Abstract
Obesity is the result of an imbalance between energy intake and energy expenditure. Using high-density DNA microarrays and Northern analyses, we demonstrated that the activation of a nutrient-sensing pathway, the hexosamine biosynthesis pathway (HBP), rapidly decreased the expression of a cluster of nuclear-encoded mitochondrial genes involved in skeletal muscle oxidative phosphorylation. Conversely, the expression of uncoupling protein-1 and of the same mitochondrial genes was increased in brown adipose tissue. Most important, these transcriptional changes were accompanied by a marked decrease in whole-body energy expenditure. Short-term overfeeding replicated this transcriptional pattern, suggesting that this adaptation to nutrient abundance occurs under physiological conditions. Thus, the activation of the HBP by nutrients represents a biochemical link between nutrient availability, mitochondrial proteins, and energy expenditure, and it is likely to play an important role in the regulation of energy balance.
Introduction
The increasing prevalence of obesity in industrialized societies is largely due to excessive caloric intake. However, there are major variations in the extent to which an individual alters energy metabolism and gains weight in response to increased caloric intake (1, 2). Furthermore, base-line energy expenditure is a strong predictor of future weight gain (3). The physiological basis for individual variability in energy expenditure is still poorly understood. A sustained increase in food intake activates adipostatic responses, which tend to limit lipid storage by enhancing energy expenditure (4) and inhibiting appetite (5). However, consistent with Neel’s “thrifty genotype” hypothesis, the increased availability of nutrients may also have biological effects designed to increase the efficiency of energy storage by limiting fuel oxidation and ATP production (6). Indeed, cells can monitor nutrient availability via the activation of nutrient-sensing pathways such as the hexosamine biosynthesis pathway (HBP) (7, 8). The final step in HBP is the formation of UDP-N-acetylglucosamine (UDP-GlcNAc), which is a product of intracellular glucose metabolism and is a main substrate for glycosylation of proteins. Many cytoplasmic and nuclear proteins are glycosylated on their serine and/or threonine residues by the addition of a single molecule of O-linked β-N-acetylglucosamine (O-GlcNAc) (9). In particular, several transcriptional factors undergo this type of rapid modification, which causes changes in their activity and/or in their stability (Figure 1a) (10–13). A prominent role of HBP in the regulation of energy balance is suggested by its role in the modulation of leptin expression and insulin action in adipose tissue and skeletal muscle (7, 8, 14–17).
Figure 1.
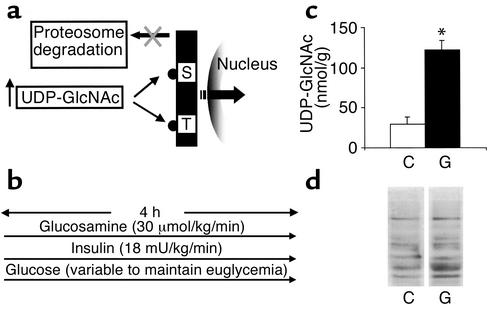
Activation of HBP increases O-linked glycosylation of nucleocytoplasmic proteins. (a) The enzyme OGTase links GlcNAc moieties to serine or threonine residues of transcriptional factors. O-GlcNAc induces conformational changes that render proteins refractory to proteosomal degradation and more available for modulation of transcription in the nucleus. (b) Schematic representation of experimental design. Acute infusion of glucosamine (30 μmol/kg/min) was performed during 4 hours of a hyperinsulinemic clamp involving the infusion of high-dose insulin (18 mU/kg/min) and glucose at variable rates to keep animals euglycemic. (c) Intracellular levels of UDP-GlcNAc in skeletal muscle of rats infused with saline (C) or glucosamine (G). *P < 0.005. (d) Western blot analysis of O-GlcNAc modification in skeletal muscle. Extracts from glucosamine-treated animals (G) and saline-infused animals (C) are compared.
Methods
Animals.
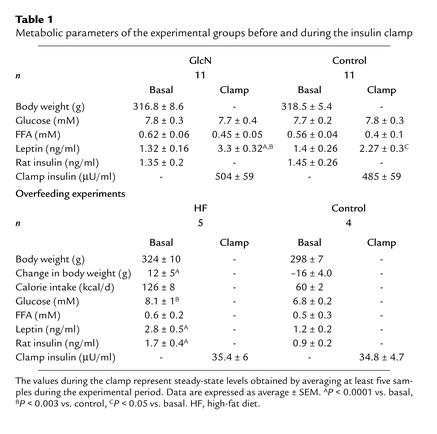
Male Sprague-Dawley rats (Charles River Laboratories, Wilmington, Massachusetts, USA) were housed in individual cages and subjected to a standard 12-hour light-dark cycle. All rats received standard chow ad libitum (catalog no. 5001; Purina Mills Inc., St. Louis, Missouri, USA), with 59% calories provided by carbohydrates, 20% by protein, and 21% by fat. Seven days prior to the in vivo studies, all rats underwent insertion of indwelling catheters into the right internal jugular vein and the left carotid artery as described previously (14). In the short-term overfeeding experiments, rats were allowed to eat ad libitum a palatable high-fat diet (catalog no. 9389; Purina Mills Inc.) with 45% calories provided by carbohydrates, 33% by fat, and 22% by protein for 3 days prior to the clamp studies as previously described (18). Control rats received a daily allowance of palatable high-fat diet restricted at 80% of their pre-intervention caloric intake.
Clamp studies.
All in vivo studies were performed in awake, unstressed rats fasted for 6 hours, using a euglycemic-hyperinsulinemic clamp technique, as described previously (14). GlcN (30 μmol/kg/min, n = 11) or saline (1.2 ml/h) was infused for 4 hours. Plasma samples for the determination of plasma glucose were obtained at 10-minute intervals throughout the insulin infusion. Samples for the measurement of plasma insulin, leptin, and other plasma substrates were obtained at 0, 60, 120, 180, and 240 minutes. The total volume of blood sampled was about 5 ml per animal; to prevent volume depletion and anemia, a solution (1:1 vol/vol) of about 6 ml of fresh blood (obtained by heart puncture from a littermate of the test animal) and heparinized saline (10 units/ml) was infused. At the end of the in vivo studies, rats were anesthetized (intravenously with 60 mg of pentobarbital per kg) and tissue samples were removed, quickly frozen in liquid nitrogen, and stored at –80°C for subsequent analysis.
DNA microarray.
Microarray slides were prepared by the Albert Einstein College of Medicine Microarray Facility (http://sequence.aecom.yu.edu/bioinf/funcgenomic.html and ref. 19). Approximately 9000 mouse cDNA clones, obtained from the I.M.A.G.E. Consortium (Integrated Molecular Analysis of Genomes and their Expression Consortium, Livermore, California, USA), were screened using hybridization protocols previously described (20). Analysis of microarray hybridizations was carried out with ScanAlyze software (ScanAlyze2, available at http://rana.lbl.gov/EisenSoftware.htm; Stanford University, Stanford, California, USA) initially created by Eisen and Brown (20). Equal amounts of cRNA from three samples were used for preparation of each probe. All data sets were normalized to total fluorescence, which represents the total amount of cRNA hybridized to a microarray. All data analyses were performed by using an average fold change, derived from three independent studies, of 1.7 or greater, since these changes could be detected and validated by Northern analysis.
Western blots.
For O-GlcNAc analysis, rat skeletal muscle was pulverized in liquid nitrogen and homogenized in 4 volumes of ice-cold buffer (10 mM Tris-HCl pH 7.9, 10 mM MgCl2, 0.5 mM PMSF, 1 mg/ml each of aprotinin, leupeptin, pepstatin, and 5 mM benzamidine). The homogenate was centrifuged at 2000 g for 10 minutes. Thirty micrograms of supernatant was loaded on a 10% SDS-PAGE and blotted on nitrocellulose filters. Immunoblots were performed with a mouse mAb specific for O-GlcNAc (Covance Research Products Inc., Richmond, California, USA).
Northern analyses.
Results of microarray analysis for selected individual genes were verified using Northern analysis. RNA was isolated from muscle and adipose tissue with Trizol (GIBCO BRL; Life Technologies Inc., Rockville, Maryland, USA), and 20 μg of total RNA was fractionated in denaturing gels and transferred to nylon membranes. DNA probes were labeled with 32P-dCTP using a random-priming technique (Stratagene, La Jolla, California, USA). After hybridization and high-stringency washing, membranes were exposed to radiographic films. The intensity of the autoradiographic bands was quantified by densitometry (ImageQuant software; Molecular Dynamics, Sunnyvale, California, USA). β-Actin hybridization was used to normalize for RNA loading.
Calorimetry.
Animals undergoing indirect calorimetry were acclimated to the respiratory chambers for 3–4 days before the clamp studies. Calorimetry was performed on 12 rats with indwelling catheters, randomized in two groups to receive either GlcN or saline infusion respectively, during a euglycemic hyperinsulinemic-clamp (see Clamp studies above). One day before the in vivo study, all rats were individually housed in the calorimeter cages, and data on base-line gas exchanges, activity, and food intake were collected. Immediately after the clamp studies, the rats were returned to the calorimeter for the post-clamp monitoring of gas exchanges and activity, in the absence of food. Indirect calorimetry was performed with a computer-controlled open-circuit calorimetry system (Oxymax; Columbus Instruments Co., Columbus, Ohio, USA). Our instrument comprises four respiratory chambers with a stainless steel elevated wire floor. Each chamber is equipped with water bottle, food tray connected to a balance, and activity monitor. Oxygen consumption and carbon dioxide production were measured for each rat at 6-minute intervals. Outdoor air reference values were measured after every ten measurements. Gas sensors were calibrated daily with primary gas standards containing known concentrations of O2, CO2, and N2 (Tech Air, White Plains, New York, USA). A mass flow meter was used to measure and control air flow. Oxygen was measured by an electrochemical sensor based on limited-diffusion, metal-air battery. CO2 was measured with a spectrophotometric sensor. Respiratory quotient (RQ) was calculated as the ratio of CO2 production (liters) to O2 consumption (liters). Energy expenditure was calculated as EE = (3.815 + 1.232 × VCO2/VO2) × VO2.
All values are presented as the mean ± SE. Unpaired Student’s t test was used for statistical analysis. The study protocol was reviewed and approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine.
Results
Glucosamine infusion increased skeletal muscle UDP-GlcNAc levels and protein glycosylation by O-GlcNAc. The in vivo protocol was designed to generate a significant increase in carbon flux through HBP in order to examine the impact of this nutrient-sensing pathway on gene transcription. We also wished to simulate the increase in the flux of nutrients into skeletal muscle that occurs during short-term overfeeding, while maintaining carefully controlled hormonal and metabolic conditions. To this end, we infused either glucosamine (GlcN) or vehicle for 4 hours in conscious rats in the presence of insulin clamp conditions (Figure 1b). The two experimental groups were matched for body weight and endocrine/metabolic parameters (Table 1). This experimental protocol resulted in a significant increase (fourfold) in the skeletal muscle concentration of UDP-GlcNAc, the end product of HBP (Figure 1c). To document that the increased availability of intracellular UDP-GlcNAc resulted in increased protein glycosylation by O-GlcNAc, we performed Western blots of total muscle extracts using antibodies specific for O-GlcNAc–modified serine/threonine residues (21). GlcN infusion increased this type of glycosylation in several muscle proteins (Figure 1d). The latter increase in O-GlcNAc modifications was quite variable depending on the specific band, with increases over vehicle ranging from about twofold to more than tenfold.
Table 1.
Metabolic parameters of the experimental groups before and during the insulin clamp
Activation of HBP decreases expression of genes for oxidative phosphorylation in skeletal muscle.
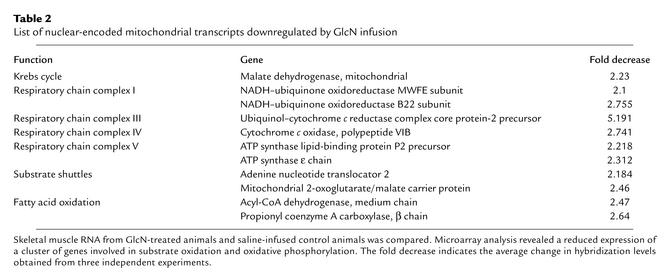
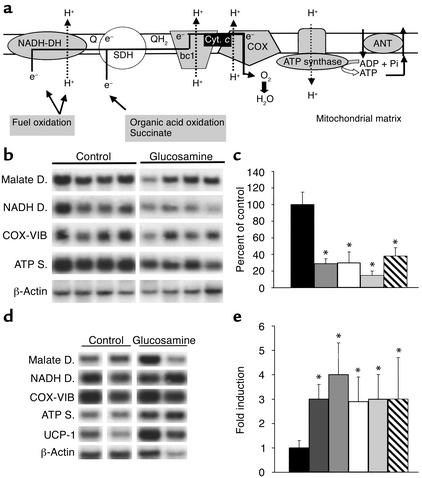
We used high-density DNA microarrays to characterize the changes in gene expression elicited by the activation of HBP in skeletal muscle. Among 8975 mouse genes and expressed sequence tags present on the chip, 97 transcripts were increased and 169 were decreased by GlcN compared with vehicle. Importantly, GlcN significantly decreased the expression of nuclear-encoded mitochondrial proteins involved in oxidative phosphorylation (Table 2). In mitochondria, the reduced NADH produced by the oxidation of nutrients generates energy by donating electrons down an array of carrier molecules called the electron transport chain (Figure 2a). The latter is composed of a series of four respiratory enzyme complexes localized in the mitochondrial inner membrane. The energy released by the electron transport is used to form a proton gradient across the mitochondrial inner membrane, which is in turn used by the ATP synthase complex to generate ATP from ADP and phosphate. A carrier protein (ANT [adenine nucleotide translocator]) exports ATP in exchange for ADP. Oxidative phosphorylation is the major cellular mechanism for the generation of energy in the form of ATP. Activation of HBP decreased the expression of one or more subunits from all but one of the respiratory chain complexes, as shown in Table 2 and Figure 2a. Additionally, GlcN also downregulated the expression of selective genes involved in fatty acid oxidation, mitochondrial substrate shuttles, and Krebs cycle. To validate the results obtained by analysis of microarrays, we next measured the levels of mRNA for selective mitochondrial proteins by Northern blot analysis (Figure 2b). Mitochondrial malate dehydrogenase (Krebs cycle) and the cytochrome c oxidase subunit VIB (respiration chain) were decreased by two- to threefold, while NADH dehydrogenase and the ε subunit of ATP synthase (respiration chain) were decreased by three- to fourfold (Figure 2b). Northern analysis of uncoupling protein-3 (UCP-3) mRNA did not reveal any significant change with HBP activation (data not shown).
Table 2.
List of nuclear-encoded mitochondrial transcripts downregulated by GlcN infusion
Figure 2.
In vivo effect of GlcN on the expression of nuclear-encoded mitochondrial transcripts. (a) Schematic illustration of respiratory chain complexes and oxidative phosphorylation. Electrons (e–) donated from fuel oxidation are sequentially transferred from ubiquinone (Q and QH2) to cytochrome c (Cyt. c), and finally to oxygen (O2). Simultaneously, protons (H+) are translocated across the inner mitochondrial membrane and create an electrochemical gradient whose energy is eventually used by ATP synthase for ATP formation. An adenine nucleotide translocator (ANT) carries ATP out of the mitochondria in exchange for ADP. The complexes indicated in gray contain one or more subunits downregulated by glucosamine. COX, cytochrome c oxidase; SDH, succinate dehydrogenase. (b) Northern analysis of mitochondrial transcripts in skeletal muscle from glucosamine versus saline infusion (control) with probes encoding mitochondrial malate dehydrogenase, NADH dehydrogenase, cytochrome c oxidase (COX-VIB), ATP synthase, and β-actin. (c) Quantitation, by densitometry, of Northern blots shown in b. Data are expressed as percent of control hybridization and are normalized with a β-actin probe. (d) Northern analysis of mitochondrial transcripts in BAT from glucosamine versus saline infusion (control) studies. Probes are the same as in b, except for the BAT-selective UCP-1. (e) Quantitation of Northern blot shown in d. Legend to bar graphs: Black, control; dark gray, UCP-1; medium gray, ATP synthase; white, cytochrome c oxidase VIB; light gray, NADH dehydrogenase; hatched, malate dehydrogenase. *P < 0.01.
The effect of HBP on the expression of genes for oxidative phosphorylation is limited to skeletal muscle.
Since brown adipose tissue (BAT) plays an important role in the regulation of energy expenditure, we next asked whether the activation of HBP also altered the expression of oxidative phosphorylation genes in BAT. Northern blot analysis performed at the completion of the GlcN infusion revealed that the expression of uncoupling protein-1 (UCP-1), which dissipates energy into heat by uncoupling oxygen consumption from ATP synthesis, and of genes of oxidative phosphorylation was actually increased in BAT. Mitochondrial malate dehydrogenase and cytochrome c oxidase were induced two- to threefold, while NADH dehydrogenase, UCP-1, and the ε subunit of ATP synthase were increased four- to fivefold (Figure 2, d and e). These findings suggest that increased dissipation of energy in BAT may partly compensate for the marked decrease in mitochondrial respiration in skeletal muscle.
Activation of HBP in skeletal muscle decreases whole-body oxygen consumption and energy expenditure. If the above transcriptional regulation leads to physiologically relevant changes in mitochondrial function, one should be able to measure alterations in in vivo respiratory exchanges. Thus, we asked whether a prolonged activation of HBP in skeletal muscle decreases whole-body oxygen consumption and energy expenditure. To this end, we measured respiratory exchanges by indirect calorimetry before and after GlcN infusion.
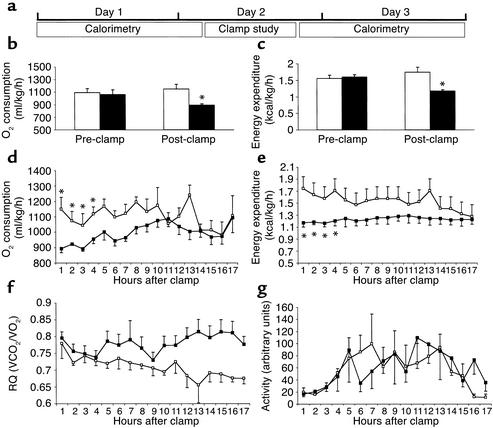
Sprague-Dawley (S-D) male rats were matched for food intake, body weight, and base-line energy expenditure and were randomized to receive either GlcN or vehicle infusion for 4 hours during insulin clamp studies (Figure 3a). After the completion of the clamp study, all rats were placed back into individual metabolic cages and their movements and gas exchange data were recorded. The post-clamp calorimetry was performed 3 hours before the onset of the light-dark cycle to allow the collection of gas exchanges at rest.
Figure 3.
Effect of GlcN on whole-body O2 consumption and energy expenditure. (a) Scheme of experimental design. Indirect calorimetry was performed on rats before and immediately after the completion of clamp studies. (b and c) Resting VO2 (b) and energy expenditure (c) are decreased in GlcN-treated animals (black bars) as compared with saline-injected animals (white bars). (d–g) Time course of VO2 (d), energy expenditure (e), RQ (f), and activity (g), after completion of clamp studies in GlcN-treated (black squares) and saline-injected animals (white squares). *P < 0.05.
Prior to infusions, oxygen consumption and energy expenditure were similar in the two groups (Figure 3, b and c). However, activation of HBP by GlcN infusion markedly decreased oxygen consumption by 22% (892.8 ± 26.2 ml/kg/h vs. 1150.3 ± 77.5 ml/kg/h of vehicle) and energy expenditure by 33% (1.17 ± 0.05 kcal/h vs. 1.75 ± 0.15 kcal/h of vehicle) compared with either base-line or vehicle (Figure 3, b and c).
Total oxygen consumption and energy expenditure remained significantly lower in the GlcN-infused rats for 4 hours after the end of GlcN infusion compared with vehicle (Figure 3, d and e). The RQ did not change significantly immediately after the clamp studies (Figure 3f). However, during the last 5 hours of the experiment, the RQ of the control group decreased from 0.78 ± 0.04 at 1 hour to 0.66 ± 0.03 at 17 hours from the end of the clamp study. By contrast, the RQ of the GlcN group did not change significantly (0.79 ± 0.02 at 1 hour vs. 0.77 ± 0.02 at 17 hours). This lack of transition to a lower RQ during fasting may reflect the downregulation of genes involved in fatty acid oxidation. Alternatively, HBP may activate net lipid biosynthesis, which would also result in a higher RQ. The effects of HBP on O2 consumption and energy expenditure could not be ascribed to differences in physical activity during the gas exchange measurements (Figure 3g).
Thus, activation of HBP leads to marked decreases in whole-body energy expenditure, consistent with the downregulation of genes involved in oxidative phosphorylation and fatty acid oxidation in skeletal muscle.
Overfeeding downregulates the expression of genes for oxidative phosphorylation in skeletal muscle.
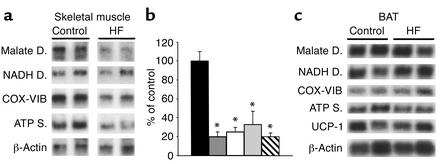
Activation of HBP rapidly enhances cellular glycosylation of proteins by O-GlcNAc and decreases the expression of key mitochondrial proteins involved in oxidative phosphorylation. In order to evaluate this link between nutrient sensing and energy expenditure in a more physiological setting, we next asked whether short-term voluntary overfeeding also induces a downregulation of genes for oxidative phosphorylation in skeletal muscle. S-D rats were given a palatable high-fat diet for 3 days. This resulted in an approximately twofold increase in caloric intake compared with controls rats who were eating regular chow (Table 1) (18). However, while body weight was markedly increased (Table 1), energy expenditure and oxygen consumption were only modestly higher (by ∼7%; data not shown) in response to short-term overfeeding. This nutritional manipulation also resulted in moderate increases in skeletal muscle (∼35%) and liver (∼60%) levels of UDP-GlcNAc. Using Northern analysis, we next measured the expression of the transcripts downregulated by HBP in muscle (Figure 4a). In this animal model, 3 days of voluntary overfeeding markedly downregulated the expression of genes encoding mitochondrial proteins (Figure 4, a and b). In particular, overfeeding decreased the skeletal muscle mRNA for mitochondrial malate dehydrogenase and the ε subunit of ATP synthase four- to fivefold, and the mRNA for NADH dehydrogenase and cytochrome c oxidase VIB subunit two- to threefold. However, short-term overfeeding failed to alter the expression of these genes and of UCP-1 in BAT (Figure 4c).
Figure 4.
Effect of short-term overfeeding on the expression of genes of oxidative phosphorylation. (a and b) Northern blot (a) and quantitation (b) in skeletal muscle of animals overeating a high-fat diet (HF) compared with control animals. (c) Northern analysis of genes for oxidative phosphorylation in BAT of overeating rats versus control. Legend to bar graphs: Black, control; medium gray, ATP synthase; white, cytochrome c oxidase VIB; light gray, NADH dehydrogenase; hatched, malate dehydrogenase. *P < 0.001.
Discussion
Mammals rapidly adapt to changes in caloric intake via compensatory changes in substrate oxidation and energy expenditure (4, 22). Thus, energy expenditure increases in response to voluntary overfeeding in an apparent attempt to limit energy “overflow” and storage. On the other hand, there are considerable interindividual variations in the effects of short-term overfeeding on energy expenditure (1). These variations appear to be genetically determined and partly due to variable expression and/or penetrance of thrifty genes, which limit the compensatory increase in energy expenditure in response to nutrient excess (2, 3). At the cellular level, long-term ATP production is maintained within a relatively tight range in most tissues, including resting skeletal muscle. It is hypothesized herein that a robust activation of a nutrient-sensing pathway is a signal of energy overflow. This signal in turn reduces the efficiency of ATP production in an attempt to limit marked excursions in the rate of cellular respiration.
Consistent with this hypothesis, here we show that overstimulation of HBP inhibits the expression of nuclear-encoded mitochondrial proteins involved in oxidative phosphorylation and substrate oxidation in skeletal muscle. Most important, this in turn leads to a significant decrease in whole-body oxygen consumption and energy expenditure. However, it should be noted that the changes in energy metabolism in response to activation of HBP are not limited to skeletal muscle. This signal of fuel abundance also resulted in a moderate increase in circulating leptin concentrations and in the induction of UCP-1 and of nuclear-encoded mitochondrial transcripts in BAT. The latter effects of GlcN infusion appear to represent attempts to dispose of the perceived abundance of fuel in skeletal muscle by increasing energy dissipation elsewhere (in BAT) and by activating leptin-driven neuroendocrine responses.
Despite these adaptive responses to HBP activation, whole-body energy expenditure was markedly reduced, suggesting that the inhibition of oxygen consumption in muscle outweighed the changes in mitochondrial bioenergetics in BAT. Thus, we speculate that the effects of HBP activation on energy metabolism are tissue-specific. In skeletal muscle, this signal of nutrient abundance limits oxidative stress by decreasing substrate oxidation and oxygen consumption, while in BAT it attempts to dissipate excess energy via the uncoupling of oxidative phosphorylation. The moderate increase in circulating leptin induced by GlcN infusions may play a role in the compensatory increase in the expression of UCP-1 (23) and selective mitochondrial proteins in BAT. Alternatively, the molecular mechanism(s) by which HBP modulates the expression of mitochondrial proteins in a tissue-specific manner may in part relate to the control of SP1 activity via O-GlcNAc modification (13, 24, 25). In fact, UCP1 and the other genes encoding for proteins in the oxidative phosphorylation and substrate oxidation pathways affected by HBP contain SP1-binding sites in their promoters (25–28). Interestingly, SP1 can function as both a positive and a negative regulator of transcription (25, 26).
If this link between nutrient sensing and mitochondrial function is relevant to physiological states of nutrient excess, it should also operate during voluntary increases in caloric intake. Indeed, in the present study, short-term voluntary overfeeding reproduced the effects of HBP activation on nuclear-encoded mitochondrial genes. Northern analyses indicate that the extent of transcriptional changes in skeletal muscle following 3 days’ overfeeding was quite comparable with that obtained with GlcN infusions.
Short-term overfeeding results in variable increases in body weight and fat mass in different individuals and in various strains of laboratory animals (1–5). This variability is partly due to differences in the compensatory increase in energy expenditure normally designed to match the marked increase in calorie intake. For example, the S-D rats in the present study increased their caloric intake approximately twofold, but they only modestly increased their energy expenditure (∼7%). This defective or delayed response to increased calorie intake is partly responsible for the rapid increase in body weight in this animal model (29). Thus, we hypothesize that the increased activation of HBP is likely to limit the compensatory increase in energy expenditure during voluntary overfeeding.
In line with the thrifty genotype hypothesis, activation of HBP in the presence of enhanced nutrient availability may have conferred a selective survival advantage by favoring the storage of excess nutrients as fat during periods of sporadic food availability. On the other hand, increased caloric intake for 3 days failed to enhance the expression of UCP-1 and other mitochondrial proteins in BAT. This is at odds with the effects of short-term activation of HBP, which significantly induced these genes in BAT. This may be due to the short duration of overfeeding in the present experiments. Alternatively, it is conceivable that rapid onset of leptin resistance may partly account for the lack of compensatory activation of UCP-1 and genes for oxidative phosphorylation in BAT. In fact, we recently reported that voluntary hyperphagia in obesity-prone rats rapidly induced insulin and leptin resistance (18). The latter was characterized by a marked decrease in leptin’s ability to modulate both liver metabolism and food intake.
Thus, in keeping with Neel’s thrifty genotype hypothesis (6), a sustained increase in the availability of nutrients may trigger biological actions designed to increase the efficiency of energy storage by limiting fuel oxidation and ATP production. This biochemical link between nutrient excess and energy expenditure may play a significant role in the pathophysiology of obesity.
Acknowledgments
This work was supported by grants from the National Institutes of Health (DK 48321 and DK 45024 to L. Rossetti), from the AECOMDiabetes Research &Training Center (DK 20541). S. Obici was the recipient of a post-doctoral fellowship and a Junior Faculty Award from the American Diabetes Association.
Footnotes
See the related Commentary beginning on page 1537.
Silvana Obici and Jiali Wang contributed equally to this work.
References
- 1.Levine JA, Eberhardt NL, Jensen MD. Role of nonexercise activity thermogenesis in resistance to fat gain in humans. Science. 1999;283:212–214. doi: 10.1126/science.283.5399.212. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard C, et al. The response to long-term overfeeding in identical twins. N Engl J Med. 1990;322:1477–1482. doi: 10.1056/NEJM199005243222101. [DOI] [PubMed] [Google Scholar]
- 3.Ravussin E, et al. Reduced rate of energy expenditure as a risk factor for body-weight gain. N Engl J Med. 1988;318:467–472. doi: 10.1056/NEJM198802253180802. [DOI] [PubMed] [Google Scholar]
- 4.Leibel RL, Rosenbaum M, Hirsch J. Changes in energy expenditure resulting from altered body weight. N Engl J Med. 1995;332:621–628. doi: 10.1056/NEJM199503093321001. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz MW, Woods SC, Porte D, Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 6.Neel JV. The “thrifty genotype” in 1998. Nutr Rev. 1999;57(Suppl. II):S2–S9. doi: 10.1111/j.1753-4887.1999.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 7.Marshall S, Bacote V, Traxinger R. Discovery of a metabolic pathway mediating desensitization of the glucose transport system: role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 8.McClain DA, Alexander T, Cooksey RC, Considine RV. Hexosamines stimulate leptin production in transgenic mice. Endocrinology. 2000;141:1999–2002. doi: 10.1210/endo.141.6.7532. [DOI] [PubMed] [Google Scholar]
- 9.Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291:2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- 10.Sayeski PP, Kudlow JE. Glucose metabolism to glucosamine is necessary for glucose stimulation of transforming growth factor-alpha gene transcription. J Biol Chem. 1996;271:15237–15243. doi: 10.1074/jbc.271.25.15237. [DOI] [PubMed] [Google Scholar]
- 11.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276:10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 12.Hanover JA, Cohen CK, Willingham MC, Park MK. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J Biol Chem. 1987;262:9887–9894. [PubMed] [Google Scholar]
- 13.Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossetti L, Hawkins M, Chen W, Gindi J, Barzilai N. In vivo glucosamine infusion induces insulin resistance in normoglycemic but not in hyperglycemic conscious rats. J Clin Invest. 1995;96:132–140. doi: 10.1172/JCI118013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veerababu G, et al. Overexpression of glutamine: fructose-6-phosphate amidotransferase in the liver of transgenic mice results in enhanced glycogen storage, hyperlipidemia, obesity, and impaired glucose tolerance. Diabetes. 2000;49:2070–2078. doi: 10.2337/diabetes.49.12.2070. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Liu R, Hawkins M, Barzilai N, Rossetti L. A nutrient-sensing pathway regulates leptin gene expression in muscle and fat. Nature. 1998;393:684–688. doi: 10.1038/31474. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, et al. The effects of leptin on Lep expression is tissue-specific and nutritionally regulated. Nat Med. 1999;5:895–899. doi: 10.1038/11335. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, et al. Overfeeding rapidly induces leptin and insulin resistance. Diabetes. 2001;50:2786–2791. doi: 10.2337/diabetes.50.12.2786. [DOI] [PubMed] [Google Scholar]
- 19.Cheung VG, et al. Making and reading microarray. Nat Genet. 1999;21:15–19. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]
- 20.Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 21.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal Biochem. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 22.Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 23.Cusin I, et al. Chronic central leptin infusion enhances insulin-stimulated glucose metabolism and favors the expression of uncoupling proteins. Diabetes. 1999;47:1014–1019. doi: 10.2337/diabetes.47.7.1014. [DOI] [PubMed] [Google Scholar]
- 24.Du XL, et al. Hyperglycemia-induced mitochondrial superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proc Natl Acad Sci USA. 2000;97:12222–12226. doi: 10.1073/pnas.97.22.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, et al. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci USA. 2001;12:6611–6616. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaid A, et al. On the role of the general transcription factor Sp1 in the activation and repression of diverse mammalian oxidative phosphorylation genes. J Bioenerg Biomembr. 1999;31:129–135. doi: 10.1023/a:1005499727732. [DOI] [PubMed] [Google Scholar]
- 27.Zaid A, Hodny Z, Li R, Nelson BD. Sp1 acts as a repressor of the human adenine nucleotide translocase-2 (ANT2) promoter. Eur J Biochem. 2001;268:5497–5503. doi: 10.1046/j.1432-1033.2001.02453.x. [DOI] [PubMed] [Google Scholar]
- 28.Scarpulla RC. Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr. 1997;2:109–119. doi: 10.1023/a:1022681828846. [DOI] [PubMed] [Google Scholar]
- 29.Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]