The past two decades have witnessed a dramatic increase in the prevalence of asthma worldwide (1). Asthma is a chronic disease characterized by variable airway obstruction, airway hyperresponsiveness (AHR), and airway inflammation and remodeling. Histological studies show that airways of asthmatic patients contain a chronic inflammatory infiltrate composed of lymphocytes, eosinophils, and mast cells. This infiltrate is usually accompanied by desquamation of the bronchial epithelial layer, goblet cell hyperplasia, and thickening of the submucosa. In most cases, the asthmatic inflammatory process results from inappropriate immune responses to common environmental antigens in a genetically susceptible individual (2). These inappropriate immune responses are orchestrated by a subset of CD4+ T helper cells termed T helper 2 (Th2) cells.
Cytokines play a pivotal role in the development of asthma by regulating the expansion of Th2 cells and by mediating many of the Th2 effector functions that underlie the pathogenic events of an asthmatic response. Much effort has recently been placed in elucidating the pathways used by cytokines to mediate their actions. These studies have revealed that cytokine-mediated signals are primarily transduced by the Jak-Stat signaling cascade (3). In this review we will highlight the recent advances made in dissecting the roles of this signaling pathway in the pathogenesis of asthma.
Jak-Stat signaling in Th1 and Th2 differentiation
The two major subsets of CD4+ Th cells, termed Th1 and Th2, secrete mutually distinct profiles of cytokines and thereby coordinate different classes of immune response (4). Th1 cells secrete IL-2, IFN-γ, and TNF-β, whereas Th2 cells produce IL-4, IL-5, IL-6, IL-10, and IL-13. It is now well accepted that Th1 cells are critically involved in the generation of delayed-type hypersensitivity responses, whereas Th2 cells can direct B cells to mount strong humoral responses. Polarization of an immune response toward a Th2 phenotype, while extremely useful in the clearance of parasites, may prove harmful if directed against an otherwise innocuous environmental antigen, as occurs in the pathogenesis of allergic diseases like asthma.
The Th2 cytokines, primarily IL-4, IL-5, and IL-13, control all the major components that characterize an inflammatory asthmatic response, including IgE isotype switching, mucus production, and the recruitment and activation of eosinophils. The involvement of Th2 cells in the pathophysiology of asthma has been corroborated by studies in both humans and mice. The population of Th2 cells is notably expanded in the airways of asthmatic subjects, and presence of these cells correlates with AHR and airway eosinophilia (2). Work in murine models of AHR confirms this correlation and demonstrates that adoptively transferred antigen-specific Th2, but not Th1, cells can mediate airway eosinophilia, mucus hypersecretion, and AHR when recipient mice are exposed to inhaled antigen (5).
The cytokines IL-12 and IL-4 direct the differentiation of Th1 and Th2 cells, respectively, from naive T helper cells (6, 7). Genetic studies have confirmed the physiologic importance of these cytokines in vivo. Mice deficient in either IL-12 or the IL-12 receptor (IL-12R) are unable to mount Th1 responses, while mice lacking IL-4 or the IL-4 receptor α (IL-4Rα) chain display defects in the generation of Th2 responses (8, 9). Indeed, IL-12 and IL-4 not only drive the expansion of their corresponding Th subset but simultaneously block the generation of the opposing subset. Given the importance of IL-4 and IL-12 in controlling Th differentiation, much effort has been placed over the past few years in dissecting the mechanisms by which these cytokines mediate their actions. These studies have revealed that, like most other cytokines, IL-4 and IL-12 activate the Jak-Stat signaling cascade discussed elsewhere in this Perspective series. In this signaling pathway, binding of a cytokine to its receptor leads to the activation of members of the JAK family of receptor-associated kinases. These kinases subsequently activate, via tyrosine phosphorylation, preexistent cytoplasmic factors termed Stats (signal transducer and activator of transcription). Tyrosine phosphorylation allows the Stat proteins to dimerize and translocate to the nucleus, where they mediate changes in gene expression by binding specific DNA elements.
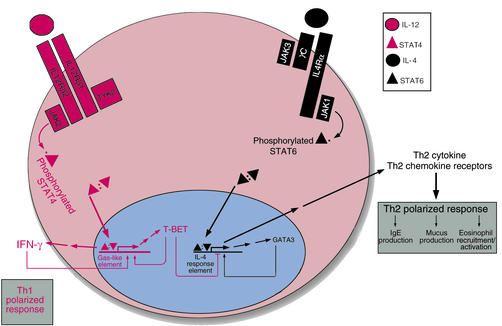
Although both IL-4 and IL-12 follow this basic signaling framework, the two cytokines differ in the specific Jak and Stat components that they activate (10). IL-4 stimulates Jak1 and Jak3 to activate Stat6. In contrast, interaction of IL-12 with its receptor leads to the activation of Jak2 and Tyk2 and the subsequent phosphorylation of Stat4. Activation of Stat6 and Stat4 are thus critical events in the signaling cascades of IL-4 and IL-12, respectively. Given the pivotal roles of these two cytokines in skewing Th cells toward either a Th2 or a Th1 phenotype, it is not surprising that Stat6 and Stat4 control multiple aspects of the Th differentiation programs, as shown in Figure 1 and detailed below.
Figure 1.
JAK-STAT signaling and the generation of Th1 and Th2 cells. Following antigen presentation, a naive CD4+ T cell will differentiate along the Th1 or Th2 pathway, depending on the nature of the cytokines it contacts. Signaling through the IL-12 receptor (red pathway) and its associated Jak and Stat proteins culminates in the expression of Th1-specific gene products, particularly the cytokine interferon-γ (IFN-γ) Conversely, IL-4 activates a distinct receptor complex, containing different Jaks and Stats, and favors the expression of Th2 cytokines and other gene products (black pathway). These pathways can regulate one another at the transcriptional level. In particular, the transcription factor GATA3, whose expression is induced by IL4 signaling, is essential for the transactivation of other Th2-associated genes, such as those for Th2 cytokines and chemokine receptors. Another transcription factor, T-BET, is induced by IFN-γ (and therefore indirectly by IL-12 stimulation) and also favors expression of this cytokine — thus establishing a positive feedback loop that supports Th1 polarization. In addition, T-BET silences expression of Th2-associated genes. Polarization toward the Th2 response leads to the expression of various cytokines — IL-4, IL-5, IL-6, IL-10, and IL-13 — that contribute to the pathologies seen in asthma.
Stat6 in Th differentiation
The importance of Stat6 in Th2 cell differentiation has been confirmed by the generation of Stat6-deficient mice (11–13), which fail to mount Th2 responses either in vitro — in Th differentiation systems — or in vivo, upon infection with parasitic pathogens that elicit Th2 responses. Th2 memory cell development and survival are dependent on Stat6 (14, 15), whereas Stat6-independent pathways of IL-4 production seem to contribute only to the initial responses mounted by naive T cells, rather than to the later regulation of the Th2 population. Stat6 appears to be not only necessary but also sufficient to drive Th2 differentiation, since introduction of constitutively active forms of Stat6 results in the expression of Th2-type cytokines even when this form of Stat6 is expressed in cells already differentiating toward a Th1 phenotype (16, 17).
Stat6’s role in Th2 differentiation is not confined to activating and silencing the expression of specific cytokines but extends to other aspects of the distinctive pattern of gene expression found in Th2 cells, including the heightened expression in these cells of the chemokine receptors CCR4 and CCR8 (18). Stat6-deficient T cells are significantly impaired in their ability to expand upon IL-4 stimulation, thus implicating this transcription factor in IL-4–driven proliferation as well as differentiation (17, 19). This defect is due to a block in the progression from the G1 to the S phase of the cell cycle and correlates with an impaired ability of Stat6-deficient T cells to downregulate the cell cycle–dependent kinase inhibitor p27kip in response to IL-4 stimulation. In parallel with its induction of Th2-specific gene expression programs, Stat6 appears to suppress Th1-specific pathways. Indeed, generation of mice deficient in both Stat4 and Stat6 has revealed that in the absence of Stat6, Stat4-deficient CD4+ T cells can differentiate into Th1 cells (20). This effect is not detected in mice deficient for Stat4 alone, suggesting that the presence of Stat6 blocks the ability of Th cells to acquire the Th1 phenotype.
The mechanisms by which Stat6 controls Th2 differentiation are complex and involve subtype-specific induction of specific transcription factors, as well as changes in the chromatin structure and the pattern of cytosine methylation at the IL4 locus. Th2-specific factors like GATA3 and c-maf synergize with NFAT and AP-1, which are more broadly expressed (6, 7), to activate the characteristic Th2 pattern of gene expression. While both the transcriptional and the epigenetic changes depend on Stat6, it remains unclear whether either of these effects is direct. Some reports also suggest that Stat6 can bind to specific sites within the IL4 promoter and to a site in the 3′ untranslated region of IL4 that may function as a Th1-specific silencer (21–23). The significance of Stat6 targeting to these regulatory regions for Th2 commitment has not been established.
Stat4 in Th differentiation
While activation of Stat6 in response to IL-4 is critical for the generation of Th2 responses, activation of a different Stat, Stat4, skews Th cells toward the Th1 phenotype following IL-12 stimulation. Phenotypic analysis of Stat4-deficient mice shows that activation of Stat4 is critical to this process. Stat4-deficient T cells are unable to produce high levels of the Th1 cytokine IFN-γ after exposure to IL-12 (20, 24). Stat4-independent pathways of Th1 differentiation also exist, but, as mentioned above, their effects are seen most clearly in the absence of Stat6 (20, 24). Recent studies suggest that Stat4 activation is particularly critical for the sustained, rather than the initial, production of Th1-type cytokines (15). Stat4 also mediates the downregulation of the cell-cycle inhibitor p27kip in IL-12–treated Th1 cells (19, 25), allowing it (like Stat6) to control Th proliferation as well as differentiation.
As with Th2 cytokines, induction of Th1-specific cytokine gene expression requires epigenetic remodeling, as well as the induction of Th1-specific transcription factors like the recently described T-bet (26). The precise mechanisms employed by Stat4 to mediate its effects on Th1 differentiation have not been fully delineated. In particular, it is unclear whether Stat4 activation participates in the induction of the Th1-specific factor T-bet, whose deficiency in mice has recently been shown to lead to the spontaneous development of airway changes characteristic of human asthma (15, 25, 27, 28). Stat4 is reported to transactivate IFN-γ directly (29), although additional factors are likely to be required for full expression of this cytokine (7). Activated Stat4 also blocks differentiation along the Th2 pathway by repressing expression of the Th2-specific factor Gata3 (30), establishing yet another parallel between the role of Stat4 in Th1 development and that of Stat6 in Th2 development. Further progress toward understanding the physiological basis of the actions of these two Stat proteins will require the identification of the key targets of these two transcription factors.
The Jak-Stat signaling cascade in IgE regulation
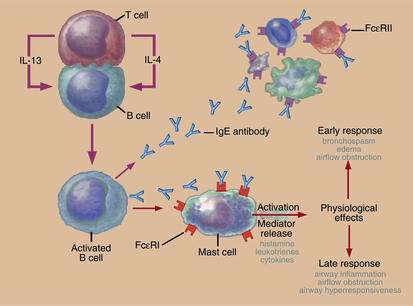
Levels of IgE correlate with the incidence of atopic asthma in humans. The recent success of anti-IgE therapy in the treatment of atopic asthmatics also highlights the importance of this class of antibodies in human asthma (31) (Figure 2). Work over the past ten years has detailed how cytokines (IL-4 and IL-13, in particular) induce the production of IgE and how other cytokines, such as IFN-γ, block this induction. IL-4 initiates signaling by oligomerizing the heterodimeric IL-4 receptor, which, in hematopoietic cells, is composed of the ligand-specific IL-4Rα chain and the common γ chain (γC) (32). This oligomerization initiates signaling by activating Jak1 and Jak3, which associate constitutively with the cytoplasmic tail of cytokine receptor subunits (Jak1 with IL-4Rα and Jak3 with γC). IL-13 likewise binds IL-4Rα and can activate signaling through Jak1, but this cytokine also binds a more specific receptor subunit, the IL-13Rα1 chain, which associates with the Jak-family kinase Tyk2.
Figure 2.
IgE in the pathogenesis of asthmatic responses. Production of Th2-type cytokines (IL-4 and IL-13) by T cells in response to antigens like airborne allergens will drive IgE synthesis by B cells. IgE can then bind to high-affinity IgE receptors on mast cells. Cross-linking of the bound IgE molecules upon reexposure to the antigen provokes mast cell degranulation with the subsequent release of a variety of mediators, which trigger both early and late inflammatory asthmatic responses.
After binding of either IL-4 or IL-13, the activated Jaks phosphorylate tyrosines within the cytoplasmic domain of the IL-4Rα, which act as docking sites for Stat6. Phosphorylated Stat6 homodimerizes, translocates to the nucleus, and activates transcription of genes involved in B cell differentiation, including the germline Iε and Iγ1 genes in mice and germline Iε and Iγ4 genes in humans (33). Induction of these germline promoters, and expression of the corresponding germline “sterile” transcripts, have been shown in mice to be essential for Ig class switching, recombination events that are required for the production of the various classes of secreted antibodies (33). Stat6 contributes to class switching to produce IgE and IgG1 (11–13). In mice, IFN-γ, which acts via Stat1 to activate the transcription of many early response genes, inhibits the transcription of germline Iε and Iγ1 and thereby blocks B cell production of IgE and IgG1 (34). This regulation appears to be mediated by the suppressor of cytokine signaling-1 (Socs-1), one of a new family of inhibitory Socs molecules defined by the presence of conserved SH2 domains and a novel motif termed a “Socs box” (35). These proteins are induced by cytokines and appear to function in a negative feedback loop. Socs-1 can bind to all the Jaks and inhibit Stat activation. The ability of IFN-γ to inhibit IL-4 signaling seems to be due to the induction of Socs-1 and the resulting suppression of Stat6 activity (36, 37).
A second inhibitor of Jak-Stat signaling implicated in atopic immune responses is Bcl-6, the product of a putative protooncogene that is rearranged or mutated in non-Hodgkin lymphomas. Normally, Bcl-6 functions as a transcriptional repressor, and it has been shown to bind to Stat6-binding sites, such as one found in the Iε promoter. Mice lacking Bcl-6 develop an inflammatory disease characterized by increased levels of IgE, Th2 cells, and mast cell infiltrates. Their B cells produce high levels of IgE (38, 39), a phenotype that requires Stat6 expression by these cells (40, 41). The importance of these regulators of Jak-Stat signaling in human asthma is still undefined.
Stat6 in asthma pathophysiology
In light of the extensive evidence that JAK-STAT signaling controls many of the physiologic events that are deregulated in asthma, several groups have pursued the role of this pathway in murine models of pulmonary inflammation and AHR. Most such studies have focused on Stat6, given its involvement in directing Th2 responses and IgE production (42–46). Following allergen provocation Stat6 deficient-mice fail to develop a pulmonary Th2 response or AHR, and they exhibit no detectable increase in IgE production or in the number of mucus-containing cells. Because intravenous administration of IL-5 to Stat6-deficient mice restores the development of eosinophilia and AHR after antigen sensitization (46), it appears that one essential role of Stat6 in the development of AHR is to drive Th2 responses and IL-5 production. Interestingly, different groups have reported distinct effects of Stat6 deficiency on the inflammatory infiltrate. Mice of the BALB/c background show only a 50% decrease in the eosinophilic infiltrate, whereas antigen-induced eosinophilia is completely blocked in Stat6-deficient C57BL/6 mice (42, 43). Whether distinctions in sensitization protocols and/or strain background contribute to the differences reported remains to be established.
Recent reports suggest that the development of allergic pulmonary inflammation requires the activation of Stat6 not only in T cells, but also in the parenchymal cells of the lung (47). Indeed, adoptively transferred antigen-specific Th2 cells from Stat6+/+ mice fail to mediate allergic inflammation in Stat6-deficient mice. These defects are believed to be due to impairments of these Th2 cells to traffic to the lung, possibly because of defects in the production of chemokines like eotaxin, which control recruitment of Th2 cells. Another feature of asthma that appears to require Stat6 is mucus production: Stat6–/– mice do not demonstrate goblet cell hyperplasia and have decreased mucus secretion in the ovalbumin-asthma challenge asthma model (42, 43, 48). Again, transfer of antigen-specific Stat6+/+ T cells fails to complement this defect, thus implicating Stat6 within the lung as a key mediator of mucus production (47). Similar findings in terms of AHR have also been reported. It will be interesting to determine whether cell type–specific transcription factors can influence the effects of Stat6 within specific cell types, given that IL-13 has recently been shown to activate different gene profiles in distinct human airway cell types (49). Taken together, all these studies strongly support the notion that Stat6 plays a central role in the pathogenesis of asthma.
Several groups have also investigated the expression and activation of STAT6 in asthmatic individuals. Peripheral blood lymphocytes from asthmatic and allergic patients do not display significant differences in the level of STAT6 activity relative to healthy controls (50), but these patients do have a higher density of STAT6-expressing cells in their airways (51). Intriguingly, the density of these STAT6-expressing cells is significantly higher in atopic than in nonatopic asthmatics, although there is no significant difference in expression of the other characteristic Th2 transcription factors GATA3 and c-maf between these groups. One recent study of subjects with severe asthma confirms that such patients show significantly elevated airway levels of STAT6 and also identifies the major STAT6-expressing cell type in this tissue as the bronchial epithelial cell (52). Hence, the microenvironment of asthmatic airways may contribute to deregulating STAT6 expression. Further studies will be needed to investigate the functional consequences of the enhanced expression of STAT6 in asthmatic patients and to determine whether deregulated Stat6 expression is indeed a critical distinction between atopic and nonatopic asthma.
References
- 1.Beasley R, Crane J, Lai CKW, Pearce N. Prevalence and etiology of asthma. J Allergy Clin Immunol. 2000;105:S466–S472. doi: 10.1016/s0091-6749(00)90044-7. [DOI] [PubMed] [Google Scholar]
- 2.Wills-Karp M. Immunologic basis of antigen-induced airway hyperresponsiveness. Annu Rev Immunol. 1999;17:255–281. doi: 10.1146/annurev.immunol.17.1.255. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 4.Seder R, Paul W. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 5.Cohn L, Ray A. T-helper type 2 cell-directed therapy for asthma. Pharmacol Ther. 2000;88:187–196. doi: 10.1016/s0163-7258(00)00091-7. [DOI] [PubMed] [Google Scholar]
- 6.Rengarajan J, Szabo SJ, Glimcher LH. Transcriptional regulation of Th1/Th2 polarization. Immunol Today. 2000;21:479–483. doi: 10.1016/s0167-5699(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 7.Murphy KM, et al. Signaling and transcription in T helper development. Annu Rev Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 8.Kopf M, et al. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 9.Noben-Trauth N, et al. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci USA. 1997;94:10838–10843. doi: 10.1073/pnas.94.20.10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19:2577–2584. doi: 10.1038/sj.onc.1203485. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan MH, Schindler U, Smilery ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 12.Takeda K, et al. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 13.Shimoda K, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 14.Finkelman FD, et al. Stat6 regulation of in vivo IL-4 responses. J Immunol. 2000;164:2303–2310. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- 15.Grogan JL, et al. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 16.Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11:677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4’s role in Th2 differentiation and cell expansion. J Immunol. 2001;166:7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Lukacs NW, Lawless VA, Kunkel SL, Kaplan MH. Differential expression of chemokines in Th1 and Th2 cells is dependent on Stat6 but not on Stat4. J Immunol. 2000;165:10–14. doi: 10.4049/jimmunol.165.1.10. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan MH, Daniel C, Schindler U, Grusby MJ. Stat proteins control lymphocyte proliferation by regulating p27Kip1 expression. Mol Cell Biol. 1998;18:1996–2003. doi: 10.1128/mcb.18.4.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 21.Curiel RE, et al. Identification of a Stat6-responsive element in the promoter of the human interleukin-4 gene. Eur J Immunol. 1997;27:1982–1987. doi: 10.1002/eji.1830270823. [DOI] [PubMed] [Google Scholar]
- 22.Georas SN, et al. Stat6 inhibits human interleukin-4 promoter activity in T cells. Blood. 1998;92:4529–4538. [PubMed] [Google Scholar]
- 23.Kubo M, et al. T-cell subset-specific expression of the IL-4 gene is regulated by a silencer element and STAT6. EMBO J. 1997;16:4007–4020. doi: 10.1093/emboj/16.13.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierfelder WE, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 25.Mullen AC, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 26.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 27.Lighvani AA, et al. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci USA. 2001;98:15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finotto S, et al. Development of spontaneous changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Sun YL, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang W, et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 31.Milgrom H, et al. Treatment of allergic asthma with monoclonal anti-IgE antibody. rhuMAb-E25 Study Group. N Engl J Med. 1999;341:1966–1973. doi: 10.1056/NEJM199912233412603. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J Allergy Clin Immunol. 2000;105:1063–1070. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]
- 33.Coffman RL, Lebman DA, Rothman P. Mechanism and regulation of immunoglobulin isotype switching. Adv Immunol. 1993;54:229–270. doi: 10.1016/s0065-2776(08)60536-2. [DOI] [PubMed] [Google Scholar]
- 34.Linehan LA, Warren WD, Thompson PA, Grusby MJ, Berton MT. STAT6 is required for IL-4-induced germline Ig gene transcription and switch recombination. J Immunol. 1998;161:302–310. [PubMed] [Google Scholar]
- 35.Chen XP, Losman JA, Rothman P. SOCS proteins, regulators of intracellular signaling. Immunity. 2000;13:287–290. doi: 10.1016/s1074-7613(00)00028-5. [DOI] [PubMed] [Google Scholar]
- 36.Venkataraman C, Leung S, Salvekar A, Mano H, Schindler U. Repression of IL-4-induced gene expression by IFN-gamma requires Stat1 activation. J Immunol. 1999;162:4053–4061. [PubMed] [Google Scholar]
- 37.Dickensheets HL, Donnelly RP. Inhibition of IL-4-inducible gene expression in human monocytes by type I and type II interferons. J Leukoc Biol. 1999;65:307–312. doi: 10.1002/jlb.65.3.307. [DOI] [PubMed] [Google Scholar]
- 38.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 39.Ye B, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 40.Harris MB, et al. Transcriptional repression of Stat6-dependent interleukin-4-induced genes by BCL-6: specific regulation of iepsilon transcription and immunoglobulin E switching. Mol Cell Biol. 1999;19:7264–7275. doi: 10.1128/mcb.19.10.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dent AL, Doherty TM, Paul WE, Sher A, Staudt LM. BCL-6-deficient mice reveal an IL-4-independent, STAT6-dependent pathway that controls susceptibility to infection by Leishmania major. J Immunol. 1999;163:2098–2103. [PubMed] [Google Scholar]
- 42.Kuperman D, Schofield B, Wills-Karp M, Grusby MJ. Signal transducer and activator of transcription factor 6 (Stat6)-deficient mice are protected from antigen-induced airway hyperresponsiveness and mucus production. J Exp Med. 1998;187:939–948. doi: 10.1084/jem.187.6.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akimoto T, et al. Abrogation of bronchial eosinophilic inflammation and airway hyperreactivity in signal transducers and activators of transcription (STAT)6-deficient mice. J Exp Med. 1998;187:1537–1542. doi: 10.1084/jem.187.9.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyata S, et al. STAT6 deficiency in a mouse model of allergen-induced airways inflammation abolishes eosinophilia but induces infiltration of CD8+ T cells. Clin Exp Allergy. 1999;29:114–123. doi: 10.1046/j.1365-2222.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- 45.Herrick CA, MacLeod H, Glusac E, Tigelaar RE, Bottomly K. Th2 responses induced by epicutaneous or inhalational protein exposure are differentially dependent on IL-4. J Clin Invest. 2000;105:765–775. doi: 10.1172/JCI8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomkinson A, et al. The failure of STAT6-deficient mice to develop eosinophilia and airway hyperresponsiveness is overcome by interleukin-5. Am J Respir Crit Care Med. 1999;160:1283–1291. doi: 10.1164/ajrccm.160.4.9809065. [DOI] [PubMed] [Google Scholar]
- 47.Mathew A, et al. Signal transducer and activator of transcription 6 controls chemokine production and T helper cell type 2 cell trafficking in allergic pulmonary inflammation. J Exp Med. 2001;193:1087–1096. doi: 10.1084/jem.193.9.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomkinson A, et al. Temporal association between airway hyperresponsiveness and airway eosinophilia in ovalbumin-sensitized mice. Am J Respir Crit Care Med. 2001;163:721–730. doi: 10.1164/ajrccm.163.3.2005010. [DOI] [PubMed] [Google Scholar]
- 49.Lee JH, et al. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am J Respir Cell Mol Biol. 2001;25:474–485. doi: 10.1165/ajrcmb.25.4.4522. [DOI] [PubMed] [Google Scholar]
- 50.Miller RL, Eppinger TM, McConnell D, Cunningham-Rundles C, Rothman P. Analysis of cytokine signaling in patients with extrinsic asthma and hyperimmunoglobulin E. J Allergy Clin Immunol. 1998;102:503–511. doi: 10.1016/s0091-6749(98)70141-1. [DOI] [PubMed] [Google Scholar]
- 51.Christodoulopoulos P, et al. Th2 cytokine-associated transcription factors in atopic and nonatopic asthma: evidence for differential signal transducer and activator of transcription 6 expression. J Allergy Clin Immunol. 2001;107:586–591. doi: 10.1067/mai.2001.114883. [DOI] [PubMed] [Google Scholar]
- 52.Mullings RE, et al. Signal transducer and activator of transcription 6 (STAT-6) expression and function in asthmatic bronchial epithelium. J Allergy Clin Immunol. 2001;108:832–838. doi: 10.1067/mai.2001.119554. [DOI] [PubMed] [Google Scholar]