Abstract
Although many bacterial chromosomes require only one replication initiator protein, e.g., DnaA, most plasmid replicons depend on dual initiators: host-encoded DnaA and plasmid-encoded Rep initiator protein for replication initiation. Using the plasmid pSC101 as a model system, this work investigates the biological rationale for the requirement for dual initiators and shows that the plasmid-encoded RepA specifically interacts with the replicative helicase DnaB. Mutations in DnaB or RepA that disrupt RepA–DnaB interaction cause failure to load DnaB to the plasmid ori in vitro and to replicate the plasmid in vivo. Although, interaction of DnaA with DnaB could not substitute for RepA–DnaB interaction for helicase loading, DnaA along with integration host factor, DnaC, and RepA was essential for helicase loading. Therefore, DnaA is indirectly needed for helicase loading. Instead of a common surface of interaction with initiator proteins, interestingly, DnaB helicase appears to have at least a limited number of nonoverlapping surfaces, each of which interacts specifically with a different initiator protein.
Keywords: preinitiation complex, replication initiation
Since its discovery by Cohen and Chang (1), pSC101, a tetracycline-resistant plasmid replicon, has become a favorite system for the analysis of the control of plasmid replication and stability (2–8). The plasmid replicon consists of a cis-acting ori sequence and a plasmid-encoded, trans-acting initiator protein called RepA. We have reported the purification of the RepA protein and its mode of interaction with the three iterons and the two inverted repeats of the plasmid ori (Fig. 1; refs. 3 and 4). Although initiation of replication of several prokaryotic replicons, e.g., Escherichia coli, phage λ, Bacillus subtilis, and the eukaryotic replicon simian virus 40 requires single initiator proteins (9), pSC101 (and several other plasmids) requires both the host-encoded DnaA protein and the plasmid-encoded RepA protein (10, 11).
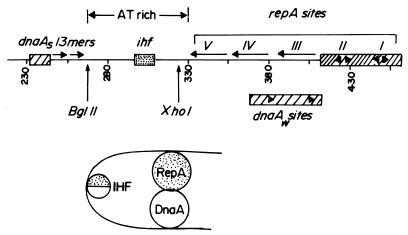
Figure 1.
Diagram of the replication origin of pSC101. (Upper) The ori consists of (left to right) the strong DnaA box (DnaAs), two 13-mers that might be the sequences that are initially melted, ihf site, three direct repeat (iterons V, IV, and III) and two inverted repeats II and I that bind to the RepA protein. The DnaAw sites are contacted by DnaA protein when IHF bends the DNA at the ihf site. The bending by IHF is believed not only to stabilize the ori–DnaA complex but also to promote contact between DnaA and RepA proteins (Lower).
An examination of the minimal ori of the plasmid reveals a single DnaA box (binding site dnaAS) that is separated from the RepA-binding iterons by a naturally bent AT-rich segment. Embedded in the AT-rich region is a binding site for the host-encoded DNA bending protein called integration host factor (IHF); the interaction of IHF with the cognate site further bends the DNA and the interaction is essential for plasmid replication (Fig. 1; refs. 12 and 13). IHF appears to enhance the binding of DnaA to the ori in vitro, and its principal role is believed to be bending of the DNA and thereby promoting contact between DnaA and RepA proteins that are bound to physically separated cognate sites and to weaker dnaAw sites in the iterons (11). Recently, we have discovered that RepA and DnaA specifically interact with each other in the absence of DNA and IHF and that the interaction is essential for initiation of pSC101 replication (H.J.D. and D.B., unpublished work). Cohen and Biek (14, 15) have shown that the requirement for IHF can be bypassed by either mutations in repA or in the host-encoded topA genes. Thus, suppresser mutations either cause alternative origin conformations that lead to DnaA–RepA interactions in the absence of IHF or else the mutations obviate the need for such an interaction.
This work was initiated to address two interrelated and interesting questions. First, besides origin binding, what other biochemical functions (e.g., protein–protein interactions) can be attributed to the plasmid initiator proteins? Second, because the recruitment of the replicative helicase is a key step of initiation, is it DnaA or RepA or both initiators that recruit the DnaB helicase to the ori and does this process involve pairwise interactions between DnaB and the two initiators?
In this paper we present evidence showing specific protein–protein interaction between RepA and DnaB. We have identified the surfaces of both DnaB and RepA that participate in the interaction and have isolated noninteracting mutants that affect both surfaces. In vivo analysis showed that the interaction was essential for pSC101 replication. In vitro experiments showed that four purified proteins (IHF, DnaA, RepA, and DnaC) were needed to load DnaB helicase onto the plasmid replication origin and a mutant that disrupted DnaB–RepA, but not DnaA–DnaB, interaction, had a detrimental effect on helicase loading. Thus, without the critical protein–protein interaction between RepA and DnaB, DnaA by itself could not load the helicase at the plasmid ori.
MATERIALS AND METHODS
DNA Constructs, Plasmids, and Bacterial Strains.
The plasmid pCV2 contains the minimal ori of pSC101 (in a HincII fragment), cloned into the vector pUC9 (4). The DnaB tagged with the kinase site was a gift from Mike O’Donnell (The Rockefeller University, New York; refs. 16 and 17). The bacterial strains DH5α [λσθβ]supE-44 lacU169 (φ80 lacZ Δ M15) hsdR17 recA1 endA1 gyrA96 thi-1 rel A1] and BL21 (DE3) [E. coli B: F− ompT hsdS (rk−mk+) gal(λDE3)] were used for standard cloning and protein expression, respectively.
Protein Purification.
The RepA, DnaA, IHF, and DnaB proteins were purified as described (12, 18–20). DnaC was purified from a overproducer strain that coexpressed DnaB and DnaC (a gift from N. Dixon, University of Sydney, Sydney, Australia). The strain yielded free DnaB and DnaC and a 1:1 complex of B-C that were separated on a monoQ column. The DnaC, present in the flow-through, was further purified through a blue dextran Sepharose column.
Site-Directed Mutagenesis.
Site-directed mutagenesis was carried out by using two complementary mutagenic oligonucleotides that were hybridized to double-stranded plasmid DNA and extended by PCR using the Pfu DNA polymerase. The input template strands were selectively removed by digestion with DpnI according to a commercial protocol (Stratagene). The procedure yielded ≈100% mutants.
Protein–Protein Interaction.
The procedures for affinity chromatography and ELISA to measure protein–protein interactions have been published (19, 21).
ABC Primosome Assay.
Using a single-stranded circular DNA template containing a DnaA binding site located in a hairpin, the assay was carried out as described (22, 23).
In Vitro Loading of DnaB to the ori of pSC101.
Supercoiled DNA substrates (200 ng) were incubated with 85 ng of single-stranded DNA-binding protein (SSB), 15 ng of IHF, 40 ng of DnaC, 120 ng of DnaA, 250 ng of RepA, and 80 ng of 32P-labeled DnaB in 50 μl of loading buffer (40 mM Hepes-KOH, pH 7.6/2 mM ATP/11 mM MgAcetate/500 μM each of UTP, CTP, and GTP/50 μg/ml of BSA/6 mM phosphokinase/100 μg/ml of creatine phosphate) at 37°C for 10 min and loaded onto 0.5 × 5 cm Biogel 1.5 M columns. The columns were run at 4°C by using the loading buffer mentioned above, and 100-μl fractions were collected. The radioactivity in the excluded and the included fractions was measured. Later experiments showed that SSB can be excluded from the reaction mixture without eliciting any negative effect. Control experiments were similarly performed to measure the loading of the normal and mutant forms of helicase to oriC of the host.
DNA-Filter Binding.
Binding of the wild-type and mutant forms of RepA protein to a labeled DNA fragment containing the ori of pSC101 and to a control fragment without the ori sequence was carried out in 50 mM Tris⋅Cl, pH 7.5/70 mM KCl/30 mM NaCl/1 mM EDTA/1 mM β-mercaptoethanol. Sonicated salmon sperm DNA (5 μg/ml) was used as a carrier. Incubations were carried out at room temperature for 20 min, and the mixture was slowly filtered through nitrocellulose filters. The filters were washed in the binding buffer, dried, and counted.
RESULTS
Demonstration of Specific RepA–DnaB Interaction and Mapping of the Interaction Surface on DnaB.
To detect possible interaction between DnaB and RepA, we constructed recombinant clones that expressed full-length and the following partial peptides of DnaB from a T7 promoter. The peptides were fused at the N-terminal end, in-frame with a hexahistidine moiety that made it convenient to purify the proteins on Ni-nitrilotriacetic acid-agarose affinity columns (Ni columns). The peptide AgeI was encoded in the DNA fragment that extended from the first ATG codon of DnaB to its 208th codon (that is marked by an AgeI restriction site; hence the name AgeI). The peptide BC2 extended from the first codon to the 785th codon (Fig. 2A). The full-length DnaB, the AgeI, and BC2 peptides were purified and immobilized on Ni-agarose. 35S-labeled RepA protein was generated by coupled in vitro transcription translation and bound to the full-length DnaB and the AgeI and BC2 peptide columns and to control Ni-agarose columns without bound DnaB or its peptides. The bound protein was eluted and analyzed by SDS/PAGE. It should be noted that the coupled transcription translation of the complete RepA gene not only generated a full-length peptide but also a partial peptide from an internal methionine (Tr.RepA, Fig. 3, lane 1). Labeled luciferase and π protein of R6K (Fig. 3, lane 2) were used as negative and positive controls, respectively.
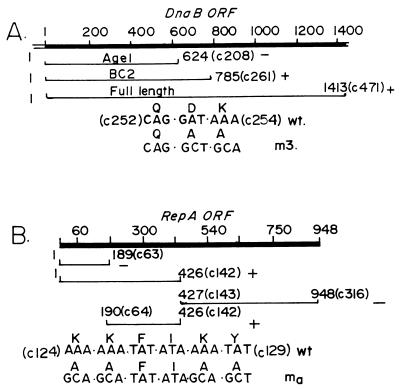
Figure 2.
Summary maps of DnaB and RepA that were used in the RepA–DnaB interaction experiments. (A) The ORF of dnaB showing the nucleotide coordinates at the top. The coordinate 1 is the first residue of the ATG initiator codon. The coordinates of the two N-terminal peptides AgeI and BC2 are shown in nucleotides and within brackets in codon numbers (e.g., c1-c208 for AgeI). The location of the m3 double mutant D253A, K254A are shown. The + and − symbols indicate positive and negative protein–protein interaction, respectively, between immobilized dnaB and RepA in solution. (B) The ORF of RepA is shown with nucleotide coordinates at the top. The nucleotide 1 corresponds to the first residue of the ATG initiator. The N-terminal and C-terminal peptides are identified by nucleotide coordinates and by codon number designation in parenthesis. The + and − symbols mark the peptides showing positive and negative interaction, respectively, with DnaB. The ma quadruple mutant of RepA (K124A, K125A, K128A, and Y129A) and the nucleotide substitutions that were used to generate the mutant are shown.
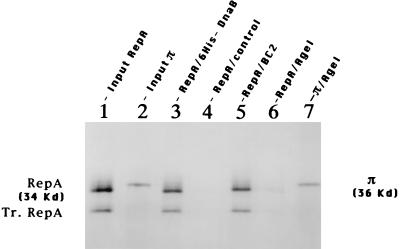
Figure 3.
Autoradiograms of SDS-10% PAGE showing the specific binding of DnaB to RepA protein. Lane 1, input full-length RepA (and a truncated form Tr.RepA, produced by internal translation initiation). Lane 2, input π. Lane 3, RepA bound to full-length immobilized DnaB. Lane 4, control Ni-Agarose. Lane 5, immobilized DnaB-BC2 peptide. Lane 6, immobilized DnaB–AgeI peptide. Lane 7, π bound to immobilized DnaB–AgeI peptide.
Autoradiograms of the gels revealed that immobilized full-length DnaB, but not the control Ni-agarose matrix, readily retained RepA (Fig. 3, lanes 3 and 4, respectively). Luciferase did not bind to either the immobilized DnaB or to the control matrix (data not shown). While the immobilized BC2 peptide retained RepA (Fig. 3, lane 5), the shorter AgeI peptide failed to retain the labeled protein (Fig. 3, lane 6). However, as previously reported (19), the immobilized AgeI peptide bound to labeled π initiator protein of R6K (Fig. 3, lane 7). We have similarly mapped the binding site of DnaA to a region immediately downstream of the BC2 peptide (H.J.D. and D.B., unpublished work). Thus, interestingly, three different initiator proteins (RepA, π, and DnaA), instead of sharing a common binding site on DnaB, bound to three linearly contiguous, but nonoverlapping, regions. We have confirmed RepA–DnaB interaction by performing reciprocal binding experiments in which full-length RepA was immobilized on an agarose matrix and was challenged with labeled full-length DnaB and the AgeI and the BC2 peptides. The full-length DnaB and the BC2 peptide, but not the AgeI peptide, bound to the RepA matrix (data not shown).
Mapping the Surface of RepA that Interacts with DnaB.
We have performed reciprocal binding experiments to map the surface of RepA that binds to DnaB as follows. We constructed recombinant clones that encoded a full-length RepA and its various peptides, fused with glutathione S-transferase (GST) and prepared affinity matrices using the fusion peptides. Labeled, full-length DnaB, fused at the N terminus with the protein kinase site, was prepared and labeled with protein kinase and γ32P[ATP] and incubated with the affinity matrices containing immobilized full-length and the various peptides of RepA described below (16). DnaB readily bound to the full-length RepA matrix but failed to bind to the control Ni-agarose without immobilized RepA (Fig. 4, lanes 2 and 3, respectively). DnaB bound to the immobilized RepA N-terminal peptide comprising codons 1–142 but not to the C-terminal one from the codons 143–316 (Fig. 4, lanes 4 and 6, respectively). However, labeled DnaB bound to the peptide extending from codons 64 to 142 (Fig. 4, lane 7) but not to the N-terminal peptide c1-c63 (Fig. 4, lane 5). Thus, the region of RepA that interacted with DnaB was localized to the 78-aa-long peptide segment encoded by the DNA from nucleotide coordinates 192 to 426. The binding results are summarized in Fig. 2B.
Figure 4.
Autoradiograms of SDS-10% PAGE showing the mapping of the surface of RepA that interacts with DnaB. Lane 1, DnaB input. Lane 2, DnaB binding to full-length immobilized RepA. Lane 3, control GST matrix. Lane 4, immobilized peptide c1-c142. Lane 5, N-terminal c1-c63. Lane 6, C-terminal c143-c316. Lane 7, N-terminal c64-c142 peptide of RepA.
Isolation of Noninteracting Mutants of DnaB.
If the in vitro interactions reported above are truly specific, one should be able to isolate mutations on both of the interacting surfaces that would abolish or reduce the specific protein–protein interactions. During our previous studies of DnaB–DnaG interaction, we had isolated several mutant forms of DnaB, one of which named m3 (D253A and K254A; see Fig. 2A), did not significantly affect the helicase activity or the ability of DnaB to interact with DnaG or to synthesize RNA primers (21). The m3 mutant was located in the segment of DnaB that interacted in vitro with RepA and therefore was a likely candidate to be the desired RepA noninteracting mutation.
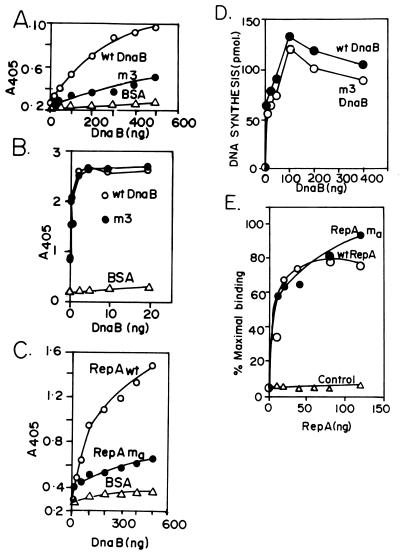
We purified the m3 mutant form of DnaB and investigated its ability to interact with RepA by both ELISA and by the GST affinity procedures. The results from both approaches were consistent with each other; for the sake of brevity, only the ELISA data are shown. The m3 mutant form of DnaB showed a ≈60% reduction in its ability to bind to RepA in comparison with wild-type DnaB (Fig. 5A). We measured the binding of m3 to immobilized DnaA and found no significant difference in its binding in comparison with the wild-type DnaB (Fig. 5B). We further analyzed the mutant form of DnaB for its ability to initiate replication in an ABC primosome system (22, 23). The ABC primosome system mimics the lagging strand replication initiated from a hairpin sequence containing the DnaA-binding site present in a single-stranded circular template. The reaction requires purified DnaA, DnaB, DnaC, DnaG, reconstituted core polymerase III, and the β sliding clamp. We found no reduction in the extent of replication catalyzed by the m3 helicase in comparison with that of the wild-type DnaB (Fig. 5D). Thus the biochemical analysis of the m3 mutation further supported the conclusion that the interaction between DnaB and RepA was specific and physiologically significant.
Figure 5.
Experiments showing characterization of the m3 and ma proteins. (A) ELISA binding of the wild-type and m3 form of DnaB in solution to immobilized RepA (full length) showing that the mutant form of DnaB has suffered a ≈70% reduction in binding to the RepA matrix in comparison with wild-type DnaB. BSA control that does not bind to DnaB is shown. (B) ELISA showing that both the wild-type and the mutant form of DnaB, however, bind equally well to immobilized DnaA but not immobilized BSA (control). (C) The ma mutant form of RepA showed a ≈50% reduction in binding to DnaB in comparison with wild-type RepA. The wild-type and the mutant form of RepA were (fused to GST) immobilized on plastic surface of microtiter plates and DnaB was applied in solution to the immobilized RepA (both the wild-type and mutant forms of RepA bound equally well to DnaA; not shown). (D) ABC primosome replication catalyzed by wild-type and m3 DnaB of a single-stranded, circular template (0.1 μg) that had the dnaA site present as a hairpin. Replication reaction (25 μl) contained SSB (800 ng), DnaC (50 ng), DnaG (50 ng), DNA polymerase III core reconstituted from the individual peptides (100 ng), the β sliding clamp (75 ng), and wild-type and m3 DnaB (100 ng) in 40 mM Hepes-KOH, pH 8.0/40 mM potassium glutamate/40 mM 4% sucrose/100 μg/ml BSA/4 mM DTT/250 μM each CTP, GTP and UTP/2 mM ATP/20 μM each dNTPs, and 3H dATP added to 200–400 cpm/pmol (incubated at 30°C for 10 min). (E) Filter binding experiment showing that both the wild-type RepA and the ma mutant form bind almost equally well to a DNA fragment containing the ori of pSC101 but not to a control fragment without the ori sequence. The binding of the wild-type RepA to a control DNA fragment lacking the ori sequence also is shown. The mutant also failed to bind to the control DNA.
A RepA Mutant That Did Not Interact with DnaB in Vitro Failed to Promote pSC101 Replication in Vivo.
We isolated a series of alanine-scanning mutants in the region of RepA that interacted with DnaB in an attempt to examine further the specificity and possible biological significance of the interaction. Two or more contiguous charged amino acid residues in the region were substituted with alanine, and the mutant forms of the protein were purified and analyzed for binding to DnaB and their ability to support pSC101 replication in vivo. Five mutant forms of RepA, each with multiple amino acid substitutions in the region of interaction with DnaB, were isolated and examined. One of the five RepA mutants, named ma (K124A, K125A, K128A, and Y128A, quadruple mutant; see Fig. 2B) consistently showed a ≈60% reduction in binding to DnaB, in both GST affinity matrices (not shown) and ELISA assays, in comparison with wild-type DnaB (Fig. 5C).
We purified the mutant (ma) form of RepA and compared its ability to specifically bind to the ori of pSC101 in comparison with the wild-type protein by using a nitrocellulose filter binding assay. A binding titration showed that both the wild-type and mutant forms of the protein specifically bound to a labeled DNA fragment containing the ori of pSC101 (which included the three iterons and two inverted repeats) but failed to bind to a control fragment of pUC18 of equivalent length (Fig. 5E). This result along with the observed similar chromatographic properties (e.g., solubility, precipitability in ammonium sulfate, salt concentration needed to elute from phosphocellulose, etc.) suggested that the ma mutation did not cause a global defect in RepA but rather that the effect was more local, a conclusion also supported by the observation that the mutant form as well as the wild-type RepA protein bound equally well to DnaA (H.J.D. and D.B., unpublished work).
We introduced the ma mutation into a chimeric plasmid called pCV2 (3, 4) that contained the minimal replicon of pSC101 cloned into the pUC8 vector and examined the ability of the pSC101 replicon, which does not need polA for replication, to be maintained in an E. coli polA12 temperature-sensitive host in which the ColE1 ori of the vector was nonfunctional at the restrictive temperature. The pCV2 (wild-type pSC101 RepA) and the vector DNAs were used in positive and negative control experiments, respectively.
Each of the three plasmid DNAs was transformed into E. coli polA12 at the permissive temperature (30°C), and the transformants were isolated on ampicillin plates. The polA12 cells bearing the three plasmids also were plated on Luria agar plates after growth at 42°C, and the numbers of colonies for equal volume of culture were very similar, indicating no loss of cell viability during growth in Luria broth at 42°C (not shown). To determine the degrees of maintenance of the plasmids in the host cells, three ampicillin resistance transformants from each transformation were picked at random and grown for multiple generations (>12 hr) in Luria broth at the nonpermissive temperature (42°C) without selection for ampicillin resistance. The cells then were plated on ampicillin-Luria agar plates at 42°C. Table 1 presents the average of three separate sets of experiments. The vector plasmid, as expected, was lost during growth in polA12 cells at 42°C. The chimeric pCV2 plasmid containing the wild-type RepA gene as expected replicated at 42°C in polA12 cells. The pCV2 ma plasmid that contained the ma mutant form of RepA was more readily lost from polA12 cells at 42°C in comparison with the pCV2 control (Table 1).
Table 1.
In vivo effect of RepA ma mutation on replication of pSC101 in E. coli strain polA12ts
| Plasmid | No. of colonies after growth at 42°C |
|---|---|
| pUC19 | 2 |
| pCV2 | 1,070 |
| pCV2 (RepA ma mutation) | 28 |
The data represent an average of three experiments.
In Vitro Recruitment of DnaB to the ori of pSC101 Required DnaB–RepA Interaction.
DnaB protein was tagged at the N-terminal end by fusion with a muscle kinase recognition sequence and labeled with muscle kinase and γ32P[ATP]. The fusion, as reported before (16, 17), and confirmed by us (data not shown), did not affect the helicase activity of DnaB. The strategy was to incubate the labeled DnaB with purified DnaC, IHF, DnaA, RepA, SSB proteins, and supercoiled pUC19 plasmid containing the minimal ori of pSC101 (or oriC, used as a control), in the molar proportions indicated in a preceding section and measure the retention of DnaB at the ori (Fig. 6A). The purified proteins mentioned above were chosen because of their known role in the preinitiation steps of oriC replication (9). After the incubation, the reaction mixtures were fractionated in 0.5 × 2.5 cm Biogel 1.5 M gel filtration columns to separate the protein–DNA complex from the free proteins. The label appearing in the excluded protein–DNA complex peak provided a measure of the amount of DnaB recruited to the plasmid or the host ori.
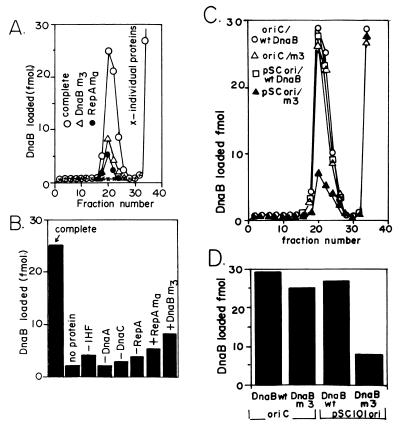
Figure 6.
Requirement for in vitro recruitment of DnaB to the ori of pSC101. (A) Wild-type and m3 forms of DnaB were tagged with protein kinase recognition sequence and labeled with 32P as described, incubated with pSC101 ori DNA along with purified IHF, DnaC, DnaA, and RepA (and SSB) proteins and passed through a gel filtration column. The profile of radioactivity in the excluded DNA–protein complex peak and in the included free protein fractions is shown. The loading of DnaB when any one of four proteins were omitted was at the background levels (x-x-x). (B) Histograms showing the extent of reduction in helicase loading in the m3 and ma mutants of dnaB and repA genes, respectively, and when one of the four proteins (DnaA, RepA, DnaC, and IHF) were omitted one at a time from the reaction mixture. (C) Gel filtration profiles of wild-type DnaB and m3 DnaB incubated with oriC and pSC101 ori in the presence of the other necessary proteins (as in A). (D) Histograms showing the comparative loading of the wild-type and the m3 form of DnaB onto an oriC and an ori pSC101 DNA. Derived from three independent experiment such as the one shown in C. Note that both the wild-type DnaB and the m3 form load almost equally well onto oriC DNA whereas the m3 loads poorly onto pSC101 ori.
The loading of DnaB to the pSC101 ori required all four purified proteins (RepA, DnaA, DnaC, and IHF), and omission of any one of the four proteins from the reaction mixture almost completely abolished the helicase recruitment (Fig. 6A). Although SSB was used initially in the reaction mixture, subsequent work showed that its presence was unnecessary for helicase loading. As a control, pUC19 DNA without the pSC101 ori sequence was used in the reaction, and the small amount of background radioactivity was deducted from each experimental data point. The pUC19 control experiment showed that the recruitment of DnaB depended on the pSC101 ori sequence. The data in Fig. 6A represent the average of three independent sets of experiments. A quantitative estimate of helicase loading is shown in the histograms of Fig. 6B.
The m3 mutant form of DnaB also was labeled with 32P and by similar analysis showed a significant reduction of DnaB loading. The ma mutant form of RepA that showed reduced physical association with DnaB also was analyzed and it also showed a significant decrease in loading of DnaB to the ori (Fig. 6 A and B). Thus, a reduction in physical interaction between DnaB and RepA was accompanied by a corresponding reduction in the ability of the mutant forms of the proteins to recruit DnaB helicase to the ori. We used the oriC plasmid in control experiments and performed similar helicase loading experiments (Fig. 6C), and the data (average of three experiments) are summarized in Fig. 6D. Wild-type DnaB and the m3 protein loaded almost equally well onto oriC (30 fmol and 25 fmol of loading, respectively; Fig. 6D), whereas the mutant form consistently showed a significant reduction in loading to the plasmid ori (27 fmol versus 8 fmol of loading promoted by the wild-type and the ma mutant form of RepA, respectively; Fig. 6D). Thus the ma mutation selectively and significantly diminished the loading of DnaB helicase to the plasmid ori without significantly altering the loading to the host oriC.
DISCUSSION
Because the realization that initiation of replication of several plasmid systems requires dual initiator proteins, namely DnaA encoded by the host and the other encoded by the plasmid replicon, the relative role of DnaA in plasmid replication as contrasted with its role in the initiation of replication of oriC of the host chromosome had remained obscure (10, 18, 24, 25). We have endeavored to investigate the biological roles of the twin initiators by systematically identifying the interactions between two proteins (and other replisomal proteins) and attempting to determine the physiological significance of such interactions. We previously have reported the interaction between π protein of R6K and DnaB (19) but the physiological role of the interaction remains to be determined.
The results described in this paper provide an important insight into the relative roles of RepA and DnaA proteins in pSC101 replication, i.e., RepA–DnaB interaction plays an indispensable role in helicase recruitment to the plasmid ori, and this requirement cannot be bypassed by DnaA–DnaB interaction. Three other proteins (DnaA, DnaC, and IHF) also are needed for the loading reaction. The following is a brief consideration of the possible roles played by each of these proteins.
What role does DnaA play in helicase recruitment? Despite the known interaction between DnaA and DnaB (26), DnaA by itself does not seem to be capable of sequestering the helicase at the plasmid ori as revealed by the inability of the m3 form of DnaB to be loaded onto the ori. It is worth recalling in this context that the m3 helicase, although defective in its interaction with RepA, retained normal interaction with DnaA. Furthermore, both the wild-type and the m3 mutant forms of DnaB were almost equally capable of carrying out lagging strand replication of oriC in a DnaA-mediated ABC primosome system, thereby suggesting that the mutation specifically disrupted the plasmid but not host replication.
We previously have presented evidence that supported interaction between RepA and DnaA at the replication origin of pSC101 (11). Mutant forms of DnaA that support oriC replication, but not pSC101, replication have been reported, and these mutants also imply a functional interaction between the two proteins (27). We recently have discovered a direct physical interaction between DnaA and RepA (H.J.D. and D.B., unpublished work) in the absence of DNA and hypothesize that this interaction, which is promoted by the DNA bending action of IHF (see Fig. 1), is critical for the initial unwinding at the plasmid ori. This initial unwinding probably generates the single-stranded DNA that facilitates helicase entry into the ori. Although this postulated two-step mechanism for helicase loading awaits verification by the isolation and analysis of mutants of RepA that no longer interact with DnaA, we have isolated similar mutants in the π initiator protein of R6K. Recently, we have reported that DnaA protein specifically interacts with the π initiator of R6K, and mutational disruption of this interaction leads to loss of ori unwinding in vitro and plasmid replication in vivo (23). The preceding observations extrapolated from the R6K system would suggest that DnaA–RepA interaction, which is facilitated by the DNA bending protein IHF, is needed for pSC101 ori unwinding. This initial unwinding probably generates the single-stranded region needed for the entry of DnaB. The collaboration between DnaA and a plasmid-encoded initiator for ori unwinding also has been observed in another system (28).
The role of DnaC in the helicase loading is not clear at the present time. ATP alone has been reported to load the toroidal, hexameric ring of DnaB onto naked single-stranded DNA circles in the absence of DnaC (17). DnaC is known to form a 1:1 complex with DnaB (29) and perhaps this interaction stabilizes the helicase complex and thus helps the loading process. Isolation of mutants of DnaB and DnaC that prevent the protein–protein interaction and biochemical analysis of the mutants might shed light on the role of DnaC in preinitiation.
How general might be this mechanism of helicase recruitment? We expect it to be common to other plasmid systems where the plasmid initiator interacts with the host helicase (e.g., R6K, ref. 19). However, in the broad host range plasmid RK2, the initiator protein TrfA does not seem to interact with DnaB and the host-encoded DnaA by itself seems to recruit the helicase to the ori (30). In phage λ, helicase recruitment involves protein–protein interaction between the phage-encoded O and the P proteins and between P and DnaB. P serves as a bridge between the O and DnaB proteins (31). Thus several different pathways of helicase loading seem to operate in nature.
The DNA-binding domain of RepA protein appears to be located toward the C terminus (32), whereas most of the specific protein–protein interaction seems to involve the N-terminal region (this paper and H.J.D. and D.B., unpublished work). The investigations of protein–protein interactions between replisomal proteins continues to provided important insights into the mechanism of DNA replication and its control. Recent work shows that interaction with DnaB distinguishes the leading strand DNA polymerase III holoenzyme from the one that synthesizes the lagging strand and the interaction of DnaB with the τ subunit of the DNA polymerase III holoenzyme heterodimer coordinates the rate of polymerization with that of DNA unwinding (17, 33). We recently have discovered that RepA protein (and DnaA and π) also interacts specifically with DnaG primase and with the τ subunit of DNA polymerase III (H.J.D. and D.B., unpublished work). Do initiator proteins also catalyze chain elongation? This question and other interesting ones remain to be answered.
Acknowledgments
This work was supported by a Merit Award to D.B. from the National Institute of Allergy and Infectious Diseases and a grant from the National Institute of General Medical Sciences.
ABBREVIATIONS
- IHF
integration host factor
- SSB
single-stranded DNA-binding protein
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Cohen S N, Chang A C Y. Proc Natl Acad Sci USA. 1973;70:1293–1297. doi: 10.1073/pnas.70.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tucker W T, Miller C A, Cohen S N. Cell. 1984;38:191–201. doi: 10.1016/0092-8674(84)90540-3. [DOI] [PubMed] [Google Scholar]
- 3.Vocke C, Bastia D. Proc Natl Acad Sci USA. 1983;80:6557–6561. doi: 10.1073/pnas.80.21.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vocke C, Bastia D. Cell. 1983;35:495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- 5.Vocke C, Bastia D. Proc Natl Acad Sci USA. 1985;82:2252–2256. doi: 10.1073/pnas.82.8.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchward G, Linder P, Caro L. Nucleic Acids Res. 1983;11:5645–5659. doi: 10.1093/nar/11.16.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamaguchi K, Yamaguchi M. Gene. 1984;29:211–219. doi: 10.1016/0378-1119(84)90181-1. [DOI] [PubMed] [Google Scholar]
- 8.Manen D, Upegui-Gonzalez L C, Caro L. Proc Natl Acad Sci USA. 1992;89:8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornberg A, Baker T. DNA Replication. San Francisco: Freeman; 1992. [Google Scholar]
- 10.Hasunuma K, Sekiguchi M. Mol Gen Genet. 1977;154:225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- 11.Stenzel T T, MacAllister T, Bastia D. Genes Dev. 1991;5:1453–1463. doi: 10.1101/gad.5.8.1453. [DOI] [PubMed] [Google Scholar]
- 12.Stenzel T T, Patel P, Bastia D. Cell. 1987;49:709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- 13.Gamas P, Burger A C, Churchward G, Caro L, Galas D, Chandler M. Mol Gen Genet. 1986;204:85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- 14.Biek D P, Cohen S N. J Bacteriol. 1989;171:2056–2065. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biek D P, Cohen S N. J Bacteriol. 1989;171:2066–2074. doi: 10.1128/jb.171.4.2066-2074.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelman Z, Naktinis V, O’Donnell M. Methods Enzymol. 1995;262:430–442. doi: 10.1016/0076-6879(95)62034-6. [DOI] [PubMed] [Google Scholar]
- 17.Yuzhakov A, Turner J, O’Donnell M. Cell. 1996;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 18.MacAllister T W, Kelley W L, Miron A, Stenzel T T, Bastia D. J Biol Chem. 1991;266:16056–16062. [PubMed] [Google Scholar]
- 19.Ratnakar P V A L, Mohanty B K, Lobert M, Bastia D. Proc Natl Acad Sci USA. 1996;93:5522–5526. doi: 10.1073/pnas.93.11.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nash H, Robertson C, Flamm E, Weisberg R A, Miller H I. J Bacteriol. 1987;169:4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Y, Ratnakar P V A L, Mohanty B K, Bastia D. Proc Natl Acad Sci USA. 1996;93:12902–12907. doi: 10.1073/pnas.93.23.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masai H, Nomura N, Arai K-I. J Biol Chem. 1990;265:15134–15144. [PubMed] [Google Scholar]
- 23.Lu Y-B, Datta H J, Bastia D. EMBO J. 1998;17:5192–5200. doi: 10.1093/emboj/17.17.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasunuma K, Sekiguchi M. J Bacteriol. 1979;137:1095–1099. doi: 10.1128/jb.137.3.1095-1099.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaylo P J, Turjman N, Bastia D. J Bacteriol. 1987;169:4703–4709. doi: 10.1128/jb.169.10.4703-4709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marszalek J, Kaguni J M. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- 27.Sutton M D, Kaguni J M. J Bacteriol. 1995;177:6657–6665. doi: 10.1128/jb.177.22.6657-6665.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konieczny I, Doran K S, Helinski D R, Blasina A. J Biol Chem. 1997;272:20173–20178. doi: 10.1074/jbc.272.32.20173. [DOI] [PubMed] [Google Scholar]
- 29.Wickner S, Hurwitz J. Proc Natl Acad Sci USA. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doran K S, Konieczny I, Helinski D R. J Biol Chem. 1998;273:8447–8453. doi: 10.1074/jbc.273.14.8447. [DOI] [PubMed] [Google Scholar]
- 31.Mallory J B, Alfano C, McMacken R. J Biol Chem. 1990;265:13297–13307. [PubMed] [Google Scholar]
- 32.Manen D, Caro L. Mol Microbiol. 1991;5:233–237. doi: 10.1111/j.1365-2958.1991.tb02103.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Dallman G H, McHenry C S, Marians J K. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]