Abstract
Mammals have nine differentially regulated isoforms of G protein-responsive transmembrane-spanning adenylyl cyclases. We now describe the existence of a distinct class of mammalian adenylyl cyclase that is soluble and insensitive to G protein or Forskolin regulation. Northern analysis indicates the gene encoding soluble adenylyl cyclase (sAC) is preferentially expressed in testis. As purified from rat testis cytosol, the active form of sAC appears to be a fragment derived from the full-length protein, suggesting a proteolytic mechanism for sAC activation. The two presumptive catalytic domains of sAC are closely related to cyanobacterial adenylyl cyclases, providing an evolutionary link between bacterial and mammalian signaling molecules.
Adenylyl cyclase (AC) is the effector molecule of one of the most widely used signal transduction pathways. Its product, cAMP, mediates cellular responses to nutritional conditions and extracellular signals in organisms from prokaryotes to higher eukaryotes. In metazoans, a seemingly ubiquitous membrane-associated AC activity is encoded by a family of transmembrane adenylyl cyclases (tmACs) that mediate cellular responses to external stimuli. In mammals, nine distinct tmAC genes differing in their patterns of expression and regulatory properties have thus far been identified. Their catalytic activities are differentially regulated by G proteins and other signaling molecules in response to stimuli such as hormones and neurotransmitters (1, 2).
In addition, another type of AC activity had been described in mammals. A soluble enzymatic activity was detected in cytosolic extracts from mammalian testis (3). Soluble AC activity appeared to be biochemically and chromatographically different from the tmACs and soluble guanylyl cyclases previously described in testis (4–6). Unlike the known tmACs, its biochemical activity depended on the divalent cation Mn2+ (3), was insensitive to G protein regulation (6), and displayed approximately 10-fold lower affinity for substrate, ATP (Km ≈1 mM) (4, 7, 8) than the tmACs (Km ≈100 μM) (9). Based on these studies, this soluble form of AC was predicted to be molecularly distinct from tmACs (8, 10).
A soluble form of AC would define a novel means for generating cAMP and would imply that the second messenger could be generated at a distance from the membrane, closer to its required site of action. However, the molecular evidence confirming that soluble AC represents a distinct form of AC had been lacking. We now describe purification, molecular cloning, and functional expression of the cDNA encoding the soluble form of AC (sAC). The full-length cDNA predicts a protein of 187 kDa, whereas the catalytically active purified form of the enzyme is only 48 kDa, suggesting a proteolytic mechanism of activation for sAC. Two distinct regions within the catalytically active portion of sAC are very similar to catalytic portions of ACs from cyanobacteria and myxobacteria, providing a link between bacterial and mammalian signaling systems.
MATERIALS AND METHODS
Cyclase Assay.
In vitro adenylyl cyclase assay was performed as described previously (11, 12), except that the standard assay conditions for sAC activity included 5 mM MnCl2 in place of MgCl2 and contained 5 mM [α-32P]ATP (specific activity = ≈4 × 104 cpm/nmol).
sAC Purification.
sAC (≈3 μg) was purified from 950 rat testes by sequential column chromatography by using the following scheme (see also Table 1): (i) Frozen rat testes (950) (Pel-Freez Biologicals) (in batches consisting of 50 testes) were homogenized and sonicated in 20 mM Tris⋅HCl, pH 7.5, in the presence of DTT and proteinase inhibitors. After debris and nuclei were removed by low-speed centrifugation (3000 × g for 10 min), a high-speed supernatant (>100,000 × g for 60 min) was prepared. (ii) Total cytosolic protein (52 g) was dialyzed and separated (as 19 equal portions consisting of 50 testes each) over DE-52 cellulose anion exchange columns (Whatman; 80 ml bed volume; 20 mM Tris⋅HCl, pH 7.5) by using a linear NaCl gradient. All sAC activity bound and eluted as one peak between 0.15 to 0.2 M NaCl. (iii) sAC activity recovered from DE-52 (4 g protein divided into 11 aliquots of 8 mls each) was separated by using an Ultrogel AcA54 gel filtration column (LKB; 4 × 100 cm/20 mM Tris⋅HCl, pH 7.5; flow rate 1.0 ml/min). sAC activity reproducibly eluted in a single peak with an apparent mass of 50–60 kDa. (iv) All sAC peak fractions from AcA54 gel filtration were pooled (1 g protein) and applied to a reactive Red 120-Agarose column (Sigma; 50 ml bed volume; 20 mM Tris⋅HCl, pH 7.5; linear gradient 0.1–1.0 M NaCl; flow rate 2 ml/min; 600 ml total). Cyclase activity eluted between 0.45 and 0.55 M NaCl. (v) Active fractions (66 mg protein) were pooled, dialyzed, and applied to a Source Q anion exchange column (Pharmacia; 15 ml bed volume; 20 mM Tris⋅HCl, pH 7.5; linear gradient 0–0.3 M NaCl; flow rate 0.5 ml/min; 150 ml total). sAC activity eluted between 0.10 and 0.15 M NaCl. (vi) Active fractions (9 mg protein) were pooled, concentrated, and applied to a reactive Green 19-Agarose column (Sigma; 9 ml bed volume; 20 mM Tris⋅HCl, pH 7.5; linear gradient 0.1–1.0 M NaCl; flow rate 0.6 ml/min; 80 ml total). Cyclase activity eluted between 0.40 and 0.50 M NaCl. (vii) Active fractions (1.8 mg) were pooled, concentrated, and loaded onto a semipreparative HydroCell QA 1000 HPLC anion exchange column (Biochrom, Terre Haute, IN; 50 × 4.6 mm; 20 mM Tris⋅HCl, pH 7.4; linear gradient 0–0.3 M NaCl over 30 min; flow rate 2 ml/min). Cyclase activity eluted between 0.07 and 0.10 M NaCl. (viii) Active fractions (0.6 mg) were pooled and loaded onto an analytical QA 1,000 HPLC anion exchange column (HydroCell Biochrom; 150 × 2.3 mm; 20 mM Tris⋅HCl, pH 6.8; linear gradient 0–0.1 M NaCl over 25 min; flow rate 1.5 ml/min; 0.5 ml/fraction). Cyclase activity eluted between 0.04 and 0.06 M NaCl. (ix) Active protein fractions were separated on SDS/PAGE, stained with Coomassie Blue G-250, and the 48- and 62-kDa bands were excised. Protein sequence data were obtained at the Rockefeller University Protein/DNA Technology Center (13, 14) from ≤5 μg of recovered 48-kDa protein.
Table 1.
Purification of sAC from 950 rat testis
| AC activity
|
||||
|---|---|---|---|---|
| Protein, mg | Total units, nmol/min | Specific activity, nmol/min/mg × 100 | Fold enrichment | |
| Cytosol | 51,900 | 2,400 | 4.6 | 1 |
| Preparative DE-52 | 4,015 | 3,000 | 75 | 16 |
| Gel filtration AcA54 | 1,074 | 2,100 | 200 | 43 |
| Reactive Red | 66 | 1,200 | 1,800 | 390 |
| Source Q | 8.8 | 1,100 | 12,500 | 2,700 |
| Reactive Green | 1.8 | 380 | 21,000 | 4,600 |
| Semipreparative QA, pH = 7.4 | 0.6 | 310 | 52,000 | 11,300 |
| Analytical QA, pH = 6.8 | ||||
| #18 | 0.003 | 90 | 3,000,000 | 650,000 |
| #19 | 0.010 | 92 | 920,000 | 200,000 |
Protein concentrations determined by OD280. Units refer to nmol of cAMP formed per minute. Fold enrichment represents specific activity after each step compared to the starting specific activity. See Materials and Methods for detailed description of each purification step.
Molecular Cloning.
Fully degenerate oligonucleotide primers designed to recognize the amino acid sequences of peptides derived from the 48-kDa purified polypeptide (Fig. 2, double underlined) were synthesized for use in PCR amplification of rat testis first-strand cDNA. A 1-kilobase (kb) PCR fragment was generated that had a single ORF extending throughout its length and that contained sequences corresponding to all three peptides. This 1-kb PCR fragment was used as probe to screen a rat testis cDNA library constructed in our laboratory (λZap, Stratagene). From over 7.5 × 105 plaques, we obtained four overlapping cDNA clones. Among these, one represented a complete full-length cDNA clone.
Figure 2.
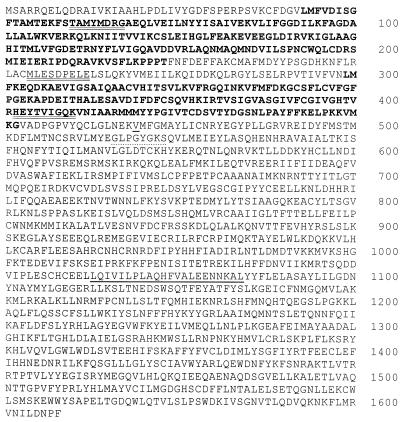
sAC amino acid sequence. Predicted amino acid sequence of rat sAC. Amino acids in bold indicate presumptive catalytic domains, C1 and C2. Double-underlined amino acids correspond to sequences of tryptic peptides derived from the purified 48-kDa protein. Dotted underlined amino acids conform to a consensus P loop sequence, and underlined sequences are predicted to form a leucine zipper. Valine 469 is underlined and is the last sAC amino acid in the catalytically active heterologously expressed truncation.
The nucleotide sequence of the full-length cDNA was determined on both strands by dye termination-automated DNA sequencing (Cornell University DNA sequencing Core Facility, Ithaca, NY) and confirmed by comparison to single-stranded sequence determined from at least one other independent cDNA clone.
Sequence and database searching was performed on-line by means of blast (http://www.ncbi.nlm.nih.gov/blast/) or psort II (http://psort.nibb.ac.jp:8800/).
Hybridizations.
Southern and Northern blots were probed with random-primed [32P]dCTP-labeled 1-kb PCR-generated fragment under standard conditions (15). Southern blot was hybridized at 65°C overnight and washed three times in [1× SSC (0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS] for 15 minutes at 55°C (low stringency) or three times in [0.5× SSC/0.1% SDS] for 15 minutes at 65°C (high stringency).
Heterologous Expression.
The full-length and truncated sAC cDNAs were expressed from the library vector (pBK-CMV) under the control of the cytomegalovirus promoter after deletion of the intervening bacterial promoter sequences (as an NheI - SpeI fragment). The truncated sAC cDNA represents a library clone that was missing an exon. The resultant protein shifted reading frame after valine469 (Fig. 2, underline), introducing two incorrect amino acids (serine and cysteine) followed by a stop codon. Expression constructs were transiently introduced into HEK293 by lipofectamine- (Life Technologies, Grand Island, NY) mediated transfection. One or two days after transfection, cells were harvested, resuspended in lysis buffer [50 mM Tris⋅HCl, pH 7.5/1 mM EDTA/1 mM DTT/0.1 mg/ml Leupeptin/1 mM phenylmethylsulfonyl fluoride] and disrupted by microtip probe sonication on ice. Whole-cell sonicates were ultramicrocentrifuged at >100,000 × g for 10 minutes. Supernatants were cleared by a second centrifugation to yield ‘cytosolic’ extracts. Pellets were resuspended in lysis buffer by passage through a 27.5-gauge needle to generate ‘particulate fraction.’
RESULTS
Purification of sAC.
We first confirmed the presence of Mn2+-dependent AC activity in cytosolic extracts from frozen rat testis. The soluble enzymatic activity we detected was unresponsive to either forskolin or the nonhydrolyzable GTP analogue, GTPγS, two general tmAC activators, and it displayed a Km for ATP of 1.2 mM in the presence of MnCl2 (data not shown). These data indicate we were assaying the previously described soluble AC (8).
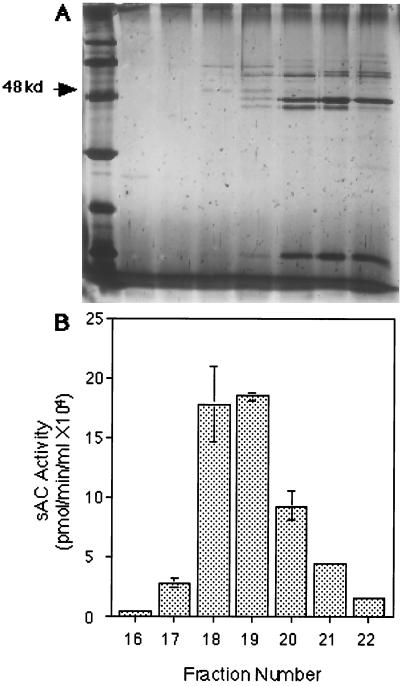
We purified sAC activity using a combination of classical chromatographic methods (Table 1) and identified a 48-kDa candidate protein band (Fig. 1). The final chromatographic separation (step viii; analytical QA 1,000 HPLC) achieved greater than 60-fold enrichment (Table 1, fraction #18) even though it used the identical QA anion exchange matrix as the HPLC column preceding it (step vii; semipreparative QA 1,000 HPLC). By varying the buffer pH (pH = 7.4 for the semipreparative QA vs. pH = 6.8 for the analytical QA), sAC activity eluted before the majority of contaminating proteins during this final chromatographic separation.
Figure 1.
Purification of sAC. (A) Silver stained gel and (B) activity profile of selected fractions from Analytical QA column chromatography. Ten-microliter aliquots of selected fractions were separated on a 12% SDS/PAGE gel and silver stained. The first lane is low molecular weight silver stain marker, and each subsequent lane corresponds to the fraction assayed for sAC activity beneath it. Bars represent the average sAC activity in duplicate assays for each fraction. Error bars indicate the standard deviation from the mean.
A silver-stained 12% SDS/PAGE gel of the active fractions from the final chromatographic step of the purification revealed two protein bands (of approximately 48 and 62 kDa) whose intensities coeluted with enzyme activity (Fig. 1). During pilot purification studies, analytical gel filtration of partially purified cytosol predicted sAC to be 45–55 kDa (data not shown), suggesting the more likely candidate was the 48-kDa protein. We initially attempted to clone cDNAs corresponding to both polypeptides; unfortunately, peptide sequences derived from the 62-kDa species were insufficient to permit its molecular isolation. The limited amino acid sequence information we did obtain from this polypeptide reveal it is completely unrelated to the sAC gene.
Cloning of sAC.
The amino acid sequences of three tryptic peptides derived from the 48-kDa candidate polypeptide were not present in the databases of known proteins, indicating it represented a novel protein. The cDNA encoding this polypeptide was isolated by PCR followed by screening a rat testis cDNA library. All isolated cDNAs appeared to derive from one transcript whose nucleotide sequence revealed a single long ORF predicting a protein of 187 kDa (Fig. 2), which is significantly larger than the size (48 kDa) estimated by SDS/PAGE of the purified sAC protein (Fig. 1). The peptides derived from the 48-kDa purified polypeptide reside completely within the amino terminal portion of the full-length protein (Fig. 2, double underline), suggesting the purified polypeptide represents a proteolytically processed active form of the protein.
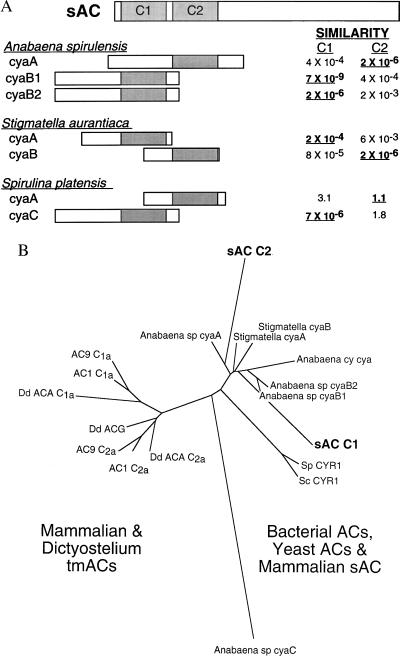
Comparison of this ORF with known protein sequences revealed two distinct regions of the putative sAC protein that display significant amino acid homology to various adenylyl cyclase catalytic domains (Fig. 2, bold type). Both sAC domains, C1 and C2, reside within its amino terminal 50 kDa and are therefore likely to be contained within the purified catalytically active processed form. The most closely related protein sequences in GenBank are the catalytic domains from a number of different cyanobacterial (Anabaena spirulensis cyaB1, cyaB2, and cyaA; and Spirulina platensis cyaC) and myxobacterial (Stigmatella aurantiaca cyaA and cyaB) adenylyl cyclases (Fig. 3A). These species have multiple AC genes with each isoform having a single catalytic domain. Interestingly, the catalytic domain of one AC isoform in each bacterial species is more similar to C1, whereas the catalytic domain of a second isoform from that species more closely resembles C2. This and the fact that C1 and C2 are not very similar to each other may suggest that during its evolution, mammalian sAC resulted from a fusion of distinct bacterial proteins rather than through duplication of a single catalytic domain.
Figure 3.
sAC has two presumptive catalytic domains most closely related to bacterial AC catalytic domains. (A) Diagram of presumptive catalytic domains of sAC aligned with catalytic portions of various bacterial ACs. The relative similarities are “expect values” taken directly from a blast search of the sAC protein vs. the nonredundant GenBank database. These values estimate the statistical significance of the match by specifying the number of matches expected to occur by chance. Relative locations of the catalytic domains within the bacterial ACs are represented as shaded boxes (24–26) and are aligned under the sAC presumptive catalytic domain with greater similarity. (B) Phylogenetic relationship between catalytic domains from a variety of ACs aligned by using clustalw (dna*) represented as an unrooted dendogram constructed by using fitch (phylip 3.5) (27) with Anabaena spirulina cyaC used as the outgroup. Accession numbers for the aligned sequences are AC1 (bovine Type I: M25579), AC9 (mouse Type IX: Z50190), Dd ACA (Dictyostelium ACA: Q03100), Dd ACG (Dictyostelium ACG: Q03101), Anabaena sp. cyaA (2126532), Anabaena sp. cyaB1 (1754638), Anabaena sp. cyaB2 (1754640), Anabaena sp. cyaC (2575807), Anabaena cy. cya (2126532), Stigmatella cyaA (729248), Stigmatella cyaB (729250), Sc CYR1 (Saccharomyces cerevisiae CYR1: M12057), and Sp CYR1 (Schizosaccharomyces pombe CYR1: M24942).
There is also significant similarity between the two presumptive sAC catalytic domains and other AC catalytic domains. Alignment of C1 and C2 with the catalytic domains from related bacterial ACs, yeast ACs, Dictyostelium tmACs, and representative mammalian tmACs (Fig. 3B) reveals that sAC C1 and C2 are more closely related to the catalytic portions of bacterial ACs than to the catalytic domains of any other cyclases. This similarity provides an evolutionary link between bacterial and mammalian signaling systems and suggests that the C1 and C2 catalytic domains in mammalian sAC are likely to have evolved independently from those in eukaryotic tmACs (C1a and C2a).
The C-terminal portion beyond the AC homologous regions revealed no significant homology to any known protein in the databases, and the hydropathy profile of the full-length protein indicated no obvious potential transmembrane-spanning domains. Sequences that could represent a nucleotide-binding P Loop (Fig. 2, dotted underline) or that could form a leucine zipper interacting domain (Fig. 2, single underline) were detected within the region unrelated to the AC catalytic domains; however, the physiological significance of these sequence motifs in sAC is not yet known.
sAC Appears to Be a Single Copy Gene.
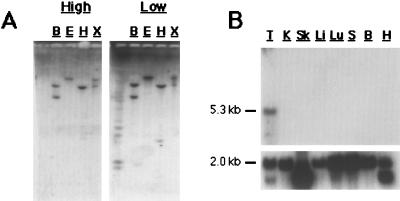
Southern hybridization to rat genomic DNA, along with numerous database searches, indicates the presumptive sAC gene does not appear to be the progenitor of a gene family of sAC-like molecules in mammals. A sAC coding sequence probe hybridized at high and low stringency to parallel rat genomic Southern blots recognized identical genomic fragments (Fig. 4A), indicating the lack of closely related sequences in the genome. Additionally, we have identified the corresponding human and mouse sAC genes by database searches and reverse transcription—PCR (RT-PCR), respectively (data not shown). The human sAC locus has been sequenced as part of the Genome Project. It is encoded by more than 30 exons that are spread across two overlapping PAC (P1-derived artificial chromosome) clones mapping to 1q24.
Figure 4.
sAC is a single copy gene preferentially expressed in testis. (A) Southern blots of 10 μg rat genomic DNA digested with BamHI (B), EcoRI (E), HindIII (H), and XhoI (X) probed at high (Left) or low (Right) stringency by using the 1-kb PCR-generated sAC fragment containing both presumptive catalytic domains. (B) Rat multiple tissue Northern blot (CLONTECH) representing approximately 2 μg poly-A+ RNA from testis (T), kidney (K), skeletal muscle (Sk), liver (Li), lung (Lu), spleen (S), brain (B), and heart (H) probed with 1-kb PCR-generated sAC fragment (Upper) or with actin control (Lower). The sAC transcript is approximately 5.3 kb and, in most tissues, actin is approximately 2.0 kb.
sAC Is Preferentially Expressed in Testis.
In mammals, soluble AC activity had been detected only in testis (3, 4, 8). Northern analysis of a limited number of tissues indicates expression of the presumptive sAC gene is detectable only in testis (Fig. 4B), supporting the previously determined biochemical restriction. However, sAC can be detected in other tissues by RT-PCR (M.L.S., M. Mattia, J.B., and L.R.L., unpublished observations).
Heterologous Expression and Activity.
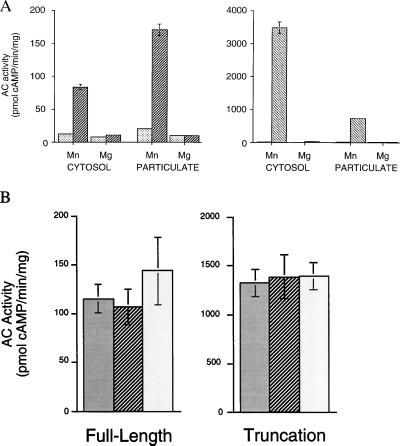
To confirm that the gene we isolated encodes an adenylyl cyclase, we heterologously expressed the full-length cDNA in HEK293 cells. Vector-transfected cells had no detectable soluble and very little unstimulated particulate AC activities. In contrast, cells transfected with the full-length sAC gene displayed substantial Mn2+-dependent soluble AC activity (Fig. 5A Left). Mn2+-dependent activity was also elevated in the particulate fraction from transfected cell sonicates. It is possible that this particulate activity accounts for the membrane-associated sAC-like enzymatic activity previously described in mammalian sperm (3).
Figure 5.
sAC is active in HEK293 cells. (A) Adenylyl cyclase activity in the presence of 5 mM MnCl2 or 5 mM MgCl2 is shown for cytosolic and particulate extracts from HEK293 cells transiently transfected with pBK-CMV vector (░⃞), full-length sAC gene (▨), or truncated sAC gene (▧). Values are expressed as pmols of cAMP formed per minute per mg protein and represent averages of triplicate determinations. Error bars indicate the standard deviation from the mean. Note the 20-fold difference in scale of the ordinate between left and right panels. (B) sAC activity in whole-cell sonicates from HEK293 cells transfected with full-length sAC (Left) or truncated sAC (Right) assayed in the absence of any activators (░⃞) or in the presence of 100 μM forskolin (▨) or 5 mM GTPγS (□). Values represent the averages of quadruplicate determinations with vector transfected HEK293 values for each condition subtracted. Error bars indicate the standard deviation from the mean.
Because the sAC polypeptide purified from rat testis was approximately 48 kDa (Fig. 1), we sought to determine whether a truncated version of sAC retained catalytic activity. An expression construct consisting of the amino terminal 53 kDa of sAC (Fig. 2) encompassing both presumptive catalytic domains was fully active, displaying an extremely high level of Mn2+-dependent soluble AC activity (Fig. 5A Right). Therefore, the sAC purified from rat testis cytosol very likely represents a proteolytically processed form resembling this N-terminal truncation. The extremely high level of cytosolic activity in cells expressing the truncated form compared with those expressing the full-length protein is consistent with processing being required for catalytic activity; the activity in cells transfected with full-length sAC may be limited by the availability of activating enzymes.
When originally detected, testis sAC was thought to be molecularly distinct from the tmACs because it appeared to be insensitive to G protein and forskolin regulation (8). We initially confirmed these results in partially purified testis cytosol; however, the probable lack of G proteins in these soluble extracts makes their interpretation problematic. Therefore, we examined whether heterologously expressed sAC responded to these known stimulators of tmAC activity. When assayed in whole-cell sonicates from transfected HEK293 cells, which should contain the full complement of endogenous G proteins, both full-length and truncated forms of sAC were completely insensitive to forskolin and to the nonspecific G protein activator, GTPγS (Fig. 5B).
DISCUSSION
In this report, we demonstrate the existence of a unique signaling molecule in mammals. The sAC gene we have identified in rat, mouse, and human encodes a cytosolic form of adenylyl cyclase that is distinct from the previously characterized mammalian tmACs. Not only is sAC not a transmembrane protein, but its catalytic domains are more closely related to the catalytic portions of bacterial ACs than they are to the catalytic domains of any other eukaryotic cyclase. In contrast, the mammalian tmACs, which are distantly related to these bacterial ACs and sAC, more closely resemble other invertebrate (Drosophila) and lower eukaryotic (Dictyostelium) ACs. Computational methods at our disposal did not reveal a one-to-one relationship between the catalytic domains of sAC (C1 and C2) and those of the tmACs (C1a and C2a). However, each catalytic domain in sAC shows greater similarity to a different AC within individual cyanobacterial and myxobacterial species, implying that sAC represents an evolutionary fusion of distinct bacterial enzymes.
sAC can also be distinguished from the tmACs by its biochemical regulation. Unlike tmACs, sAC is insensitive to G protein and forskolin modulation, and its in vitro catalytic activity depends on the divalent cation Mn2+. Additionally, sAC requires higher concentrations of ATP than the tmACs. This decreased affinity for substrate may reflect a specialization because of its physiological role. Biochemical (3, 10) and molecular (Fig. 4B) evidence indicate sAC is preferentially expressed in testis and is very likely in post-meiotic germ cells (6, 16, 17) where the very high endogenous ATP concentration would support sAC activity. With its molecular isolation and functional heterologous expression, we now have the necessary tools to explore the biochemical regulation of sAC activity and ultimately determine its biological role.
Also unlike tmACs, sAC contains a consensus P loop or nucleotide binding sequence. However, this region is not contained within the heterologously expressed active truncation and is therefore not necessary for catalytic activity. Whether this region binds nucleotide, perhaps as a modulatory mechanism reminiscent of the membrane guanylyl cyclases (18, 19), remains to be determined. Also in the C-terminal portion of sAC not required for cyclase activity is a potential leucine zipper or coiled–coiled interaction domain of unknown significance. It is intriguing to speculate that this region serves as a protein–protein interaction domain, possibly tethering sAC to the cytoskeleton at specific locations within the cell.
The N-terminal 50 kDa of sAC is sufficient for enzymatic activity and approximately corresponds to the size of the protein purified from rat testis cytosol. Because all the cDNAs isolated from rat testis fell into a single class encoding the 187-kDa polypeptide, the 48-kDa purified protein should result from posttranslational cleavage. Truncating the sAC gene increased cyclase activity 10–20 fold in tissue culture cells, suggesting that the shorter molecule approximates an activated form. In fact, the full-length protein may not have measurable cyclase activity; the activity detected in cells transfected with the full-length cDNA could result from a cleaved molecule. If true, the cyclase activity in full-length sAC transfected cells would be limited by the availability of a posttranslational processing mechanism. Regardless of the relative activity of the full-length protein, it remains to be determined whether the proteolytic cleavage of sAC in testis cytosol represents a physiologically relevant maturation from a precursor protein or a fortuitous degradation caused by experimental manipulation.
Its restricted localization suggests sAC contributes to male fertility. Sperm functions thought to be mediated by cAMP (reviewed in ref. 20) include maturation, motility, and the acrosome reaction. Multiple tmACs are known to be expressed in testis, but sAC provides the intriguing ability to generate cAMP precisely where it is needed in the cytoplasm. Two distinct cAMP-dependent protein kinase anchoring proteins (AKAPs) localize to the sperm flagellum, one to the mitochondrial sheath (21) and the other to the fibrous sheath (22). Unlike plasma membrane-localized tmACs, cytosolic sAC could generate second messenger directly at the site of these AKAP-localized cAMP-dependent protein kinases.
Given the ubiquitous nature of the tmACs, it is very likely that some cells in the testis express both transmembrane and cytosolic forms of adenylyl cyclase. It is possible that cAMP generated by sAC and by the tmACs could cooperate in modulating a single signal or could propagate separate signals. Furthermore, the activities of the G protein-responsive tmACs and sAC are regulated by distinct mechanisms, but it will be interesting to examine whether the cAMP they generate modulates distinct effectors. For example, would cytoplasmically generated cAMP activate membrane spanning cyclic-nucleotide gated ion channels?
Whereas much remains to be determined about its biochemistry and biological function, sAC clearly defines a novel means for generating cAMP. Models describing cAMP signaling in mammalian cells include only membrane proximal generation of the second messenger. Because its major effector can be anchored at various places in the cell by AKAP proteins (reviewed in ref. 23), cAMP must diffuse through the cytoplasm to propagate its signal. However, recent suggestions that signal transduction occurs in microdomains seem to contradict this theory. We now demonstrate that cAMP can be synthesized anywhere in the cytosol by the soluble form of adenylyl cyclase, removing the membrane-proximal limitation on cAMP generation and revealing new aspects of what was previously thought to be a very well characterized signaling pathway.
Acknowledgments
We thank Mayya Maksimova for technical assistance; Dr. Vadim Iourgenko, Dr. Arlene Rifkind, Dr. Marcus Reidenberg, Dr. Mark J. Zoller, and Dr. Randall R. Reed for critical reading of the manuscript; and Dr. Sheena Mische and Dr. Joseph Fernandez of the Rockefeller University Protein Sequencing Core Facility for peptide sequence determination. Supported by National Institutes of Health Grants DK48022 and DK52797 to J.B. and GM52891 to L.R.L. and by Weill Medical College of Cornell University Research Associates Bridge Funds. J.B. is a Pew Scholar in the biomedical sciences.
ABBREVIATIONS
- AC
adenylyl cyclase
- tmACs
transmembrane adenylyl cyclases
- sAC
soluble form of AC
- RT-PCR
reverse transcription–PCR
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The cDNA sequence of rat sAC has been deposited in the GenBank database (accession no. AF081941).
References
- 1.Taussig R, Gilman A G. J Biol Chem. 1995;270:1–4. doi: 10.1074/jbc.270.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Sunahara R K, Dessauer C W, Gilman A G. Annu Rev Pharmacol Toxicol. 1996;36:461–480. doi: 10.1146/annurev.pa.36.040196.002333. [DOI] [PubMed] [Google Scholar]
- 3.Braun T, Dods R F. Proc Natl Acad Sci USA. 1975;72:1097–1101. doi: 10.1073/pnas.72.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neer E J. J Biol Chem. 1978;253:5808–5812. [PubMed] [Google Scholar]
- 5.Neer E J, Murad F. Biochim Biophys Acta. 1979;583:531–534. doi: 10.1016/0304-4165(79)90070-9. [DOI] [PubMed] [Google Scholar]
- 6.Braun T, Frank H, Dods R, Sepsenwol S. Biochim Biophys Acta. 1977;481:227–235. doi: 10.1016/0005-2744(77)90155-3. [DOI] [PubMed] [Google Scholar]
- 7.Gordeladze J O, Hansson V. Mol Cell Endocrinol. 1981;23:125–136. doi: 10.1016/0303-7207(81)90064-2. [DOI] [PubMed] [Google Scholar]
- 8.Braun T. Methods Enzymol. 1991;195:130–136. doi: 10.1016/0076-6879(91)95160-l. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R A, Shoshani I. Methods Enzymol. 1994;238:56–71. doi: 10.1016/0076-6879(94)38006-6. [DOI] [PubMed] [Google Scholar]
- 10.Neer E J. Adv Cyclic Nucleotide Res. 1978;9:69–83. [PubMed] [Google Scholar]
- 11.Levin L R, Han P L, Hwang P M, Feinstein P G, Davis R L, Reed R R. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- 12.Levin L R, Reed R R. J Biol Chem. 1995;270:7573–7579. doi: 10.1074/jbc.270.13.7573. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez J, Andrews L, Mische S M. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez J, DeMott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Gordeladze J O, Purvis K, Clausen O P, Rommerts F F, Hansson V. Int J Androl. 1981;4:172–182. doi: 10.1111/j.1365-2605.1981.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 17.Gordeladze J O, Abyholm T, Cusan L, Clausen O P, Hansson V. Arch Androl. 1982;8:199–204. doi: 10.3109/01485018208987040. [DOI] [PubMed] [Google Scholar]
- 18.Chinkers M, Singh S, Garbers D L. J Biol Chem. 1991;266:4088–4093. [PubMed] [Google Scholar]
- 19.Wong S K, Ma C P, Foster D C, Chen A Y, Garbers D L. J Biol Chem. 1995;270:30818–30822. doi: 10.1074/jbc.270.51.30818. [DOI] [PubMed] [Google Scholar]
- 20.Garbers D L, Kopf G S. Adv Cyclic Nucleotide Res. 1980;13:251–306. [PubMed] [Google Scholar]
- 21.Lin R Y, Moss S B, Rubin C S. J Biol Chem. 1995;270:27804–27811. doi: 10.1074/jbc.270.46.27804. [DOI] [PubMed] [Google Scholar]
- 22.Johnson L R, Foster J A, Haig-Ladewig L, VanScoy H, Rubin C S, Moss S B, Gerton G L. Dev Biol. 1997;192:340–350. doi: 10.1006/dbio.1997.8767. [DOI] [PubMed] [Google Scholar]
- 23.Lester L B, Scott J D. Recent Prog Horm Res. 1997;52:409–429. [PubMed] [Google Scholar]
- 24.Katayama M, Ohmori M. J Bacteriol. 1997;179:3588–3593. doi: 10.1128/jb.179.11.3588-3593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasahara M, Yashiro K, Sakamoto T, Ohmori M. Plant Cell Physiol. 1997;38:828–836. doi: 10.1093/oxfordjournals.pcp.a029241. [DOI] [PubMed] [Google Scholar]
- 26.Coudart-Cavalli M P, Sismeiro O, Danchin A. Biochimie. 1997;79:757–767. doi: 10.1016/s0300-9084(97)86934-9. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. phylip, Phylogeny Inference Package. Univ. of Washington, Seattle: Department of Genetics; 1993. , Version 3.5. [Google Scholar]