HSF4 6 R | AGCTGCCTCAGGACCATAAA | ||
| HSF4 intron 9 | HSF4 Intron 9 F | TGTACAACAGCCTGAACCAA | 60 |
| HSF4 Intron 9 R | TGCCTCAGTCTCCCAAGTAG | ||
| HSF4 intron 9 | HSF4 Mut 1 F | TGACTTCTCAGGGTATTCTCCAA | 60 |
| HSF4 Mut 1 R | TACATGGGCTTTAGGGGTTG | ||
| HSF4 intron 9 | HSF4 Mut 2 F | GATCTGGCAGGATGGTTCAT | 60 |
| HSF4 Mut 2 R | AGCCACCCTTCCTCTTTGTT | ||
| HSF4 intron 9 | HSF4 Mut 3 F | TAAATTAATTAAAAACAAAGAGGAAGG | 60 |
| HSF4 Mut 3 R | TGCTCTTGTCTTAACCCACTG | ||
| HSF4 intron 9 | HSF4 Mut 3.1 F | CAGTGGGTTAAGACAAGAGCA | 60 |
| HSF4 7.1 R | GCTCTGCTTCATCCGTCTCT | ||
| Intron 9 insertion | LR HSF4 1 F | CCCCTTTAGCACTCAGTGGGTTAAGACAAG | 62 |
| LR HSF4 1 R | GTCCTACCCCAGGCAAAGAGATACTCACT | ||
| HSF4 exons 10–12 | HSF4 EX10 F | CAATAGCAGTTCTTTATGCA | 55 |
| HSF4 EX10 R | ATTCAGACCGTGATGGCTTC | ||
| HSF4 intron 12 | HSF4 Intron 12 F | TGACGGTCAAGGAGTTGAAT | 60 |
| HSF4 Intron 12 R | GCCTGGACATCTAGCATGAG | ||
| HSF4 exon 13 | HSF4 7.6 F | GGAACTCTGGGCCTGGAT | 60 |
| (RT) HSF4 3 R | GGCTTTTTCAGAGGGATGCAG | ||
| HSF4 upstream | HSF4 PROM 1 F | TCCGTCCCCTCTGTACACTC | 60 |
| HSF4 PROM 1 R | GGGCTCGGAAAGTCCTAGTT | ||
| HSF4 5′ cDNA | (RT) HSF4 1 F | CTTCCTCGGCAAGCTATGG | 60 |
| (RT) HSF4 1 R | TTGGCTCCTGTACTGCTG | ||
| HSF4 mid-cDNA | (RT) HSF4 8.1 F | GAGTTTCAGCATCCGAGCTT | 55 |
| (RT) HSF4 9.1 R | CTGTAGCTCCCTTTCCCTTC | ||
| HSF4 3′ cDNA | (RT) HSF4 7.5 F | GAGAGTCTGCTACCCCCAAT | 58 |
| HSF4-ET chimeric cDNA | HSF4 Intron 12 R | GCCTGGACATCTAGCATGAG | |
| Splice 1.1 F | GAAGGGAAAGGGAGCTACAG | 58 | |
| Splice 1.1 R | TCTCTGCCATTCTTCAGGTC |
RT-PCR and Northern blot
Total RNA was isolated from whole P21 and P1 eyes using TRIzol reagent (Invitrogen) and was electrophoresed (20 μg/lane), blotted, and hybridized following a standard protocol [18]. RT-PCR was performed as previously described [19] using primers summarized in Table 1. For hybridization, the 5′ end of the Hsf4 cDNA was generated via RT-PCR from C57BL/6 mouse eyes using primers from Table 1. To generate an RT-PCR product of the alternatively spliced Hsf4–ET cDNA primers were selected to anneal in Hsf4 exon 9 and ETnII (Accession No. Y17106) as indicated in Table 1. As a control a glyceraldehyde-3-phosphate dehydrogenase (Gapdh) partial cDNA probe was generated via RT-PCR from C57BL/6 mouse kidneys using primers 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′ and 5′-CATGTAGGCCATGAGGTCCACCAC-3′. The probe was radiolabeled with [α-32P]dCTP (Amersham Biosciences, Piscataway, NJ, USA) using the Random Primed DNA Labeling Kit (Invitrogen) following the manufacturer's protocol. For each subsequent hybridization the probe was stripped and rehybridized as described previously [19].
Southern blot and long-range PCR (LR-PCR)
Genomic DNA (10 μg) from lop11/+ and lop11/lop11 was digested with HindIII and PstI, electrophoresed on 0.9% agarose, blotted, and hybridized as described previously [19]. The 3′ end Hsf4 cDNA probe was RT-PCR generated (Table 1) and radiolabeled with [α-32P]dCTP (Amersham Biosciences) using the Random Primed DNA Labeling Kit (Invitrogen) following the manufacturer's protocol. For LR-PCR primers were designed to amplify a region in Hsf4 intron 9 (Table 1). LR-PCR was performed using the GeneAmp kit (Applied Biosystems). The reactions were carried out in 100-μl volumes containing 100 ng genomic DNA and a 900 nM concentration of each primer following the manufacturer's protocol. Reactions were carried out as follows: 94°C (1 min); 35 cycles of 94°C (30 s), 62°C (30 s), 72°C (3 s); final extension 72°C (10 min). PCR products were electrophoresed on 1% agarose gels, stained with ethidium bromide, and photographed.
Western blots
Lenses from P21 eyes from C57BL/6 and lop11/lop11 were collected and protein was extracted following a standard protocol [20]. Forty micrograms of soluble protein extracts from each C57BL/6 and lop11/lop11 was SDS–PAGE electrophoresed and blotted as previously described [20]. As a positive control for Hsf4 protein 5 μg of Jurkat cell lysate (BD Biosciences, San Jose, CA, USA) was run. The filters were hybridized as previously described [20] with anti-Hsf4 monoclonal antibody (BD Biosciences) at a 1:100 dilution. Membranes were washed and hybridized with peroxidase-conjugated affinity-purified polyclonal anti-rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) at a 1:5000 dilution. To ensure even loading the blots were hybridized with β-actin monoclonal antibody (Sigma–Aldrich, St. Louis, MO, USA) at a 1:2000 dilution followed by peroxidase-conjugated affinity-purified polyclonal anti-rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories) at a 1:5000 dilution. The detection was performed using ECL Western blotting detection reagents (Amersham Biosciences) and a chemiluminescence kit as previously described [20].
Acknowledgments
This work was supported in part by NEI/NIH Grant EY015173, NEI Grant EY07758, a Core Grant for Vision Research at the Medical College of Wisconsin (P30 EY01931), The Morris Animal Foundation Grant D05CA-049, the Research to Prevent Blindness Foundation, a Vision Core Grant from the National Eye Institute to the University of Tennessee Health Science Center (EY031080), and an unrestricted grant to the Department of Ophthalmology at the University of Tennessee Health Science Center from Research to Prevent Blindness. We thank Janice Burke, Anna Fekete, and Mary Lorenzen for expert technical assistance in histology and advice on protein extractions and Western blotting.
References
- 1.Foster A. Cataract—A global perspective: output, outcome and outlay. Eye. 1999;13:449–453. doi: 10.1038/eye.1999.120. [DOI] [PubMed] [Google Scholar]
- 2.Asbell PA, Dualan I, Mindel J, Brocks D, Ahmad M, Epstein S. Age-related cataract. Lancet. 2005;365:599–609. doi: 10.1016/S0140-6736(05)17911-2. [DOI] [PubMed] [Google Scholar]
- 3.Zetterstrom C, Lundvall A, Kugelberg M. Cataracts in children. J Cataract Refract Surg. 2005;31:824–840. doi: 10.1016/j.jcrs.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Reddy MA, Francis PJ, Berry V, Bhattacharya SS, Moore AT. Molecular genetic basis of inherited cataract and associated phenotypes. Surv Ophthalmol. 2004;49:300–315. doi: 10.1016/j.survophthal.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Francis PJ, Moore AT. Genetics of childhood cataract. Curr Opin Ophthalmol. 2004;15:10–15. doi: 10.1097/00055735-200402000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Graw J. Congenital hereditary cataracts. Int J Dev Biol. 2004;48:1031–1044. doi: 10.1387/ijdb.041854jg. [DOI] [PubMed] [Google Scholar]
- 7.Pras E, et al. A missense mutation in the LIM2 gene is associated with autosomal recessive presenile cataract in an inbred Iraqi Jewish family. Am J Hum Genet. 2002;70:1363–1367. doi: 10.1086/340318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pras E, et al. A nonsense mutation (W9X) in CRYAA causes autosomal recessive cataract in an inbred Jewish Persian family. Invest Ophthalmol Visual Sci. 2000;41:3511–3515. [PubMed] [Google Scholar]
- 9.Smaoui N, et al. A homozygous splice mutation in the HSF4 gene is associated with an autosomal recessive congenital cataract. Invest Ophthalmol Visual Sci. 2004;45:2716–2721. doi: 10.1167/iovs.03-1370. [DOI] [PubMed] [Google Scholar]
- 10.Forshew T, et al. Locus heterogeneity in autosomal recessive congenital cataracts: linkage to 9q and germline HSF4 mutations. Hum Genet. 2005;117:452–459. doi: 10.1007/s00439-005-1309-9. [DOI] [PubMed] [Google Scholar]
- 11.Pras E, et al. A nonsense mutation in the glucosaminyl (N-acetyl) transferase 2 gene (GCNT2): association with autosomal recessive congenital cataracts. Invest Ophthalmol Visual Sci. 2004;45:1940–1945. doi: 10.1167/iovs.03-1117. [DOI] [PubMed] [Google Scholar]
- 12.Riazuddin SA. Mutations in betaB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Visual Sci. 2005;46:2100–2106. doi: 10.1167/iovs.04-1481. [DOI] [PubMed] [Google Scholar]
- 13.Chang B, et al. Identification of a missense mutation in the alphaA-crystallin gene of the lop18 mouse. Mol Vision. 1999;5:21. [PubMed] [Google Scholar]
- 14.Bu L, et al. The γS-crystallin gene is mutated in autosomal recessive cataract in mouse. Genomics. 2002;80:38–44. doi: 10.1006/geno.2002.6803. [DOI] [PubMed] [Google Scholar]
- 15.Jablonski MM, et al. The ldis1 lens mutation in RIIIS/J mice maps to chromosome 8 near cadherin 1. Mol Vision. 2004;10:577–587. [PubMed] [Google Scholar]
- 16.Bu L, et al. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet. 2002;31:276–278. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- 17.Chang B, Hawes NL, Smith RS, Heckenlivel JR, Davisson MT, Roderick TH. Chromosomal localization of a new mouse lens opacity gene (lop18) Genomics. 1996;36:171–173. doi: 10.1006/geno.1996.0439. [DOI] [PubMed] [Google Scholar]
- 18.Sidjanin DJ, et al. A 76-bp deletion in the Mip gene causes autosomal dominant cataract in Hfi mice. Genomics. 2001;74:313–319. doi: 10.1006/geno.2001.6509. [DOI] [PubMed] [Google Scholar]
- 19.Sidjanin DJ, et al. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum Mol Genet. 2002;11:1823–1833. doi: 10.1093/hmg/11.16.1823. [DOI] [PubMed] [Google Scholar]
- 20.Youn Y, Hong J, Burke JM. Endogenous N-cadherin in a subpopulation of MDCK cells: distribution and catenin complex composition. Exp Cell Res. 2005;303:275–286. doi: 10.1016/j.yexcr.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Brulet P, Condamine H, Jacob F. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc Natl Acad Sci USA. 1985;82:2054–2058. doi: 10.1073/pnas.82.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baust C, Gagnier L, Baillie GJ, Harris MJ, Juriloff DM, Mager DL. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J Virol. 2003;77:11448–11458. doi: 10.1128/JVI.77.21.11448-11458.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiels A, Bassnett S. Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat Genet. 1996;12:212–215. doi: 10.1038/ng0296-212. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann M, Harris M, Juriloff D, Boehm T. Spontaneous mutations in SELH/Bc mice due to insertions of early transposons: molecular characterization of null alleles at the nude and albino loci. Genomics. 1998;15:107–109. doi: 10.1006/geno.1998.5409. [DOI] [PubMed] [Google Scholar]
- 25.Min JN, Zhang Y, Moskophidis D, Mivechi NF. Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis. 2004;40:205–217. doi: 10.1002/gene.20087. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto M, et al. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23:4297–4306. doi: 10.1038/sj.emboj.7600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somasundaram T, Bhat SP. Developmentally dictated expression of heat shock factors: exclusive expression of HSF4 in the postnatal lens and its specific interaction with alphaB-crystallin heat shock promoter. J Biol Chem. 2004;279:44497–44503. doi: 10.1074/jbc.M405813200. [DOI] [PubMed] [Google Scholar]
- 28.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanabe M, et al. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem. 1999;274:27845–27856. doi: 10.1074/jbc.274.39.27845. [DOI] [PubMed] [Google Scholar]


