Abstract
Translocation of exogenous DNA through the nuclear membrane is a major concern of gene delivery technologies. To take advantage of the cellular import machinery, we have synthesized a capped 3.3-kbp CMVLuciferase-NLS gene containing a single nuclear localization signal peptide (PKKKRKVEDPYC). Transfection of cells with the tagged gene remained effective down to nanogram amounts of DNA. Transfection enhancement (10- to 1,000-fold) as a result of the signal peptide was observed irrespective of the cationic vector or the cell type used. A lysine to threonine mutation of the third NLS amino acid completely abolished these remarkable features, suggesting importin-mediated translocation. Our hypothesis is that the 3-nm-wide DNA present in the cytoplasm is initially docked to and translocated through a nuclear pore by the nuclear import machinery. As DNA enters the nucleus, it is quickly condensed into a chromatin-like structure, which provides a mechanism for threading the remaining worm-like molecule through the pore. A single NLS signal is thus sufficient, whereas many signals on a gene would actually inhibit entry, the same DNA molecule being threaded through adjacent pores.
The nuclear membrane of eukaryotic cells is freely permeable to solutes of up to about 9 nm (e.g., 40- 60-kDa proteins). Transport of larger molecules through nuclear pores is signal-mediated, involves shuttle molecules, and requires energy. The basic peptide derived from the simian virus 40 large tumor antigen (PKKKRKV) is a nuclear localization signal (NLS) that mediates binding of the karyophilic protein to importin α (1). Complex formation triggers binding to importin β and the ternary complex is then carried through the nuclear pore with the help of the GTPase Ran. Macromolecules and particles up to 25 nm have been shown to enter the nucleus in this way, although the translocation mechanism is still not fully understood (2–5).
Eukaryotic DNA viruses, which replicate in the nucleus, seem to be capable of diverting the cell’s nuclear import machinery to their own benefit (6, 7). Recombinant viruses derived from these viruses are being used to carry therapeutic genes into humans. However, the host’s immune response is currently a limitation to the clinical development of viral gene therapy. Nonviral alternatives using plasmid DNA and cationic carrier molecules suffer from a different drawback, the low efficacy of gene delivery. The main barrier to transgene expression in vitro is the nuclear membrane (8–10). Because breakdown of this membrane during cell division helps nuclear localization, this obstacle is probably still more a problem in vivo, where cells can be considered to be resting with respect to the lifetime of DNA.
Several attempts to improve entry of plasmid DNA into the nucleus have been published. These include use of electrostatic binding of DNA to cationic NLS-containing proteins (11, 12) or peptides (13) or lipids (14), as well as sequence-specific binding of DNA to karyophilic proteins (15–17). Yet such “piggyback” nuclear transport relies on the unpredictable stability of the complexes within the cytoplasm. Recent work by the group of Jon Wolff (18) with digitonin-permeabilized cells demonstrated nuclear accumulation of fluorescently labeled DNA that was randomly tagged with hundreds of NLS peptides; nuclei of intact cells did not take up the modified DNA. Our approach took advantage of a chemically controlled pathway to irreversibly link a single NLS-peptide to one end of a gene. Moreover, following Heisenberg’s principle, we avoided taking a too close look at nuclear import using potentially disturbing techniques. The goal being gene delivery, the level of transgene expression after transfection was chosen as the most pertinent test of success.
EXPERIMENTAL PROCEDURES
Chemicals, Enzymes, and Oligonucleotides.
4-(N-Maleimidomethyl)cyclohexane-1-carboxylic acid N-hydroxysuccinimide ester (SMCC) was purchased from Sigma. The SAL 34-mer 5′-d(TCGATGTCCGCGTTGGCTTXTGCCAACGCGGACA) oligodeoxynucleotide was synthesized by Appligene by using an amino-modified deoxythymidine (X; amino-modified dT, Glen Research, Sterling, VA). The XMA 34-mer 5′-d(CCGGCTACCTTGCGAGCTTTTGCTCGCAAGGTAG) oligodeoxynucleotide, the NLS peptide NH2-PKKKRKVEDPYC, and the mutated-NLS peptide NH2-PKTKRKVEDPYC with C-terminal amidation were synthesized by Genosys (The Woodlands, TX). Hairpin structures composed of a loop of four thymines, a stem of 13 bp, and a sticky 5′ end were formed by boiling and subsequently cooling the XMA or the SAL oligonucleotides in ice. XmaI, XmnI, SalI, and BspHI restriction endonucleases, T4 polynucleotide kinase, T4 DNA ligase, and exonuclease III were purchased from New England Biolabs. Linear 22-kDa (ExGen500) and branched 25-kDa polyethylenimines (PEI) were purchased from Euromedex (Souffelweyersheim, France) and Fluka (Saint-Quentin Fallavier, France), respectively. Transfectam (dioctadecylamido glycylspermine) was synthesized as described (19).
Preparation of the Oligonucleotide-Peptide Conjugate.
Two A260 units (6.46 nmol, assuming ɛ260 = 309,800 M−1⋅cm−1) of the amino-modified SAL oligonucleotide in 20 μl of phosphate buffer [10 mM sodium phosphate (pH 7.5)] was mixed with a 40:1 molar excess of the bifunctional crosslinker SMCC in dimethylformamide (as a 30 mM stock solution) and incubated at room temperature for 2 h. Excess SMCC was removed with a Nick-Spin column (Amersham-Pharmacia) that had been equilibrated with PBS. The recovered oligonucleotide solution (100 μl) was immediately reacted with a 10-fold molar excess of NLS-peptide (or mutated-NLS-peptide) overnight at room temperature and then stored at −20°C. The oligonucleotide-peptide conjugate was purified by preparative PAGE. The coupling yield was 30%, based on quantification of the radiolabeled oligonucleotides after migration in a 20% denaturing polyacrylamide gel. To assess the peptide content, 1 pmol of radiolabeled oligonucleotide-NH2 or oligonucleotide-NLS was incubated in 10 mM Tris⋅HCl, pH 8/1 mM EDTA in the presence of proteinase K (0.5 mg/ml) for 10 min at 37°C, followed by incubation for 10 min at 65°C and subsequent loading on a denaturing polyacrylamide gel. Visualization and quantification were performed with a PhosphorImager 425 (Molecular Dynamics).
Ligation of the CMVLuc Restriction Fragment to the Hairpin Oligonucleotides.
pCMVLuc plasmid, encoding the Photinus pyralis luciferase under the control of the cytomegalovirus enhancer/promoter and followed by the simian virus 40 early polyadenylation signal (a gift from B. Demeneix, Museum National d’Histoire Naturelle, Paris) was propagated and purified as described (20). XmnI/XmaI double digestion (10 U/μg of DNA) was performed at 37°C for 2 h, followed by enzyme heat inactivation (65°C for 20 min), before SalI cleavage (37°C for 2 h; 10 U/μg of DNA). After removal of the endonucleases by DNA precipitation, BspHI digestion was performed for 2 h at 37°C (10 U/μg of DNA). Separation of the CMVLuc-containing DNA fragment (3,380 bp) from the shorter restriction fragments was performed by ultracentrifugation (30,000 rpm, SW41 rotor, 19 h at 25°C, Beckman Ultracentrifuge L8/55, France) in a 15–30% sucrose gradient.
The 5′ end of the oligonucleotide-peptide carrying the NLS or the mutated NLS peptide was radiolabeled with [γ-32P]ATP and T4 polynucleotide kinase (1 U/pmol of oligonucleotide at 37°C for 30 min). The 5′ end of the oligonucleotide-cap was phosphorylated similarly with ATP. Excess [γ-32P]ATP and ATP were removed with Microspin G-25 columns (Amersham-Pharmacia). Before ligation, the hairpin form of the oligonucleotides presenting a sticky 5′ end was obtained by boiling and subsequently cooling the sample in ice. Ligation of the CMVLuc fragment with the oligonucleotide-cap and the oligonucleotide-peptide was performed overnight at 13°C with a 15-fold molar excess of each oligonucleotide and T4 DNA ligase (10,000 U/μg of DNA). The excess oligonucleotide was removed with a Microspin S-400 HR column (Amersham-Pharmacia). Quantification of the ligase reaction yield (80–90%) was performed by Cerenkov counting (TRI-CARB 2100 TR Liquid Scintillation Analyzer, Packard). Capping of the CMVLuc fragment was checked by digestion with exonuclease III (10 U/μg of DNA) at 37°C. Agarose gel electrophoresis showed the uncapped and hemicapped fragments to be totally digested, whereas the capped fragment remained undigested.
Cells and Cell Culture.
NIH 3T3 murine fibroblasts were purchased from American Type Culture Collection and grown in DMEM (GIBCO/BRL). BNL CL.2 murine hepatocytes were provided by E. Wagner (Bender, Wien, Austria) and grown in DMEM, high glucose (4.5 g/liter). HeLa human cervix epitheloid carcinoma cells (provided by L. Monaco, Istituto San Raffaele, Milano, Italy) were grown in MEM with Earle’s salt (PolyLabo, Strasbourg, France). Human monocytes were isolated from the blood of a healthy donor by elutriation/cytapheresis and differentiated into macrophages by 1-week culture in RPMI 1640 medium supplemented with 100 U/ml GM-CSF. Dorsal root ganglia neurons from new born rats were obtained and grown as described (21). Cell culture media were supplemented with 10% fetal calf serum (GIBCO/BRL), 2 mM l-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml; GIBCO/BRL). Cells were maintained at 37°C in a 5% CO2 humidified air atmosphere.
Cell Transfection.
For each cell line used, 10,000 cells per well were seeded 24 h before transfection in 96-well tissue culture plates (Costar) to reach 60–70% confluence during transfection. Before transfection, cells were rinsed and 0.2 ml of fresh culture medium supplemented (transfection in the presence of serum) or not (transfection in the absence of serum) with 10% fetal calf serum was added to each well. The desired amount of DNA was diluted into 46 μl (final volume) of 0.15 M NaCl or 5% glucose. The desired quantity of ExGen500, 25-kDa PEI, or Transfectam (from a 1 mM aqueous amine nitrogen stock solution of PEI or a 2 mM ethanolic stock solution of Transfectam) was then added to the DNA-containing solutions, vortex-mixed gently, and centrifuged. After 10 min, volumes corresponding to 10, 20, or 200 ng of CMVLuc fragment (10 ng/μl of DNA) or to the gene-number-corrected amount of plasmid DNA were added to the cells. The cell culture dish was immediately centrifuged (Sigma product) for 5 min at 1500 rpm (280 × g) or 500 rpm for primary neurons. After 2–3 h, 20 μl of fetal calf serum was added to the serum-free wells. Cells were cultured for 24 h and tested for reporter gene expression. All experiments were done in duplicate.
Luciferase Assay.
Luciferase gene expression was measured by a luminescence assay. The culture medium was discarded and cell lysate was harvested after incubation of cells for 30 min at room temperature in 100 μl of Lysis Reagent 1× (Promega). The lysate was vortex-mixed gently and centrifuged for 5 min at 14,000 rpm at 4°C (Sigma 3K10). Twenty microliters of supernatant was diluted into 100 μl of luciferase reaction buffer (Promega) and the luminescence was integrated over 10 sec (Mediators, Wien, Austria). Results were expressed as light units per mg of cell protein (BCA assay, Pierce).
RESULTS
Synthesis of a Capped Gene-Peptide Conjugate, Purification, and Proof of Structure.
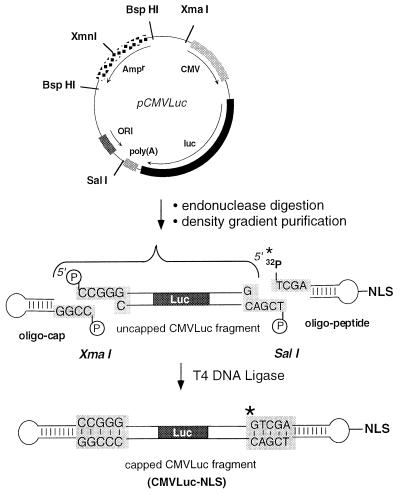
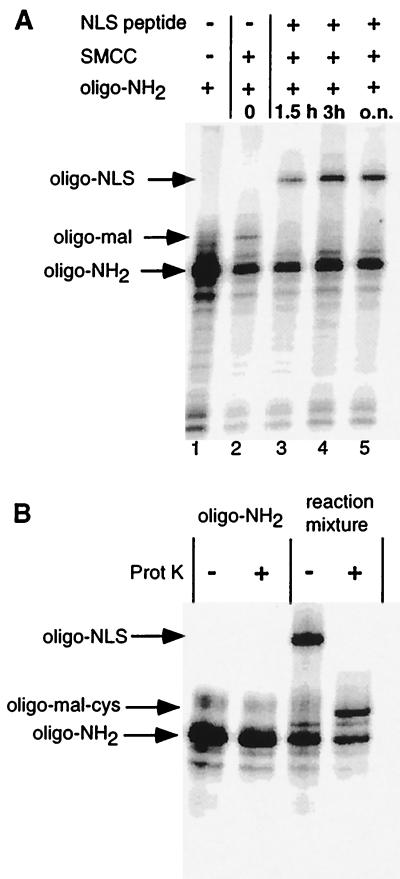
The construction was based on ligation of a pair of hairpin oligonucleotides to unique cohesive termini generated on the reporter gene (Fig. 1). Incorporation of 32P into the modified oligonucleotide allowed us to follow the reaction kinetics, to purify the fragment of interest, and to verify its structure. The firefly luciferase reporter gene (Luc) flanked by the cytomegalovirus (CMV) enhancer/promoter sequence and the simian virus 40 polyadenylation signal was excised from the pCMVLuc plasmid (Fig. 1). Quadruple endonuclease digestion ensured straightforward large-scale separation of the 3,380-bp CMVLuc fragment from the remaining <1-kbp fragments by ultracentrifugation through a 15–30% sucrose gradient. T4 loops are well suited for hairpins and the C-terminal thiol group of the PKKKRKVEDPYC peptide was conjugated to a thymine with a C(5) amino group (22) via an activated ester/maleimide bifunctional linker (SMCC) as detailed in Fig. 2. Preliminary experiments showed that the global reaction yield was highest after activation of oligonucleotide-NH2 with SMCC for 2 h (significant parasitic maleimide hydrolysis may occur with time). The 5′ 32P radiolabeling and polyacrylamide gel electrophoresis of oligonucleotides (Fig. 3A) showed the peptide conjugation reaction to be completed after 3 h, giving 30% oligonucleotide-NLS, based on the starting oligonucleotide. Proteinase K digestion converted oligonucleotide-NLS to a faster migrating compound, presumably the oligonucleotide conjugated to the C-terminal amino acid (oligonucleotide-Mal-Cys), thus establishing the chimeric nature of the conjugate (Fig. 3B). The oligonucleotide-NLS was purified by electrophoresis. The uncapped CMVLuc fragment was then simultaneously reacted with a 15-fold excess of oligonucleotide-cap and oligonucleotide-NLS by using T4 DNA ligase in previously optimized conditions. Ligation reaction yield (80–90%) was assessed by quantitative radioactivity counting and full capping of the CMVLuc fragment was checked by 3′ exonuclease digestion. CMVLuc-NLS was purified by gel permeation.
Figure 1.
Strategy for the preparation of a double-stranded DNA fragment coupled to an NLS peptide. A functional luciferase gene of 3,380 bp was cut out of pCMVLuc with XmaI and SalI. Further digestion with XmnI and BspHI cut the unwanted restriction fragment into small fragments (970, 875, 768, and 240 bp) that were removed by sucrose gradient centrifugation. The capped CMVLuc-NLS DNA was obtained by ligation of the 32P-labeled (∗) oligonucleotide-peptide and oligonucleotide-cap hairpins to the restriction fragment.
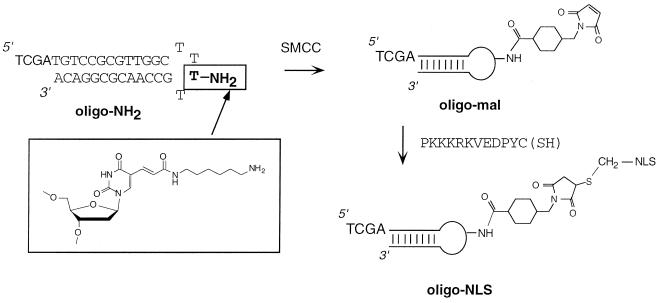
Figure 2.
Reaction scheme for the chemical coupling steps leading to the oligonucleotide-peptide conjugate (oligo-NLS). A hairpin oligonucleotide with a free alkylamino group in the T4 loop (oligo-NH2) was reacted with the heterobifunctional crosslinker SMCC to give a thiol-reactive maleimide oligonucleotide (oligo-Mal), which was in turn reacted with the C-terminal cysteinamide residue of the NLS dodecapeptide.
Figure 3.
(A) Synthesis of oligonucleotide-NLS. PAGE analysis of the formation of the oligonucleotide-peptide conjugate is shown. Aliquots of the reaction mixture were taken at different times and analyzed on a 20% denaturing gel after radiolabeling of the oligonucleotides. Lanes: 1, oligonucleotide-NH2; 2, oligonucleotide-NH2/SMCC reaction mixture after 2 h; 3–5, reaction mixture 1.5 h, 3 h or overnight after addition of the NLS-peptide to the oligonucleotide-NH2/SMCC mixture, respectively. (B) The presumed oligonucleotide-NLS conjugate is a substrate of proteinase K. Radiolabeled oligonucleotide-NH2 or the crude oligonucleotide-NLS reaction mixture were digested with proteinase K. Products were analyzed on a 20% denaturing gel and show total conversion of oligonucleotide-NLS into a faster migrating band, presumably oligo-Mal-Cys.
Transfection with Small Amounts of DNA.
The reporter gene construct was obtained by chemical/enzymatic synthesis and purified by PAGE/gel permeation, instead of being amplified in bacteria. Only limited amounts of DNA could, therefore, be obtained, and the transfection setup had to be miniaturized. A convenient 96-well microtiter plate assay developed by Felgner et al. (23) was chosen. Using several optimized cationic lipid formulations, these authors showed that transfection was best at 2–0.5 μg of plasmid and quickly fell off below 0.25 μg. We confirmed this result with pCMVLuc and two other cell types (Table 1, entry 4, and Fig. 4), thus putting our ultimate goal to obtaining an effective transfection with less than 200 ng of DNA. As a preliminary test, we looked at the relevance to cap the reporter gene at both ends, thus avoiding free DNA ends that are prone to exonuclease-mediated DNA degradation and to eliciting a DNA repair response. To this end, the uncapped, hemicapped, and fully capped CMVLuc genes (200 ng) were transfected into hepatocytes. Luciferase activities (Table 1) showed both un- and hemicapped molecules to give 25-fold less gene expression than the capped one thus justifying the choice of our chemical strategy.
Table 1.
Free DNA ends decrease efficacy
| DNA form | Symbol | Transfection efficiency, RLU/mg of protein | ||
|---|---|---|---|---|
| Uncapped CMVLuc | 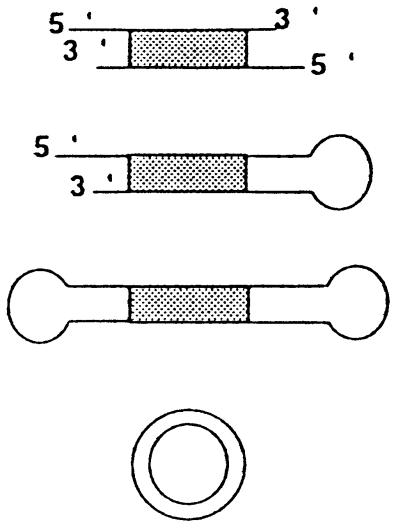 |
1 ± 0.4 × 105 | ||
| Hemi-capped CMVLuc | 9 ± 2 × 104 | |||
| CMVLuc | 2.5 ± 0.6 × 106 | |||
| Circular pCMVLuc plasmid | 2.5 ± 0.2 × 108 3 ± 1 × 104* |
BNL CL.2 hepatocytes were transfected with 200 ng of DNA per well. Complexes were formed with 25-kDa PEI (N/P = 10) in 150 mM NaCl and in the absence of serum. RLU, relative light units.
Transfection with 20 ng of plasmid.
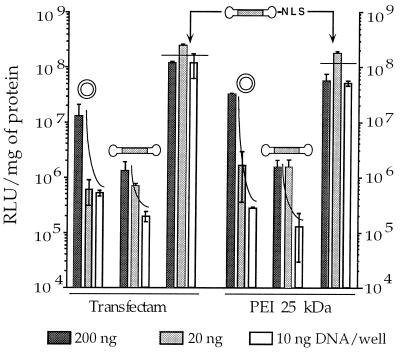
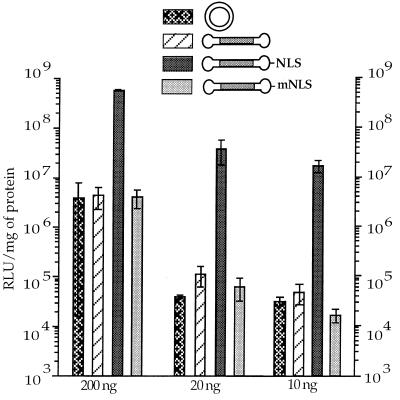
Figure 4.
NLS peptide promotes high and sustained transfection levels down to 10 ng DNA. 3T3 cells were transfected with decreasing amounts of DNA complexed in 150 mM NaCl to a cationic lipid (Transfectam, vector nitrogens over DNA phosphate, N/P = 6) or to a cationic polymer (25-kDa PEI, N/P = 10) in the absence of serum.
The NLS Peptide Allows Effective Transfection with Minute Quantities of DNA.
3T3 cells were transfected with decreasing amounts of DNA—the NLS-bearing capped gene (CMVLuc-NLS), the capped gene (CMVLuc), and the corresponding mass-corrected amount of plasmid DNA (pCMVLuc). Although efficacy very much decreased for “20-ng” plasmid DNA (in fact 38 ng), as expected (see above), transfection levels remained remarkably high and constant over the range of 200–10 ng of DNA with CMVLuc-NLS (Fig. 4). Comparison with the capped gene lacking the NLS peptide showed 100- to 1,000-fold more expression when the NLS peptide was present on the gene. Similar conclusions were obtained, by using a cationic lipid (Transfectam, Fig. 4 Left) or a cationic polymer (branched PEI, Fig. 4 Right). Further data (not shown) confirmed that other cationic carrier molecules behaved similarly but also that efficacy dropped 50-fold within the range of 10–1 ng of CMVLuc-NLS.
Improved Transfection Involves the Cellular Nuclear Import Machinery.
Comparative experiments between CMVLuc and CMVLuc-NLS, although interesting, do not really allow one put forward any hypothesis concerning the mechanism. Nuclear import of DNA and oligonucleotides has mainly been followed with fluorescent tags and fluorescence in situ hybridization, in microinjected or digitonin-permeabilized cells. Proof of active import relied on inhibition experiments with energy-decoupling molecules, nuclear pore-binding agglutinins, or mutations in the signal peptide sequence. Gene expression is an unambiguous proof of nuclear localization and transfection can be regarded as a straightforward way of introducing DNA into intact cells. One lysine to threonine mutation abolishes nuclear import (24). We therefore synthesized a capped gene (CMVLuc-mNLS) with a mutated PKTKRKVEDPYC sequence. Comparative transfection experiments with HeLa cells are shown in Fig. 5. Irrespective of the amount of DNA used, the lysine to threonine mutation brought transfection down to the level obtained with the capped gene having no peptide at all, confirming the functionality of the NLS–importin α interaction.
Figure 5.
Sustained luciferase expression levels are due to the nuclear localization peptide. HeLa cells were transfected with various amounts of DNA complexed to ExGen500 PEI (N/P = 5 in a 5% glucose solution) in the presence of 10% serum.
NLS Peptide-Mediated Transfection Enhancement Is a General Phenomenon.
A series of comparative experiments was undertaken with various cell types. Transfection enhancement due to the presence of the NLS peptide was always observed (Table 2). However, enhancement factors were spread widely with cell type (10- to 1,000-fold), with no obvious relation with tissue origin nor cell type (primary vs. transformed cells). Nondividing primary cells such as human monocyte-derived macrophages and rat dorsal root ganglia neurons showed less impressive enhancement than 3T3 and HeLa cells. However, similar values were also seen for the easily transfected and fast dividing rat hepatocyte-derived cell line BNL CL.2.
Table 2.
Average enhancement of transfection with CMVLuc-NLS
| Cells | Enhancement factor* |
|---|---|
| BNL CL.2 | 10 |
| 3T3 | 400†–1000 |
| HeLa | 200‡–300 |
| Murine DRG neurons | 30 |
| Human macrophages | 10 |
Values refer to transfection with 10–20 ng of DNA per well, in the absence of serum. DRG, dorsal root ganglion.
Estimated spread is ±30%.
In the presence of 10% serum during transfection.
In 24-well plates using 200 ng of DNA in the presence of 10% serum.
The general experimental setup for transfection included 96-well microtiter plates and no serum for 2 h after addition of the DNA–vector complexes to the cells. Several experiments were also performed on a larger scale (24-well plates) or in the presence of 10% serum during transfection. The results obtained for HeLa and 3T3 cells (Table 2) confirmed the general conclusions derived with the 96-well setup.
Time Course of the Transgene Expression.
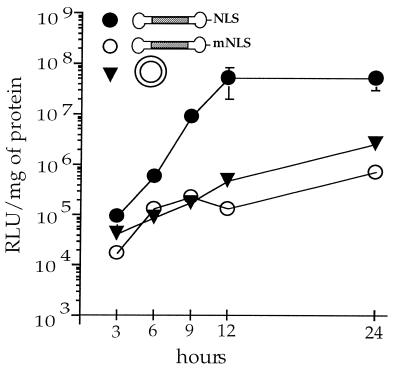
Active transport of the reporter gene via the nuclear import machinery could result in faster appearing expression. More interesting, a lower intracellular barrier to gene delivery means less DNA–vector complexes required in the cytoplasm and hence lower toxicity and more sustained expression. The kinetics of gene expression was followed in detail up to 24 h (Fig. 6) and then daily up to 3 days. Again, transfection with CMVLuc-NLS was much more effective than with CMVLuc-mNLS. Remarkably (and reproducibly), NLS-driven transfection reached its plateau after 12 h, whereas transfection levels obtained with the plasmid or the mutated NLS sequence were still increasing significantly up to 24 h. Previous experiments (25) showed both transfection and cytoplasmic injection of DNA–PEI complexes to have superimposable kinetics of transgene expression, suggesting that the slow step was intracellular trafficking/nuclear entry rather than cell entry. Using the nuclear import machinery seems to speed up the rate-limiting step. For longer time periods, expression stayed broadly constant with a low standard deviation. However, the rather large variability observed upon repeating this experiment prevented us from drawing any conclusion (the inconsistent results observed at t > 1 day may be due to the fact that cells reached confluency at day 1).
Figure 6.
Reporter protein activity appears faster with CMVLuc-NLS. HeLa cells were transfected with 10 ng of DNA complexed to ExGen 500 PEI (N/P = 5 in 5% glucose solution) in the absence of serum. Cells were lysed and luciferase activity was measured at the indicated time after transfection. The absence of error bars indicates that the SEM is smaller than the label on the graph.
DISCUSSION
The goal of this work was to improve nonviral plasmid-mediated gene delivery. When fully counterion-condensed, a single plasmid molecule collapses into a sphere of about 25 nm (26) that is already at the size-exclusion limit for signal-mediated nuclear import (4, 27). Plasmid condensation with cationic lipids or polymers generally leads to even larger multimolecular aggregates that reach the cytoplasm after binding to cell-surface anionic proteoglycans (9, 28) and eventual escape from the formed vacuoles (8, 9, 30). Because the particles are too large to cross an intact nuclear membrane, transfection of resting cells can reasonably only be accounted for by uncomplexed DNA that has been released by exchange with phosphatidylserine (31) or heparan sulfate (9) present in the vacuolar membrane. Any molecular information (e.g., NLS) borne by the cationic vector will thus be lost before reaching the nuclear membrane, hence its ineffectiveness.
In sharp contrast to a condensed DNA molecule, a free hydrated DNA double helix (3 nm) is thin enough to enter the nuclear pore by diffusion, i.e., with no energy or signal requirement. Restricted intracellular motion of DNA [the cytoplasm is equivalent to 13% dextran (32)], and a rather low nuclear pore coverage [<10% of the membrane surface (33)], however, give this event a low probability to occur: the nuclear membrane has been recognized as a major barrier to gene delivery (8, 9). Quantitative cytoplasmic microinjection of DNA (25, 34, 35) indeed led to less than 0.1% of the DNA being expressed. The probability for DNA to find and subsequently enter a nuclear pore can be increased by a bound karyophilic signal peptide able to dock DNA to the nuclear pore filaments and help an initial part of the molecule to cross the membrane. The DNA–karyophilic signal link should, however, be stable, hence the chemical strategy we chose to use in this study.
The major achievement of our approach is that the amount of DNA required to effectively transfect cells in vitro has now shifted from the microgram to the nanogram range (see also ref. 17). Although the improvement (one to three orders of magnitude) varies with cell type (showing that the nuclear membrane is not the only possible barrier), even the modest enhancements observed for primary cell cultures look interesting (1,000%) when not presented on a logarithmic scale. Is this exciting result really a consequence of the involvement of the nuclear import machinery? We choose to tackle the academic aspect of this work by using the lysine to threonine mutation, which indeed abolished the beneficial effect brought about by the DNA-bound NLS sequence.
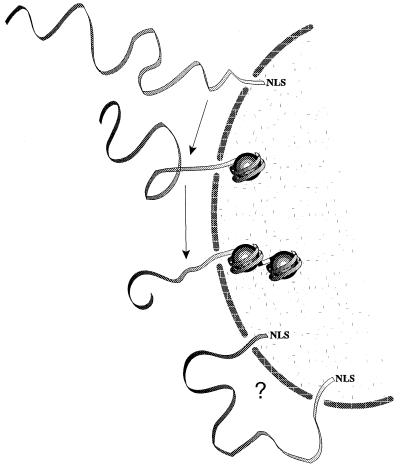
Taking our hypothesis further, the last unanswered question deals with threading of the rest of the plasmid molecule (Fig. 7). Random-walk diffusion of a micrometer-long molecule would be inefficient and slow. Fortunately, naked DNA will not remain free in the nucleus: histones (and eventually basic nuclear matrix proteins) indeed quickly assemble transfected DNA into chromatin-like structures (36, 37), thus providing a mechanism for pulling and condensing the filamentous molecule into the nucleus. Following this line of thinking, many NLS signals distributed along the DNA may actually inhibit nuclear entry if the nucleic acid is longer than the distance separating adjacent pores (Fig. 7). A straightforward calculation that uses the pore density in HeLa cells (33) shows this to become probable above 1 kbp.
Figure 7.
Hypothetical scheme of foreign DNA entering the nucleus. The vectorial movement is driven by DNA binding to chromatin-forming basic proteins. A single NLS peptide helps initial threading, whereas too many signals may inhibit full translocation.
The present work is a proof of principle rather than a new gene delivery technology. However, it will hopefully provide the impetus for searching more versatile solutions to the link between a plasmid and a NLS peptide, such as triple-helix forming (38, 39) or strand-invading (40, 41) oligonucleotide-NLS conjugates.
Acknowledgments
We thank Dr. Olivier Feugeas and Dr. Anne Feltz for providing the monocytes and neurons, respectively. We are grateful to Dr. Robert P. P. Fuchs and Dr. M. Bichara for providing technical facilities and for helpful discussions. This work was supported by grants from the Association Française contre les Myopathies, the Association pour la Recherche contre le Cancer, and the Ligue contre le Cancer. M.A.Z. received a fellowship from Transgene SA.
ABBREVIATIONS
- NLS
nuclear localization signal
- SMCC
4-(N-maleimidomethyl)cyclohexane-1-carboxylic acid N-hydroxysuccinimide ester
- PEI
polyethylenimine
- N/P
nitrogen/phosphate
References
- 1.Adam S A, Gerace L. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 2.Panté N, Aebi U. J Cell Sci Suppl. 1995;19:1–11. doi: 10.1242/jcs.1995.supplement_19.1. [DOI] [PubMed] [Google Scholar]
- 3.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 4.Ohno M, Fornerod M, Mattaj I W. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 5.Melchior F, Gerace L. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 6.Greber U F, Suomalainen M, Stidwill R P, Boucke K, Ebersold M W, Helenius A. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greber U F. In: Self-Assembling Complexes for Gene Delivery: From Laboratory to Clinical Trial. Kabanov A V, Felgner P L, Seymour L W, editors. Chichester: Wiley; 1998. pp. 89–114. [Google Scholar]
- 8.Zabner J, Fasbender A J, Moninger T, Poellinger K A, Welsh M J. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 9.Labat-Moleur F, Steffan A M, Brisson C, Perron H, Feugeas O, Furstenberger P, Oberling F, Brambilla E, Behr J P. Gene Therapy. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 10.Hagstrom J E, Ludtke J J, Bassik M C, Sebestyén M G, Adam S A, Wolff J A. J Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 11.Kaneda Y, Iwai K, Uchida T. Science. 1989;243:375–378. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- 12.Fritz J D, Herweijer H, Zhang G F, Wolff J A. Hum Gene Ther. 1996;7:1395–1404. doi: 10.1089/hum.1996.7.12-1395. [DOI] [PubMed] [Google Scholar]
- 13.Collas P, Husebye H, Aleström P. Transgenic Res. 1996;5:541–548. doi: 10.1007/BF01980210. [DOI] [PubMed] [Google Scholar]
- 14.Remy J S, Kichler A, Mordvinov V, Schuber F, Behr J P. Proc Natl Acad Sci USA. 1995;92:1744–1748. doi: 10.1073/pnas.92.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fominaya J, Wels W. J Biol Chem. 1996;271:10560–10568. doi: 10.1074/jbc.271.18.10560. [DOI] [PubMed] [Google Scholar]
- 16.Dean D A. Exp Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 17.Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K. J Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebestyén M G, Ludtke J J, Bassik M C, Zhang G, Budker V, Lukhtanov E A, Hagstrom J E, Wolff J A. Nat Biotech. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 19.Behr J P, Demeneix B, Loeffler J P, Perez-Mutul J. Proc Natl Acad Sci USA. 1989;86:6982–6986. doi: 10.1073/pnas.86.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zanta M A, Boussif O, Adib A, Behr J P. Bioconjugate Chem. 1997;8:839–844. doi: 10.1021/bc970098f. [DOI] [PubMed] [Google Scholar]
- 21.Lambert R C, Maulet Y, Dupont J L, Mykita S, Craig P, Volsen S, Feltz A. Mol Cell Neurosci. 1996;7:239–246. doi: 10.1006/mcne.1996.0018. [DOI] [PubMed] [Google Scholar]
- 22.Seibel P, Trappe J, Villani G, Klopstock T, Papa S, Reichmann H. Nucleic Acids Res. 1995;23:10–17. doi: 10.1093/nar/23.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felgner J H, Kumar R, Sridhar C N, Wheeler C J, Tsai Y J, Border R, Ramsey P, Martin M, Felgner P L. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]
- 24.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 25.Pollard H, Remy J S, Loussouarn G, Demolombe S, Behr J P, Escande D. J Biol Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 26.Blessing T, Remy J S, Behr J P. Proc Natl Acad Sci USA. 1998;95:1427–1431. doi: 10.1073/pnas.95.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 28.Mislick K A, Baldeschwieler J D. Proc Natl Acad Sci USA. 1996;93:12349–12354. doi: 10.1073/pnas.93.22.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 30.Boussif O, Lezoualch F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J P. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y H, Szoka F C. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 32.Luby-Phelps K. Curr Opin Cell Biol. 1994;6:3–9. doi: 10.1016/0955-0674(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 33.Maul G G, Deaven L. J Cell Biol. 1977;73:748–760. doi: 10.1083/jcb.73.3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirzayans R, Aubin R A, Paterson M C. Mutat Res. 1992;281:115–122. doi: 10.1016/0165-7992(92)90045-j. [DOI] [PubMed] [Google Scholar]
- 35.Dowty M E, Williams P, Zhang G, Hagstrom J E, Wolff J A. Proc Natl Acad Sci USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cereghini S, Yaniv M. EMBO J. 1984;3:1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong S, Stein A. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser H E, Dervan P B. Science. 1987;238:645. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 39.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout J, Thuong N T, Lhomme J, Hélène C. Nucleic Acids Res. 1987;15:7749. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen P E, Egholm M, Berg R H, Buchardt O. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 41.Schmid N, Behr J P. Tetrahedron Lett. 1995;36:1447–1450. [Google Scholar]