Abstract
Serum levels of HDL are inversely correlated with the risk of coronary heart disease. The anti-atherogenic effect of HDL is partially mediated by its major protein constituent apoA-I. In this study, we identify bile acids that are activators of the nuclear receptor farnesoid X receptor (FXR) as negative regulators of human apoA-I expression. Intrahepatocellular accumulation of bile acids, as seen in patients with progressive familial intrahepatic cholestasis and biliary atresia, was associated with diminished apoA-I serum levels. In human apoA-I transgenic mice, treatment with the FXR agonist taurocholic acid strongly decreased serum concentrations and liver mRNA levels of human apoA-I, which was associated with reduced serum HDL levels. Incubation of human primary hepatocytes and hepatoblastoma HepG2 cells with bile acids resulted in a dose-dependent downregulation of apoA-I expression. Promoter mutation analysis and gel-shift experiments in HepG2 cells demonstrated that bile acid–activated FXR decreases human apoA-I promoter activity by a negative FXR response element mapped to the C site. FXR bound this site and repressed transcription in a manner independent of retinoid X receptor. The nonsteroidal synthetic FXR agonist GW4064 likewise decreased apoA-I mRNA levels and promoter activity in HepG2 cells.
Introduction
Epidemiological studies have shown that low HDL cholesterol levels in serum are associated with an increased risk of coronary heart disease (1). Recently, results from the Veterans Affairs High-Density Lipoprotein Cholesterol Intervention Trial (VAHIT) indicated that pharmacological elevation of HDL concentrations may contribute to a reduction of coronary events (2). A major protein component of HDL (3), apoA-I promotes cholesterol and phospholipid efflux from tissues via the ABCA1 pathway (4), the first step of reverse cholesterol transport (5). During this process, cholesterol is transported from peripheral tissues to the liver, where it is converted into bile acids and secreted together with free cholesterol into the bile (5, 6). The demonstration that mice overexpressing human apoA-I are protected from atherosclerosis provided direct evidence for the protective action of this apolipoprotein (7). Given the pivotal role of apoA-I in the reverse cholesterol transport pathway, a better understanding of mechanisms that regulate apoA-I synthesis and thus control HDL levels is of potential therapeutic importance.
The farnesoid X receptor (FXR) (NR1H4) is a member of the nuclear receptor superfamily and is expressed in the intestine, kidney, and liver (8, 9) and binds to FXR response elements (FXREs) after heterodimerization with the retinoid X receptor (RXR). The optimal DNA-binding sequence for the FXR/RXR heterodimer is an inverted repeat composed of two AGGTCA half-sites spaced by one nucleotide (IR-1) (10). Recently, it was shown that bile acids are ligands for FXR (11–13) and that FXR negatively regulates bile acid production by decreasing transcription of Cyp7A, which encodes a rate-limiting enzyme of the bile acid synthesis pathway (14, 15). In addition, FXR activates transcription of the intestinal bile acid–binding protein (I-BABP) (16), which is a cytosolic intestinal protein that binds bile acids with high affinity and is implicated in their enterohepatic circulation (17), and also transcription of the bile salt export pump (BSEP) (18), which promotes bile acid efflux from the liver.
Recent findings have shown that FXR regulates lipoprotein metabolism. Genetic evidence came from FXR-deficient mice (19) that display elevated serum cholesterol and triglyceride levels. Moreover, FXR mediates bile acid induction of phospholipid transfer protein expression (20), which plays a role in HDL remodeling (21). Interestingly, results from previous studies using bile acid sequestrants demonstrated that disruption of bile acid enterohepatic recirculation significantly increases HDL concentrations both in humans (22, 23) and animals (24). Moreover, cholestyramine treatment increases serum concentrations of lipoprotein A-I (LpA-I) (22), which is an antiatherogenic apoA-I–containing HDL particle. In addition, it has been reported that treatment of gallstone patients, as well as patients with cerebrotendinous xanthomatosis, with chenodeoxycholic acid, a relatively hydrophobic primary bile acid and FXR agonist, lowers serum HDL levels (25, 26). Taken together, these observations point to a potential role for FXR in HDL and, possibly, apoA-I metabolism.
The goal of this study was to determine whether bile acid FXR agonists influence HDL metabolism through regulation of apoA-I gene expression. Serum apoA-I concentrations were reduced in patients with hepatocellular progressive familial intrahepatic cholestasis (PFIC) or bile duct–related cholestatic liver disease biliary atresia (BA), experiencing systemic and intrahepatic bile acid accumulation. Since the human apoA-I transgenic mouse model has proven a good model to study the in vivo regulation of human apoA-I gene expression (27), the influence of bile acid treatment on human apoA-I expression and serum HDL metabolism was subsequently studied in these mice. Our results show that bile acids downregulate apoA-I expression at both mRNA and protein levels and reduce serum HDL. Furthermore, using human hepatoblastoma HepG2 cells, it is demonstrated that bile acids, as well as the synthetic FXR agonist GW4064, repress apoA-I mRNA levels and promoter transcription via activation of FXR. The C site was identified as the negative FXRE in the human apoA-I promoter to which FXR binds independently of RXR. These findings indicate that bile acids control serum apoA-I and HDL levels.
Methods
Patients.
The apoA-I levels and liver function parameters were investigated in children with PFIC with low γ-glutamyltransferase (GGT) activity (mean 17 ± 8 U/l, n = 15), BA (n = 15), or in healthy volunteers (n = 110). Diagnosis was made on the basis of typical clinical symptoms, biochemical abnormalities, histological examination, and/or surgical exploration.
Determination of total serum bile acids and apoA-I in patients.
GGT was measured by standard methods. Total bile acid concentrations in serum were determined enzymatically as described (28). The serum concentrations of apoA-I were measured by competitive solid-phase ELISA using mAb’s (29) or in a turbidometric assay (Serapak; Bayer AG, Leverkusen, Germany). Statistical significance was calculated by ANOVA and a post-hoc test was performed using Dunnett’s T3 test for multigroup comparison (SPSS Science, Chicago, Illinois, USA).
Animals and treatment.
Female human apoA-I transgenic mice (30) in a C57BL/6 genetic background (IFFA-CREDO, L’Arbresle, France) were divided into two groups of animals (n = 4 each) randomized on serum apoA-I levels. One group received a standard rodent chow diet (control), whereas the other group received the same diet containing 0.5% (wt/wt) taurocholic acid (TCA) (Sigma-Aldrich, L’Isle d’Abeau Chesnes, France) for 5 days. Blood was obtained after a 4-hour fasting period by retro-orbital puncture under barbital-induced anesthesia, and serum was kept at –20°C until analysis. At the end of the treatment animals were sacrificed, organ weights were recorded, and livers were collected and frozen until further analysis.
Analysis of serum lipids and apoA-I levels in mice.
Serum cholesterol concentrations were determined by an enzymatic assay using commercially available reagents (Cholesterol C System; Boehringer Mannheim GmbH, Mannheim, Germany). Serum human apoA-I levels were determined by immunonephelometry using the Berhing Nephelometer Apparatus system and apoA-I standards (Behringwerke AG, Marburg, Germany). Lipoprotein cholesterol profiles were determined by fast protein liquid chromatography (FPLC). Two-hundred microliters of serum were injected into a Superose 6HR 10/30column (Pharmacia LKB Biotechnology, Uppsala, Sweden), and lipoproteins were separated at a flow rate of 0.2 ml/min. The elution profile was monitored at 280 nm and recorded using an analogue recorder chart-tracing system (Pharmacia LKB Biotechnology). The effluents were collected in 0.26-ml fractions. Cholesterol was measured in each collected fraction using commercially available enzymatic kits (Cholesterol C System; Boehringer Mannheim GmbH).
Cell culture and transfection assays.
Human primary hepatocytes were isolated as described previously (31) and were incubated 48 hours in dexamethasone-free William’s E medium without serum containing chenodeoxycholic acid (CDCA; 10 μM). HepG2 cells and HepG2 cells expressing the Na+-TCA cotransporting polypeptide (NTCP) (32) were grown in DMEM supplemented with 10% FCS, streptomycin/penicillin, sodium pyruvate, glutamine, and nonessential amino acids (Life Technologies, Cergy-Pontoise, France) at 37°C in a humidified 5% CO2 atmosphere. The medium was changed every 48 hours. HepG2 cells were treated with either CDCA or TCA at the indicated concentrations for 36 hours (dose response experiment) or with either GW4064 (5 μM) or CDCA (75 μM) for the indicated times (time course experiment). GW4064 was synthesized in the chemistry department of Genfit SA (Loos, France), according to Maloney et al. (33). HepG2 cells were transfected using Lipofectamine (Life Technologies). One microgram of the indicated firefly luciferase reporter plasmids were cotransfected with or without 300 ng of pcDNA3 hFXR, 300 ng of pSG5 mRXRα, or pSG5 (Stratagene, La Jolla, California, USA) and pcDNA3 (Invitrogen, Leek, The Netherlands) empty vectors as control. All samples were complemented with pBSK+ plasmid (Stratagene) to an identical total amount of DNA (2 μg). After 3 hours, cells were incubated with medium containing 10% FCS with or without CDCA (75 μM) or GW4064 (100 nM), and luciferase activity was assayed 36 hours later using a LB 9507 LUMAT luminometer. Transfection efficiency was monitored by cotransfecting 300 ng of a CMV-driven β−galactosidase expression plasmid. Transfection experiments were done at least three times in triplicate.
RNA analysis, RT-PCR, and Q-PCR.
Total RNA was isolated from livers or cells by the acid guanidinium thiocyanate/phenol/chloroform method (34), and Northern blot analysis was performed as described (35). Human and mouse apoA-I and 28S cDNAs were used as probes, and filters were hybridized to 106 cpm/ml of each probe. For RT-PCR analysis of FXR expression, total RNA from HepG2 or HepG2 NTCP cells was reverse transcribed using random hexamer primers and Superscript reverse transcriptase (Life Technologies, Paisley, United Kingdom) and subsequently amplified by PCR using as primers the NR1H4S (5′-GAAGTGGAACCATACTCGCAATACA-3′) and the NR1H4AS (5′-AACATAGCTTCAACCGCAGACC-3′) oligonucleotides (fragment size: 861 bp). For positive control, pcDNA3 hFXR was used. The resulting products were separated on a 1% agarose gel and stained with ethidium bromide. For quantitative PCR, reverse-transcribed SHP and 28S cDNAs were quantified by real-time PCR on a MX 4000 apparatus (Stratagene), using specific oligonucleotide primers SHP: 5′-CTCTTCCTGCTTGGGTTGGC-3′ and 5′-GCACATCGGGGTTGAAGAGG-3′, and 28S: 5′-AAACTCTGGTGGAGGTCCGT-3′ and 5′-CTTACCAAAAGTGGC CCACTA-3′ (36). PCR amplification was performed in a volume of 25 μl containing 100 nM of each primer, 4 mM MgCl2, the Brilliant Quantitative PCR Core Reagent Kit mix as recommended by the manufacturer (Stratagene), and SYBR Green 0.33X (Sigma-Aldrich). The conditions were 95°C for 10 minutes, followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C. SHP mRNA levels were subsequently normalized to 28S mRNA.
Plasmid cloning and site-directed mutagenesis.
The human apoA-I promoter was cloned as described previously (37). C3 TKpGL3 constructions were obtained by cloning three copies of the C (5′-CAGAGCTGAT CCTTGAACTCTTAAGTT-3′) site in the TKpGL3 luciferase vector. Mutations were introduced in the C site of the human apoA-I promoter using the Quick Change Site Directed Mutagenesis Kit (Stratagene) and the oligonucleotides Cmt (5′-CAGAGCTGATCCTTATATATTAAGTT-3′) and CGG (5′-CAGAGCTGATCCT TGAAGTGTTAAGTT-3′), where nucleotides in bold indicate the mutated bases. To create a FXR mutant that is defective in heterodimerization with RXR, the Leu at position 433 in FXR was mutated into Arg using the oligonucleotide Leu433Arg (5′-CCACTTCTTGATGTGCGACAAAAG TTGTGTAAG-3′). The mutated nucleotide is indicated in bold.
Electrophoretic mobility shift assays (EMSA).
Human wild-type and mutated FXR and mRXRα proteins were synthesized in vitro using the TNT Quick Coupled Transcription/Translation System (Promega, Madison, Wisconsin, USA). Double-stranded oligonucleotides were end labeled with γ-32P ATP using T4-polynucleotide kinase. FXR and/or RXR proteins (2 μl) were incubated for 15 minutes at room temperature in a total volume of 20 μl with 2.5 μg poly(dI-dC) and 1 μg herring sperm DNA in binding buffer (10 mM HEPES, pH 7.6, 2.5 mM MgCl2, 10% glycerol, 2.5 mg/ml BSA, 50 mM NaCl, and 0.5 mM DTT) before the radiolabeled probe (0.5 ng) was added. Binding reactions were further incubated for 15 minutes and resolved by 6% nondenaturing polyacrylamide gel electrophoresis in 0.25X Tris-Borate-EDTA (TBE) buffer at room temperature. For competition experiments, the following unlabeled oligonucleotides were included in the binding reaction at the indicated excess concentrations over the probe just before adding the labeled oligonucleotide: apoA-I C wild-type (Cwt: 5′-CAGAGCTGATCCTTGAACTCTTAAGTT-3′) and mutated oligonucleotides (Cmt: 5′-CAGAGCTGATCCTTATATATTAAGTT-3′ or CGG: 5′-CAGAGCTGATCCTTGAAGTGTTAAGTT-3′); a FXR IR-1 consensus response element-containing oligonucleotide (IR-1: 5′-GATCTCAAGAGGTCATTGACCTT TTTG-3); and the natural I-BABP FXR response element–containing oligonucleotide (I-BABP: 5′-CCCCAGGGTGAATAACCTCGGGG-3′). For mapping of the FXRE in the C site, labeled apoA-I C wild-type, Cmt, or CGG oligonucleotides were included in the binding reaction mixture.
Results
Low serum apoA-I concentrations in patients with low GGT-type PFIC.
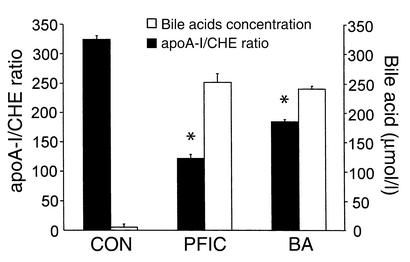
To determine the influence of different pathophysiological defects leading to cholestasis on serum apoA-I levels in humans, serum apoA-I was determined in patients with PFIC leading to hepatocellular cholestasis through BSEP deficiency (38) and BA with primarily ductular cholestasis. Serum apoA-I levels in PFIC and BA patients were low when compared with normal controls (PFIC: 0.26 ± 0.21 g/l; normal controls: mean 0.8–1.1 g/l, depending on age). To exclude alterations secondary to diminished liver synthetic function, serum apoA-I levels were normalized to cholinesterase (CHE) levels. In PFIC and BA patients the apoA-I/CHE ratio was significantly lower when compared with controls. Bile acid accumulation in serum as a quantitative measure of cholestasis was similar in both groups of patients and strongly increased compared with controls (Figure 1). These observations suggest that apoA-I expression is repressed under pathophysiological conditions of intrahepatic bile acid accumulation.
Figure 1.
Low serum apoA-I in patients with progressive familial intrahepatic cholestasis. Serum apoA-I was determined in patients with low GGT-type PFIC (n = 15) in patients with BA (n = 15) or in normal control individuals (CON; n = 110). Serum apoA-I concentrations were normalized to CHE activities; data are expressed as means ± SEM. Serum total bile acid concentrations were also determined in the different groups as a marker of cholestasis. Bile acid concentration is expressed in micromoles per liter. Statistical analysis was performed by ANOVA (P < 0.001) followed by Dunnett’s T3 test. *P < 0.001 versus control.
Bile acid treatment reduces serum protein and liver mRNA levels of apoA-I in human apoA-I transgenic mice.
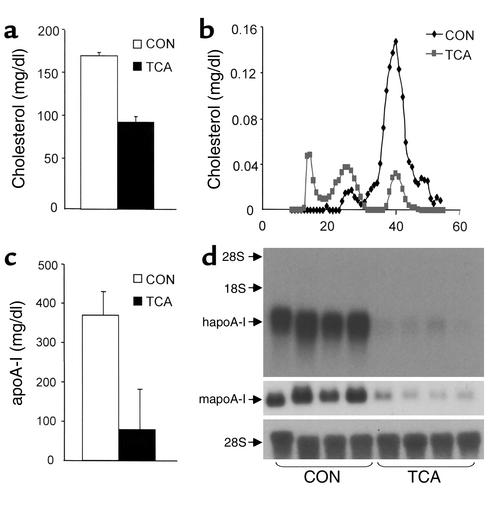
To study whether human apoA-I gene expression is indeed regulated in vivo by bile acids, human apoA-I transgenic mice were treated for 5 days with 0.5% TCA (wt/wt) or control chow diet. No changes in body weight or food intake were observed between control and treated groups (data not shown). TCA treatment led to a strong decrease of serum cholesterol levels (Figure 2a). The determination of lipoprotein cholesterol distribution demonstrated that the reduction of serum cholesterol was mainly due to a decrease in the HDL-sized fraction, whereas a slight increase of cholesterol in the VLDL- and LDL-sized fractions was observed (Figure 2b). Furthermore, serum concentrations of human apoA-I, the major apolipoprotein of HDL, were reduced in TCA-treated animals (Figure 2c). To evaluate whether the reduction of serum apoA-I levels was correlated with decreased apoA-I mRNA levels in liver in which the transgene is selectively expressed (7), Northern blot analysis was performed on liver RNA isolated from control and TCA-treated animals. Both human and mouse apoA-I mRNA levels were markedly decreased in livers of the TCA-treated mice (Figure 2d). Taken together these data indicate that both serum levels and hepatic mRNA levels of human apoA-I are downregulated by TCA feeding.
Figure 2.
TCA decreases serum HDL cholesterol, apoA-I concentrations, and liver apoA-I mRNA levels in human apoA-I transgenic mice. Human apoA-I transgenic mice (n = 4/group) were fed a diet containing 0.5% TCA (wt/wt) mixed in standard rodent chow for 5 days. Control mice received standard rodent chow (CON). ApoA-I and cholesterol concentrations were measured in serum after a 4-hour fasting period, as described in Methods. Values of serum total cholesterol (a) and human apoA-I (c) are expressed as means ± SD. Sera of control and treated mice were pooled and cholesterol profiles analyzed by FPLC (b). Human and mouse apoA-I mRNA levels in the liver (d) were analyzed by Northern blot. 28S mRNA was measured as control.
Bile acids decrease human apoA-I mRNA in human primary hepatocytes and HepG2 cells.
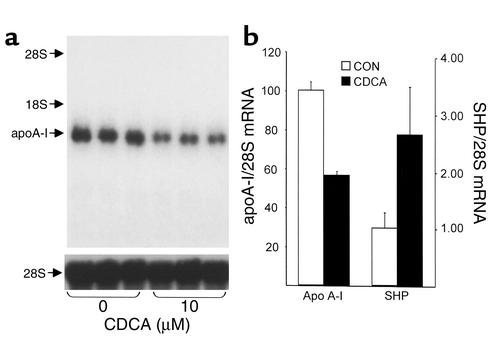
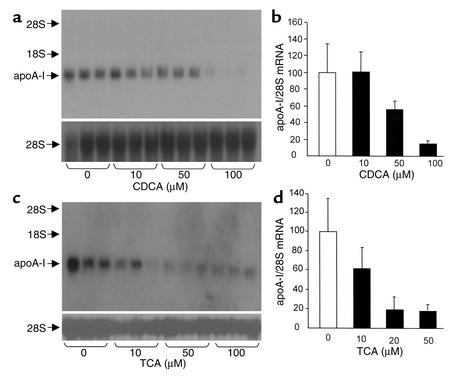
Human primary hepatocytes and human hepatoblastoma HepG2 cells were used to investigate the molecular and cellular mechanisms by which bile acids regulate human apoA-I gene expression. For this purpose, human primary hepatocytes were incubated for 48 hours with 10 μM CDCA, and apoA-I mRNA levels were determined by Northern blot analysis (Figure 3a). An approximately 50% decrease in the level of human apoA-I mRNA was observed in treated cells when compared with vehicle-treated cells (Figure 3). As a control, mRNA levels of SHP, a well-characterized FXR target gene (14, 15), were measured. Treatment with CDCA resulted in a 2.7-fold induction of SHP mRNA (Figure 3b). Then, HepG2 cells were treated for 36 hours with increasing CDCA concentrations, and apoA-I mRNA levels were determined by Northern blot analysis (Figure 4a). A strong decrease in the level of human apoA-I mRNA was observed in CDCA-treated HepG2 cells when compared with control cells treated with vehicle (Figure 4, a and b). Moreover, this reduction occurred in a dose-dependent manner. Since hydrophilic bile acids, such as TCA, require the presence of a specific transporter to be imported into cells (39), HepG2 cells stably transfected with the NTCP were treated for 36 hours with TCA at various concentrations, and apoA-I mRNA levels were determined by Northern blot analysis (Figure 4c). A reduction of apoA-I mRNA levels occurred in TCA-treated cells when compared with vehicle-treated cells (Figure 4, c and d). This reduction was already maximal at 20 μM of TCA, a concentration likely to saturate the transporter (32). Moreover, under the same conditions, an increase of SHP mRNA was observed (1.9- ± 0.4-fold of untreated control at 20 μM of TCA). Taken together, these data indicate that both hydrophobic and hydrophilic bile acids negatively regulate human apoA-I gene expression in primary hepatocytes and HepG2 cells.
Figure 3.
Bile acids decrease apoA-I mRNA levels in primary human hepatocytes. (a and b) Primary human hepatocytes were incubated for 48 hours with CDCA (10 μM) or vehicle. (a) Human apoA-I mRNA was analyzed by Northern blot. 28S mRNA was measured as control. (b) Densitometric quantification of apoA-I/28S mRNA levels. Values are expressed as a percentage of mRNA levels in control cells and are means ± SD (n = 3). SHP mRNA levels were measured by quantitative RT-PCR and normalized to 28S RNA levels as control. Values are expressed as fold induction of mRNA levels in control cells and are means ± SD (n = 3).
Figure 4.
Bile acids downregulate apoA-I mRNA levels in human hepatoblastoma HepG2 cells. (a and b) HepG2 cells were incubated for 36 hours with CDCA at the indicated concentrations or with vehicle. (c and d) HepG2 cells, stably transfected with the NTCP expression vector, were incubated for 36 hours with TCA at the indicated concentrations or with vehicle. (a and c) Human apoA-I mRNA levels were measured by Northern blot analysis. 28S mRNA was measured as internal control. (b and d) Densitometric quantification of apoA-I/28S mRNA levels. Values are expressed as percentage of mRNA levels in control cells and are means ± SD (n = 3).
FXR is expressed in HepG2 and HepG2 NTCP cells.
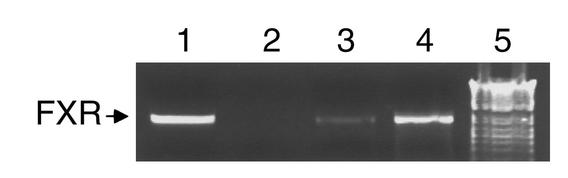
Since transcriptional regulation by bile acids may be mediated by the FXR (11–13), it was investigated whether FXR is expressed in the HepG2 cells used for these studies. RT-PCR was performed on RNA extracted from both HepG2 and HepG2 NTCP (Figure 5) cells using specific oligonucleotide primers. A 821-bp amplicon representing the FXR transcript was obtained from both the HepG2 and HepG2 NTCP cells (Figure 5, lanes 3 and 4). These results demonstrate that FXR is expressed in HepG2 cells and thus could mediate the effects of bile acids on apoA-I expression in these cells.
Figure 5.
Bile acids decrease human apoA-I expression in HepG2 cells, which express the bile acid nuclear receptor FXR. RT-PCR analysis was performed using RNA from HepG2 (lane 3) and HepG2 NTCP (lane 4) cells with oligos designed to amplify FXR. pcDNA3 hFXR was used as positive control (lane 1) and water as negative control (lane 2). The 1-kb ladder from Gibco was used as size control (lane 5) (Life Technologies).
Mapping of an FXR response element in the human apoA-I promoter.
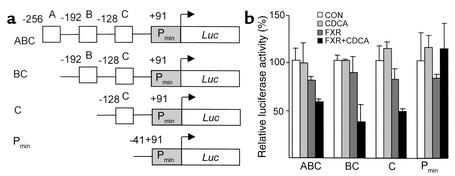
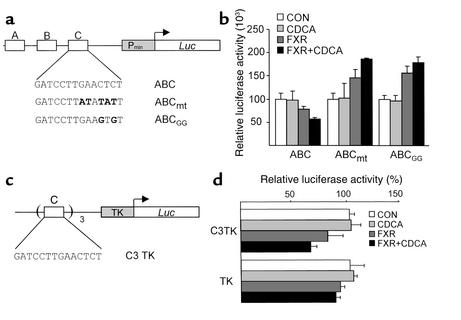
To test the hypothesis that FXR regulates apoA-I gene expression, HepG2 cells were transfected with different 5′ deletion constructs of the apoA-I promoter (Figure 6a) in the presence or absence of FXR and/or CDCA. As shown in Figure 6b, FXR slightly decreased activity of the apoA-I promoter containing the A, B, and C sites, an effect that was enhanced by the presence of CDCA. This repression was still observed on the BC and C deletion constructs, but was abolished for the Pmin-containing apoA-I promoter construct. Therefore, the C site appears to contain a site negatively regulated by FXR. To test whether this site could mediate the repression by FXR, mutations in the C site were introduced in the context of the ABC promoter construct (Figure 7a). Mutations in the C site abolished the repression of apoA-I promoter activity by CDCA-activated FXR (Figure 7b). Next, the C site was cloned in 3 copies in front of the heterologous thymidine kinase promoter–driven luciferase reporter gene (Figure 7c), and cotransfection experiments were performed. Activity of the TKpGL3 vector driven by three copies of the C site decreased in the presence of CDCA-activated FXR, whereas no change in activity of the TKpGL3 control vector was observed (Figure 7d). Taken together, these results indicate that the C site is a negative cis-acting element by which FXR represses human apoA-I promoter activity.
Figure 6.
Bile acids downregulate human apoA-I promoter activity by FXR acting on the C site. (a) Schematic representation of the human apoA-I promoter constructs. (b) HepG2 cells were transfected with the indicated human apoA-I promoter-driven luciferase (Luc) reporter plasmid (1 μg) in the presence of either the empty pcDNA3 or FXR- and RXR-containing expression vectors (300 ng). Cells were subsequently treated with CDCA (75 μM) or vehicle for 36 hours. Values are expressed as percentage of control set at 100% normalized to internal control β-galactosidase activity as described in Methods.
Figure 7.
The human apoA-I promoter C site confers negative FXR responsiveness to both the human apoA-I and the heterologous thymidine kinase promoter. (a and b) Mutations (in bold) were introduced in the human apoA-I ABC-luc vector. HepG2 cells were transfected with the wild-type or C site–mutated human apoA-I promoter constructs. Reporter plasmid (1 μg) was transfected in the presence of either the empty pcDNA3 or FXR- and RXR-containing expression vectors (300 ng). Cells were subsequently treated with CDCA (75 μM) or vehicle (b) for 36 hours. (c and d) The C site was cloned in three copies in front of the TKpGL3 vector. (c) Reporter plasmid (1 μg) was transfected in the presence of either the empty pcDNA3 or FXR- and RXR-containing expression vectors (300 ng) and cells subsequently treated with CDCA (75 μM) or vehicle. Values are expressed as percentage of control set at 100% and normalized to internal control β−galactosidase activity as described in Methods.
Mapping of the FXR-binding site in the C site.
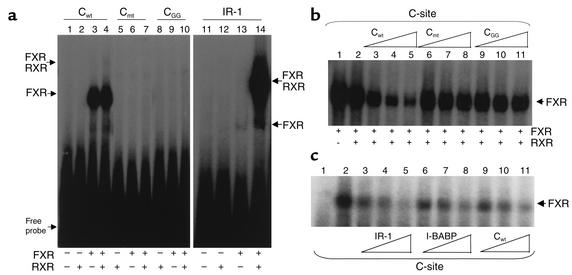
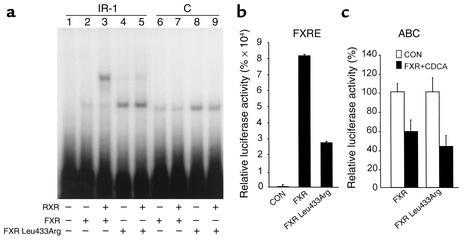
To determine whether human FXR can bind to the C site, gel shift assays were performed. FXR in the presence of mRXR bound as a heterodimer to the consensus IR-1 site (Figure 8a, lane 14). Interestingly, FXR alone already bound to the C site, whereas only a very slight binding of the FXR/RXR heterodimer was observed in the presence of RXR (Figure 8a, lanes 3 and 4). The mutations functionally tested in a transfection assay (Figure 7) were also used as probes in gel shift analysis. FXR in the absence of mRXR bound to the C site (Figure 8a, lane 3). However, FXR did not bind the oligonucleotides carrying the mutated C site, Cmt (Figure 8a, lanes 6 and 7), or CGG (Figure 8a, lane 9 and 10). The addition of increasing amounts of the unlabeled apoA-I Cwt oligonucleotide could compete for binding of FXR to the radiolabeled Cwt site (Figure 8b, lanes 3–5), whereas the apoA-I Cmt and CGG oligonucleotides did not compete for binding (Figure 8b, lanes 6–8 and 9–11, respectively). Thus, FXR alone was able to bind to the apoA-I Cwt site in a manner independent of RXR.
Figure 8.
FXR binds the human apoA-I promoter C site. (a) Gel retardation assays were performed on end-labeled Cwt (lanes 1–4), Cmt (lanes 5–7), CGG (lanes 8–10), and IR-1 consensus FXRE (lanes 11–14) oligonucleotides in the presence of in vitro–transcribed/translated unprogrammed reticulocyte lysate (lanes 1 and 11), mRXR (lanes 2, 5, 8, and 12), hFXR (lanes 3, 6, 9, and 13), or both mRXR and hFXR (lanes 4, 7, 10, and 14). (b) Gel retardation assays were performed on end-labeled wild-type C-site oligonucleotide in the presence of in vitro–transcribed/translated hFXR and mRXR. Competitions were performed by adding 100-fold (lanes 3, 6, and 9), 200-fold (lanes 4, 7, and 10), or 400-fold (lanes 5, 8, and 11) molar excess of cold Cwt wild-type (lanes 3–5), Cmt (lanes 6–8), or CGG (lanes 9–11) oligonucleotides, respectively. (c) Gel retardation assays were performed on end-labeled wild-type C-site oligonucleotide in the presence of in vitro–transcribed/translated hFXR and mRXR proteins. Competitions were performed by adding tenfold (lanes 2, 5, and 8), 50-fold (lanes 3, 6, and 9), or 100-fold (lanes 4, 7, and 10) molar excess of cold IR-1 consensus FXRE (lanes 2–4), I-BABP (lanes 5–7), or Cwt wild-type (lanes 8–10) oligonucleotides, respectively.
Finally, the relative affinity of the C site compared with consensus IR-1 and the natural FXRE from the I-BABP promoter was examined by competition gel shift analysis (Figure 8c). Binding of FXR to the C site was competed to a similar extent by cold C site as by the natural I-BABP FXRE(Figure 8c, lanes 9–11 and 6–8, respectively), and IR-1 oligonucleotide (Figure 8c, lanes 3–5). Therefore the apoA-I C site binds FXR with an affinity similar to the natural functionally active I-BABP FXRE (16).
A RXR dimerization-incompetent FXR binds the apoA-I C site and negatively regulates apoA-I promoter activity.
To unequivocally demonstrate that FXR binds to the C site in a manner independent of RXR, a mutation was introduced by site-directed mutagenesis in helix 10 of FXR at position 433, replacing Leu by Arg. Such mutation previously has been shown to abolish RXR heterodimerization of nuclear receptor such as PPARα (40). The DNA-binding properties of this mutant Leu433Arg FXR was analyzed by gel shift assay and compared with wild-type FXR (Figure 9a). As expected, wild-type FXR bound primarily as a heterodimer with RXR to the IR-1 consensus–containing oligonucleotide. By contrast, Leu433Arg FXR did not significantly heterodimerize with RXR, but bound principally as a monomer or homodimer to the probe (Figure 9a). Interestingly, Leu433Arg FXR also bound to the C site, in a manner similar to wild-type FXR (Figure 9a, lanes 8 and 9 and 6 and 7, respectively). These results demonstrate that FXR binds to the C site of the apoA-I promoter independently of RXR.
Figure 9.
The RXR heterodimerization-incompetent FXR mutant Leu433Arg binds the C site and negatively regulates apoA-I promoter activity. (a) Gel retardation assays were performed on end-labeled IR-1 consensus (lanes 1–5) and wild-type C-site (lanes 6–9) oligonucleotides in the presence of unprogrammed reticulocyte lysate (lane 1) or in the presence of in vitro–transcribed/translated wild-type hFXR (lanes 2 and 6), mutant hFXR Leu433Arg (lanes 4 and 8), or both mRXR and wild-type (lanes 3 and 7) or mutant Leu433Arg (lanes 5 and 9) hFXR proteins. (b and c) HepG2 cells were transfected with a IR-1–driven (b) or apoA-I promoter–driven (c) reporter vectors (1 μg) in the presence of either the pcDNA3 wild-type or mutated FXR- and RXR-containing expression vectors (300 ng) as indicated. Cells were subsequently treated with CDCA (75 μM) or vehicle for 36 hours. Values are expressed as percentage of control set at 100% normalized to internal control β-galactosidase activity as described in Methods. FXRE, FXR response element.
Next, the transcriptional activity of Leu433Arg FXR was determined and compared with wild-type FXR by transient transfection experiments in HepG2 cells. Interestingly, Leu433Arg activated transcription of a reporter vector driven by the consensus IR-1 FXRE to a much lesser extent than wild-type FXR (Figure 9b). By contrast, Leu433Arg FXR repressed apoA-I promoter activity to an extent similar to wild-type FXR (Figure 9c). Taken together, these results demonstrate that FXR is able to decrease apoA-I gene transcription through its C site without involvement of RXR.
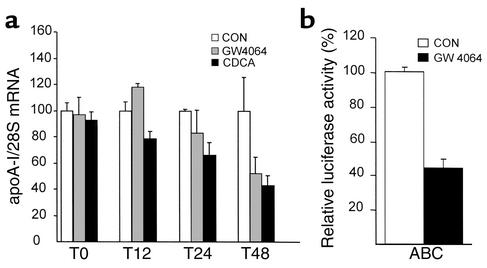
The nonsteroidal FXR agonist GW4064 represses apoA-I mRNA and promoter activity in HepG2 cells.
Since bile acids may exert FXR-independent effects by activating other signal transduction pathways (41, 42), the influence of a synthetic nonsteroidal FXR agonist on apoA-I expression was tested. Treatment of HepG2 cells with GW4064 led to a decrease of human apoA-I mRNA levels in a time-dependent manner (Figure 10a), albeit slightly delayed in time compared with CDCA. Moreover, GW4064 decreased activity of the apoA-I promoter in transient transfection assays (Figure 10b). These results demonstrate that FXR activation results in the repression of apoA-I expression.
Figure 10.
A nonsteroidal FXR agonist decreases mRNA levels and promoter activity of human apoA-I in HepG2 cells. (a) HepG2 cells were incubated for 0, 12, 24, and 48 hours with CDCA (75 μM), or GW4064 (5 μM), or vehicle. Human apoA-I mRNA levels were measured by Northern blot analysis. 28S mRNA was measured as internal control. (b) HepG2 cells were transfected with the indicated human apoA-I promoter–driven luciferase reporter plasmid (1 μg). Cells were subsequently treated with GW4064 (100 nM) or vehicle for 36 hours. Values are expressed as percentage of control set at 100% normalized to internal control β-galactosidase activity as described in Methods.
Discussion
Recent data from the VAHIT provided evidence that elevation of serum HDL levels contribute to a reduction in the incidence of coronary heart disease (2). Therefore, the identification of environmental and genetic factors controlling HDL levels remains an important objective with potential pharmacological applications. In the present study, we identified bile acids as major regulators of HDL metabolism through their inhibitory action on apoA-I expression. Primary intracellular accumulation of bile acids in hepatocytes due to deficiency of the bile acid–transporting protein, BSEP or ABCB11, at the canalicular membrane (38) or elevated bile acid concentrations secondary to impaired bile flow, such as in BA, may be the reason for the specific downregulation of serum apoA-I levels in these patient groups. Partial biliary diversion may lead to restoration of hepatocellular integrity and function with improvement of cholestasis in PFIC patients, and, as a consequence of the ensuing reduction in intracellular bile acid load, apoA-I synthesis appears to be restored (43). Therefore, it is tempting to speculate that the specific intracellular accumulation of bile acids in BSEP deficiency (PFIC) and BA may lead to suppressed apoA-I transcription.
To investigate the molecular mechanism for downregulation of apoA-I synthesis by bile acids, we used human apoA-I transgenic mice. We first demonstrated that bile acid feeding of human apoA-I transgenic mice reduces serum HDL cholesterol levels. The decrease of HDL cholesterol was associated with a strong lowering of serum human apoA-I concentrations and hepatic human and mouse apoA-I mRNA levels. Bile acid treatment also lowered apoA-I mRNA levels in human primary hepatocytes and in human hepatoblastoma HepG2 cells. Moreover, this regulation occurred in a dose-dependent manner and was observed with both relatively hydrophilic (TCA) and hydrophobic (CDCA) bile acids, as well as with the synthetic nonsteroidal GW4064 compound, which are ligands for the nuclear receptor FXR. Finally, it is shown that bile acid–activated FXR represses human apoA-I promoter activity, and a negative FXR response element was mapped in the C site of the promoter.
It has been reported previously that cholestyramine, a bile acid sequestrant, significantly increases hepatic apoA-I mRNA levels in animals (24) and serum apoA-I levels in both animals (24) and humans (22). In humans, cholestyramine treatment selectively increases the concentration of the LpA-I subfraction of HDL (44, 45), which contains only apoA-I and no apoA-II. Since cholestyramine interrupts the enterohepatic circulation of bile acids, this treatment should decrease exposure of hepatocytes to bile acids, thus potentially damping FXR activity and subsequently relieving its repressive activity on target genes such as apoA-I. Treatment of patients with CDCA for gallstone dissolution has the opposite effect and leads to reduced serum HDL levels (25). Similarly, CDCA treatment of patients with cerebrotendinous xanthomatosis, a disorder of sterol metabolism, also results in decreased serum HDL levels (26). Thus, our findings demonstrating that bile acids, which are FXR agonists, reduce human apoA-I expression and may provide a molecular mechanism for the observed effects in clinical practice of cholestyramine and of CDCA, as well as of inborn errors in bile acid transport (PFIC) on HDL and apoA-I levels (38, 43).
Recently, Sinal et al. (19) described the generation of FXR-deficient mice. Using a semiquantitative Western blot method, these authors reported a slight decrease in apoA-I levels in the FXR-deficient mice compared with wild-type mice. The effects of FXR deficiency on HDL were very modest, with no differences between wild-type and deficient mice when fed a chow diet and only a slight increase in the HDL fraction in FXR-deficient mice fed a high-cholesterol diet. Although the increase of serum HDL and phospholipid concentrations in FXR-deficient mice concurs with our findings, the reported decrease of serum apoA-I appears contradictory. One possible explanation is the existence of adaptive pathways that would counterbalance the expected induction of apoA-I levels in FXR-deficient mice. It is also important to note that the study reported only basal apoA-I and HDL levels, and the influence of bile acid treatment on these parameters in these mice was not analyzed. Since our data clearly demonstrate that bile acids not only reduce expression of the human transgene, but also that of the endogenous mouse apoA-I gene, it will be of interest to compare the influence of bile acid treatment on mouse apoA-I and HDL in wild-type and FXR-deficient mice. Finally, it is important to consider that apoA-I is expressed in both the liver and intestine (46, 47). In human apoA-I transgenic mice, the transgene is exclusively expressed in the liver (28). It may be that murine apoA-I is regulated in an opposite manner in the liver and in the intestine, which may attenuate the changes in plasma apoA-I in the FXR-deficient mice. Such a mechanism is unlikely to be operative for human apoA-I, since cholestyramine treatment results in an elevation of serum apoA-I concentration (22), whereas CDCA treatment and inherited cholestatic syndromes are associated with reduced serum apoA-I and HDL (25, 26, 43).
In the present study, it is shown that FXR can bind to the C site in the human apoA-I promoter. Moreover, mutations in the C site abolished both the FXR-DNA interaction and the repression of apoA-I promoter activity, indicating an absolute requirement of this site for the bile acid–induced repression. Interestingly, this site is conserved in the mouse and human promoters, and both human and mouse apoA-I are repressed by bile acids. Although the C site clearly mediates the negative effect of FXR agonists on the activity of the proximal apoA-I promoter, the existence of additional negative FXREs elsewhere in the regulatory region of apoA-I gene and contributing to the negative regulation of apoA-I expression by bile acids cannot be excluded. Results from cotransfection experiments and use of a nonsteroidal FXR agonist demonstrate that FXR is required for this downregulation. Together, these data strongly suggest that bile acids exert their action on the apoA-I promoter by FXR. Bile acids have been shown to negatively regulate their own synthesis by transcriptionally repressing Cyp7A expression. Several mechanisms for a negative regulation of target gene transcription by bile acids have been proposed. First, previous reports demonstrated that since no FXR binding sites were found in the Cyp7A promoter, FXR and bile acids downregulate Cyp7A in an indirect manner by inducing the nuclear receptor SHP, the small heterodimer partner (48). Induction of SHP leads to the formation of an inactive complex of SHP with LRH-1 (liver receptor homologue 1) (49–51), the latter being a major transcription factor controlling the tissue-specific expression of Cyp7A (51). SHP binding to LRH-1 impaired transactivation of the Cyp7A promoter, thus leading to a repression of Cyp7A promoter activity (14, 15). To examine whether the LRH-1/SHP pathway could regulate apoA-I through the FXRE C site, we performed cotransfection assays using the TKpGL3 vector driven by three copies of the C site (C3TK) in the presence of LRH-1. LRH-1 did not affect C3TK luciferase activity (data not shown). Moreover, cotransfection assays with an SHP expression vector did not affect apoA-I promoter activity, whereas, as a control, cotransfection of COUP-TFII, a well-known inhibitor of apoA-I transcription (52), decreased apoA-I promoter activity (data not shown). These observations strongly suggest that bile acid–activated FXR acts as a repressor of apoA-I transcription in a manner independent of the SHP/LRH-1 regulatory pathway. Second, bile acids are also able to regulate gene expression in an FXR-independent manner. For instance, it has been shown that bile acids can repress Cyp7A gene expression through activation of the c-Jun N-terminal kinase pathway (42). However, activation of this pathway occurs very rapidly (minutes) and is usually transient (a few hours). Furthermore, apoA-I mRNA, in contrast to Cyp7A mRNA (53), is quite stable, and the repression by bile acids is observed only after several hours. Moreover, our transfection results demonstrate that FXR is required for the repression of apoA-I promoter activity by bile acids and that this repression is also observed using a nonsteroidal agonist. Thus, taken together, data demonstrate that it appears unlikely that the c-Jun N-terminal kinase pathway is involved in the repression of human apoA-I gene expression by bile acids. Finally, the pregnane X receptor (PXR) has been shown to be activated by certain bile acids (41). Yet, in this study we show that TCA, a primary and taurine-conjugated bile acid that does not activate PXR (13, 41), as well as a synthetic FXR agonist, both repress apoA-I expression. Therefore, these results indicate that PXR is not involved in the human apoA-I regulation by bile acids. Nevertheless, we cannot exclude that other, as yet unknown, mechanisms may contribute to the observed repression of apoA-I expression by bile acids. The observation that apoA-I repression by CDCA occurs more rapidly than by GW4064 (Figure 10a) suggests that such additional mechanisms may be operative. Further studies using FXR-null mice crossed with human apoA-I transgenic mice should help to clarify whether such additional mechanisms exist.
Nuclear receptors exert their regulatory action by binding to DNA response elements. These elements are usually composed of 6-bp half-sites arranged as inverted, direct, or everted repeats. The nucleotide composition, the orientation, the spacing between half-sites, and the sequence flanking these half-sites are determinants for binding of a given nuclear receptor (54, 55). Most known, previously characterized FXREs are inverted repeats spaced by one nucleotide (IR-1) acting as positive sites to which FXR binds as a heterodimer with RXR (10, 16). In this study, we demonstrate that FXR negatively regulates apoA-I gene expression through binding to a negative response element: the C site. Several mechanisms could underlie the distinct behavior of FXR on differing FXREs. FXR binding to the C site could result in a distinct conformation leading to the recruitment of corepressors instead of coactivators. Our results show that FXR binds to the C site primarily as a monomer or as a homodimer, but not as heterodimer with RXR. Interestingly, transcriptional repression by thyroid hormone receptors has been shown recently to be due to binding of homodimers, and not heterodimers, with RXR, resulting in the selective recruitment of the corepressor SMRT (56). Alternatively, it cannot be excluded that FXR competes for binding with another strong transcriptional activator to the C site, thus resulting in a passive repression of transcription. Studies on the glucocorticoid receptor showed that this nuclear receptor also either activates or represses transcription, depending on the response elements implicated (57–59). Interestingly, Starr et al. identified a K461A mutant glucocorticoid receptor that activates transcription on all glucocorticoid response elements tested (60). This observation led to the conclusion that a response element contains information that is interpreted by the nuclear receptor by interaction of critical amino acids with DNA. It will be interesting to determine the structural requirement for FXR-repressive activity in the future.
In conclusion, the strong decrease in serum HDL and human apoA-I levels associated with reduced hepatic human apoA-I expression in TCA-fed human apoA-I transgenic mice identifies bile acids that are FXR agonists as important negative modulators of human apoA-I expression and HDL metabolism. Furthermore, our results indicate that the bile acid nuclear receptor FXR directly downregulates human apoA-I expression through binding of activated FXR to a negative FXR response element in the human apoA-I promoter. Several epidemiological studies have demonstrated that low levels of HDL cholesterol, also known as hypoalphalipoproteinemia, are a risk factor for coronary heart disease (1). In addition, raising HDL cholesterol concentrations has been shown to effectively reduce progression of atherosclerotic lesions and coronary events (2). In view of the reported findings, FXR antagonists could be promising compounds to raise serum HDL levels.
Acknowledgments
The technical assistance of Eric Baugé, Bruno Derudas, Christian Duhem, and Giulia Chinetti is greatly appreciated. The authors would like to acknowledge members of the B. Staels laboratory for helpful discussions. P. Young, T. Willson, and G. Kullak-Ublick are thanked for providing various expression plasmids and the NTCP HepG2 cells. We would like to thank D. Bojanowski, M. Bojanowski, and M. Burdelski for their support with patient data. This work was supported by grants from Institut National de la Santé et de la Recherche Médicale, Institut Pasteur de Lille, Genfit SA, and the University of Lille II.
References
- 1.Miller GJ, Miller NE. Plasma-high-density-lipoprotein concentration and development of ischaemic heart-disease. Lancet. 1975;1:16–19. doi: 10.1016/s0140-6736(75)92376-4. [DOI] [PubMed] [Google Scholar]
- 2.Rubins HB, et al. Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. Veterans Affairs High-Density Lipoprotein Cholesterol Intervention Trial Study Group. N Engl J Med. 1999;341:410–418. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- 3.Andersson LO. Pharmacology of apolipoprotein A-I. Curr Opin Lipidol. 1997;8:225–228. doi: 10.1097/00041433-199708000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz G, Langmann T. Structure, function and regulation of the ABC1 gene product. Curr Opin Lipidol. 2001;12:129–140. doi: 10.1097/00041433-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Miller NE, La Ville A, Crook D. Direct evidence that reverse cholesterol transport is mediated by high-density lipoprotein in rabbit. Nature. 1985;14:109–111. doi: 10.1038/314109a0. [DOI] [PubMed] [Google Scholar]
- 6.Barter PJ, Rye KA. Molecular mechanisms of reverse cholesterol transport. Curr Opin Lipidol. 1996;7:82–87. doi: 10.1097/00041433-199604000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Rubin EM, Krauss RM, Spangler EA, Verstuyft JG, Clift SM. Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature. 1991;353:265–267. doi: 10.1038/353265a0. [DOI] [PubMed] [Google Scholar]
- 8.Forman BM, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 9.Seol W, Choi HS, Moore DD. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 10.Laffitte BA, et al. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 12.Makishima M, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 13.Parks DJ, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 14.Lu TT, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell Biol. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 16.Grober J, et al. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem. 1999;274:29749–29754. doi: 10.1074/jbc.274.42.29749. [DOI] [PubMed] [Google Scholar]
- 17.Kramer W, et al. Substrate specificity of the ileal and the hepatic Na(+)/bile acid cotransporters of the rabbit. I. Transport studies with membrane vesicles and cell lines expressing the cloned transporters. J Lipid Res. 1999;40:1604–1617. [PubMed] [Google Scholar]
- 18.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 19.Sinal CJ, et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;2:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 20.Urizar NL, Dowhan DH, Moore DD. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J Biol Chem. 2000;275:39313–39317. doi: 10.1074/jbc.M007998200. [DOI] [PubMed] [Google Scholar]
- 21.Jiang XC, et al. Targeted mutation of plasma phospholipid transfer protein gene markedly reduces high-density lipoprotein levels. J Clin Invest. 1999;103:907–914. doi: 10.1172/JCI5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bard JM, Parra HJ, Douste-Blazy P, Fruchart JC. Effect of pravastatin, an HMG CoA reductase inhibitor, and cholestyramine, a bile acid sequestrant, on lipoprotein particles defined by their apolipoprotein composition. Metabolism. 1990;39:269–273. doi: 10.1016/0026-0495(90)90046-f. [DOI] [PubMed] [Google Scholar]
- 23.Hagen E, et al. Fluvastatin efficacy and tolerability in comparison and in combination with cholestyramine. Eur J Clin Pharmacol. 1994;46:445–449. doi: 10.1007/BF00191909. [DOI] [PubMed] [Google Scholar]
- 24.Staels B, Van Tol A, Fruchart JC, Auwerx J. Effects of hypolipidemic drugs on the expression of genes involved in high density lipoprotein metabolism in the rat. Isr J Med Sci. 1996;32:490–498. [PubMed] [Google Scholar]
- 25.Leiss O, von Bergmann K. Different effects of chenodeoxycholic acid and ursodeoxycholic acid on serum lipoprotein concentrations in patients with radiolucent gallstones. Scand J Gastroenterol. 1982;17:587–592. doi: 10.3109/00365528209181063. [DOI] [PubMed] [Google Scholar]
- 26.Kuriyama M, Tokimura Y, Fujiyama J, Utatsu Y, Osame M. Treatment of cerebrotendinous xanthomatosis: effects of chenodeoxycholic acid, pravastatin, and combined use. J Neurol Sci. 1994;125:22–28. doi: 10.1016/0022-510x(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 27.Berthou L, et al. Opposite regulation of human versus mouse apolipoprotein A-I by fibrates in human apolipoprotein A-I transgenic mice. J Clin Invest. 1996;97:2408–2416. doi: 10.1172/JCI118687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy GM, Billing BH, Baron DN. A fluorimetric and enzymatic method for the estimation of serum total bile acids. J Clin Pathol. 1970;23:594–598. doi: 10.1136/jcp.23.7.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bojanovski M, et al. Parameters of lipoprotein metabolism and cholestasis in healthy and cholestatic infants and children. Prog Lipid Res. 1991;30:295–300. doi: 10.1016/0163-7827(91)90027-3. [DOI] [PubMed] [Google Scholar]
- 30.Rubin EM, Ishida BY, Clift SM, Krauss RM. Expression of human apolipoprotein A-I in transgenic mice results in reduced plasma levels of murine apolipoprotein A-I and the appearance of two new high density lipoprotein size subclasses. Proc Natl Acad Sci USA. 1991;88:434–438. doi: 10.1073/pnas.88.2.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li AP, Roque MA, Beck DJ, Kaminski DL. Isolation and culturing of hepatocytes from human liver. J Tissue Cult Methods. 1992;14:139–146. [Google Scholar]
- 32.Kullak-Ublick GA, et al. Stable expression and functional characterization of a Na+-taurocholate cotransporting green fluorescent protein in human hepatoblastoma HepG2 cells. Cytotechnology. 2000;34:1–9. doi: 10.1023/A:1008152729133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maloney PR, et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem. 2000;43:2971–2974. doi: 10.1021/jm0002127. [DOI] [PubMed] [Google Scholar]
- 34.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 35.Staels B, Auwerx J. Perturbation of developmental gene expression in rat liver by fibric acid derivatives: lipoprotein lipase and alpha-fetoprotein as models. Development. 1992;115:1035–1043. doi: 10.1242/dev.115.4.1035. [DOI] [PubMed] [Google Scholar]
- 36.Bonazzi A, Mastyugin V, Mieyal PA, Dunn MW, Laniado-Schwartzman M. Regulation of cyclooxygenase-2 by hypoxia and peroxisome proliferators in the corneal epithelium. J Biol Chem. 2000;275:2837–2844. doi: 10.1074/jbc.275.4.2837. [DOI] [PubMed] [Google Scholar]
- 37.Vu-Dac N, et al. Negative regulation of the human apolipoprotein A-I promoter by fibrates can be attenuated by the interaction of the peroxisome proliferator-activated receptor with its response element. J Biol Chem. 1994;269:31012–31018. [PubMed] [Google Scholar]
- 38.Jansen PL, et al. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology. 1999;117:1370–1379. doi: 10.1016/s0016-5085(99)70287-8. [DOI] [PubMed] [Google Scholar]
- 39.Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest. 1994;93:1326–1331. doi: 10.1172/JCI117091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorla-Bajszczak A, Juge-Aubry C, Pernin A, Burger AG, Meier CA. Conserved amino acids in the ligand-binding and tau(i) domains of the peroxisome proliferator-activated receptor alpha are necessary for heterodimerization with RXR. Mol Cell Endocrinol. 1999;147:37–47. doi: 10.1016/s0303-7207(98)00217-2. [DOI] [PubMed] [Google Scholar]
- 41.Staudinger JL, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 43.Melter M, et al. Progressive familial intrahepatic cholestasis: partial biliary diversion normalizes serum lipids and improves growth in noncirrhotic patients. Am J Gastroenterol. 2000;95:3522–3528. doi: 10.1111/j.1572-0241.2000.03370.x. [DOI] [PubMed] [Google Scholar]
- 44.Puchois P, et al. Apolipoprotein A-I containing lipoproteins in coronary artery disease. Atherosclerosis. 1987;68:35–40. doi: 10.1016/0021-9150(87)90091-8. [DOI] [PubMed] [Google Scholar]
- 45.Barbaras R, Puchois P, Fruchart JC, Ailhaud G. Cholesterol efflux from cultured adipose cells is mediated by LpAI particles but not by LpAI:AII particles. Biochem Biophys Res Commun. 1987;142:63–69. doi: 10.1016/0006-291x(87)90451-7. [DOI] [PubMed] [Google Scholar]
- 46.Elshourbagy NA, et al. Expression of rat apolipoprotein A-IV and A-I genes: mRNA induction during development and in response to glucocorticoids and insulin. Proc Natl Acad Sci USA. 1985;82:8242–8246. doi: 10.1073/pnas.82.23.8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haddad IA, Ordovas JM, Fitzpatrick T, Karathanasis SK. Linkage, evolution, and expression of the rat apolipoprotein A-I, C-III, and A-IV genes. J Biol Chem. 1986;261:13268–13277. [PubMed] [Google Scholar]
- 48.Seol W, Choi HS, Moore DD. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 49.Becker-Andre M, Andre E, DeLamarter JF. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. 1993;194:1371–1379. doi: 10.1006/bbrc.1993.1976. [DOI] [PubMed] [Google Scholar]
- 50.Galarneau L, et al. The alpha1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci USA. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ladias JA, Karathanasis SK. Regulation of the apolipoprotein AI gene by ARP-1, a novel member of the steroid receptor superfamily. Science. 1991;251:561–565. doi: 10.1126/science.1899293. [DOI] [PubMed] [Google Scholar]
- 53.Pandak WM, Stravitz RT, Lucas V, Heuman DM, Chiang JY. Hep G2 cells: a model for studies on regulation of human cholesterol 7alpha-hydroxylase at the molecular level. Am J Physiol. 1996;270:G401–G410. doi: 10.1152/ajpgi.1996.270.3.G401. [DOI] [PubMed] [Google Scholar]
- 54.Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers, and heterodimers. Endocr Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 55.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 56.Yoh SM, Privalsky ML. Transcriptional repression by thyroid hormone receptors. A role for receptor homodimers in the recruitment of SMRT corepressor. J Biol Chem. 2001;276:16857–16867. doi: 10.1074/jbc.M010022200. [DOI] [PubMed] [Google Scholar]
- 57.Weiner FR, et al. The effects of dexamethasone on in vitro collagen gene expression. J Biol Chem. 1987;262:6955–6958. [PubMed] [Google Scholar]
- 58.Akerblom IE, Slater EP, Beato M, Baxter JD, Mellon PL. Negative regulation by glucocorticoids through interference with a cAMP responsive enhancer. Science. 1988;241:350–353. doi: 10.1126/science.2838908. [DOI] [PubMed] [Google Scholar]
- 59.Birnberg NC, et al. Glucocorticoids regulate proopiomelanocortin gene expression in vivo at the levels of transcriptionsecretion. Proc Natl Acad Sci USA. 1983;80:6982–6986. doi: 10.1073/pnas.80.22.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starr DB, Matsui W, Thomas JR, Yamamoto KR. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]