Abstract
Activation of T lymphocytes by specific antigen triggers a 3- to 7-day maturation process. Terminal differentiation begins late after T cell activation and involves expression of effector genes, including the chemokine RANTES and its major transcriptional regulator, RANTES factor of late-activated T lymphocytes-1 (RFLAT-1). In this article we demonstrate that RFLAT-1 expression is translationally regulated through its 5′-UTR and in a cell type–specific manner. Overexpression of the translation initiation factor eIF4E increases RFLAT-1 protein, while inhibition of Mnk1, which phosphorylates eIF4E, reduces RFLAT-1 production, indicating cap-dependent translational regulation. These events are regulated by ERK-1/2 and p38 MAP kinases and allow T cells to rapidly adjust RANTES expression in response to changes in the cellular environment, such as stress and/or growth factors. These findings provide a molecular mechanism for a rheostat effect of increasing or decreasing RANTES expression at sites of inflammation. Memory T cells, already poised to make RANTES, are finely regulated by translational control of the major transcription factor regulating RANTES expression. This is the first example of such a mechanism regulating a chemokine, but it seems likely that this will prove to be a general way for cells to rapidly respond to stress, cytokines, and other proinflammatory factors in their local environment.
Introduction
RANTES is a member of a large family of proinflammatory cytokines called chemokines (1). RANTES is a potent chemoattractant for T cells, monocytes (2), eosinophils (3, 4), basophils (5), and natural killer cells (6). RANTES activates and induces proliferation of T lymphocytes, mediates degranulation of basophils, and induces respiratory burst in eosinophils (7–9). RANTES is also associated with resistance to HIV (10). The chemokine receptor CC-CKR5, which binds RANTES and the closely related chemokines, macrophage inflammatory protein 1α (MIP-1α) and MIP-1β, functions as a coreceptor for HIV entry into target cells (11–14). Based on its role in inflammation and HIV pathogenesis, RANTES and its receptors are important therapeutic targets for immune-mediated diseases and AIDS (15).
RANTES is expressed by various cells and tissues under different conditions (16–18). In fibroblasts, epithelial cells, and monocytes/macrophages, RANTES expression increases within hours of stimulation, under the control of the Rel family of transcription factors (19). In T lymphocytes, by contrast, RANTES mRNA is induced late (3–5 days) after activation with either antigen or mitogen (1). These kinetics are similar to those of genes involved in T cell terminal differentiation including perforin, granulysin, and granzymes A and B.
We reported that expression of RANTES in T cells is largely controlled by the transcription factor RANTES factor of late-activated T lymphocytes-1 (RFLAT-1) (20). RFLAT-1 belongs to the growing family of Krüppel-like transcription factors and is also known as Krüppel-like factor 13 (KLF13) (21). It shares the most homology with basic transcription element-binding protein 1 (BTEB1/KLF9) and BTEB4. RFLAT-1 binds to the A site of the human RANTES promoter and strongly activates its transcription in T lymphocytes (20). Although steady-state levels of RFLAT-1 message remain constant throughout T cell activation, RFLAT-1 protein appears only after day 3 of activation, coincident with RANTES gene expression. These findings suggest that the expression of RFLAT-1 is regulated by a posttranscriptional mechanism.
Translational efficiency can be modulated by the 5′- and 3′-untranslated regions (UTRs) of a message (22, 23). Extensive secondary structure within the 5′-UTR can effectively inhibit translation (24). The 5′-UTR of RFLAT-1 is very GC-rich and therefore predicted to be highly structured. Three upstream open reading frames (uORFs) are also present, a characteristic feature of genes that are translationally regulated (25). Examples of “translationally repressed” genes include growth factors and cytokines, protein kinases involved in cell signaling, cell cycle regulators, and transcription factors (26). Interestingly, BTEB-1, the closest relative of RFLAT-1, is translationally regulated (27). A long GC-rich 5′-UTR containing multiple upstream AUGs (uAUGs) has been implicated in the regulation of BTEB-1 expression.
Here we report that RFLAT-1 expression is translationally regulated through its 5′-UTR. The effect of the RFLAT-1 5′-UTR on translation is specific to T lymphocytes. RFLAT-1 expression is regulated through a cap-dependent mechanism, involving eIF4E, Mnk1, and MAP kinases and allows T cells to rapidly adjust RANTES expression in response to environmental changes.
Methods
Plasmids and mutagenesis.
The full-length luciferase ORF derived from pGL2Basic, was ligated into pcDNA 3.1 V5 His Topo (Invitrogen Corp., Carlsbad, California, USA) to create pcDNA 3.1 Luc. A hybrid construct containing the 5′-UTR of RFLAT-1 and the luciferase gene was made by ligation of the PCR-amplified 5′-UTR from pcDNA3.1 RFLAT-1 into pcDNA3.1 Luc. The following 5′-UTR point mutations were introduced by PCR: UUG1, A38U; UUG2, A155U; and UUG3, A319U. PCR was used to delete the first 142 nucleotides (ΔAUG2) and the first 306 nucleotides (ΔAUG3) of the RFLAT-1 5′-UTR. The integrity of all constructs was verified by sequencing.
Ab’s and reagents.
Ab’s were obtained from the following sources: anti-eIF4E (4E) (Transduction Laboratories, Lexington, Kentucky, USA); anti-p44/42 MAP kinase (ERK-1/2), anti–phospho-p44/42 MAP kinase (Thr202/Tyr204) (P-ERK-1/2), and anti–phospho-4EBP (Ser65) Ab (Cell Signaling Technologies, Beverly, Massachusetts, USA); anti–α-actinin (Upstate Biotechnology Inc., Lake Placid, New York, USA); FITC-conjugated RANTES Ab and its IgG2B isotype control (Caltag Laboratories Inc., Burlingame, California, USA), and anti–FLAG M2 (Sigma-Aldrich St. Louis, Missouri, USA). The anti–RFLAT-1 Ab was previously described (20). Rapamycin was a gift from Wyeth-Ayerst Laboratories (Philadelphia, Pennsylvania, USA). The rIL-2 was a gift from The National Cancer Institute (NCI), Biological Resources Branch, NCI, Frederick Cancer Research and Development Center, Frederick, Maryland, USA. The MAP kinases inhibitors SB203580, SB202474, and PD98058 were purchased from Calbiochem-Novabiochem Corp. (San Diego, California, USA) .
T cell isolation and culture.
Human peripheral blood lymphocytes (PBLs) and IL-2–dependent T cells were prepared as described (28). PBLs (2 × 106/ml) stimulated with 5 μg/ml phytohemagglutin-P (PHA-P) were cultured for up to 7 days. IL-2–dependent cells (106/ml), cultured in the presence of 100 U/ml rIL-2, were stimulated with 1,000 U/ml rIL-2 for up to 6 hours. Intracellular staining was performed using the Cytofix/Cytoperm kit (PharMingen, San Diego, California, USA), and fluorescence was measured by FACScan (Becton Dickinson Immunocytometry Systems, San Jose, California, USA). A CD8+ long-term cytotoxic T lymphocyte cell line (28) (106/ml), pretreated with 100 nM rapamycin for 30 minutes at 37°C, was stimulated with 500 U/ml rIL-2 and cultured in the presence of rapamycin for 0, 2, 4, and 6 hours.
Polyribosome preparation.
Resting or activated PBLs (2 × 107/sample) were incubated with 0.1 mg/ml cycloheximide for 3 minutes at 37°C before the cells were harvested and lysed by detergent treatment. The cytoplasmic extracts were layered on 10–50% linear sucrose gradients and centrifuged at 150,000 g for 2 hours 45 minutes at 4°C in Beckman SW41 rotor. RNA from the fractions was isolated by guanidinium isothiocyanate extraction and analyzed by Northern blot using standard techniques. The RFLAT-1 probe was prepared by α-32P-labeling of the BamHI-to-XhoI fragment of the RFLAT-1 cDNA.
RNA analysis.
Lysates of Jurkat T cells transfected with Luc, and 5′-UTR Luc plasmids were prepared using Direct Protect Lysate ribonuclease protection assay (RPA; Ambion Inc., Austin, Texas, USA). RPAs were performed using antisense RNA probes synthesized from a luciferase PCR fragment corresponding to nucleotides 14 to 328 of pGL2 basic (Promega Corp., Madison, Wisconsin, USA) and pTRI-beta-actin-human (Ambion Inc.). Full protection of the luciferase riboprobe by the luciferase transcripts resulted in a 314-nucleotide fragment, while the protected actin fragment was 245 nucleotides.
Transient transfections, in vitro translation, and luciferase assays.
NIH 3T3, HEK 293, S2-6, and S2-6-4E cell lines were transfected with Lipofectin (Invitrogen Corp.), and Jurkat T cells were electroporated as described (20). Nonradioactive luciferase reactions were performed using the TnT Quick-coupled transcription translation system (Promega Corp.), according to the manufacturer’s protocol. Luciferase activity was measured as described (20). In the transient transfection assays, luciferase constructs (5 μg each) were cotransfected with pRLNULL (0.1 μg per transfection), and luciferase activity was normalized to Renilla luciferase activities to account for differences in transfection efficiency. Whole cell lysates were prepared using a modified RIPA buffer (28). For Western analysis, 20 μg of total protein from each sample was run on a 10% SDS-PAGE.
Statistical analysis.
Results from all luciferase assays are representative of at least three independent experiments performed in triplicate. The Western blots are rep-resentative of two to three independent experiments. Significance of differences between experimental and control values was calculated using ANOVA. The calculations were performed using Microsoft Excel. A P value of less than 0.05 was considered significant.
Results
Kinetic studies of RFLAT-1 expression in T lymphocytes demonstrated that its mRNA remains constant in both resting and activated cells. Nevertheless, RFLAT-1 protein is expressed only 3–5 days after activation (20). A multiple human tissue Western blot probed with anti–RFLAT-1 Ab revealed that RFLAT-1 protein is found only in adult spleen and lung and not in liver, brain, kidney, heart, and reproductive organs (data not shown), while its message is ubiquitously present in all tissues examined (20). This observation indicates that RFLAT-1 expression is regulated through a posttranscriptional mechanism.
RFLAT-1 mRNA is actively translated late after T cell activation.
Sucrose gradients were used to analyze the association of RFLAT-1 mRNA with polysomal fractions derived from resting (day 0) or activated (day 1 and day 5 after activation with mitogen) PBLs. As seen in Figure 1,a and b, a dramatic shift of RFLAT-1 mRNA from the monosomal (fraction 4) and early polysomal (fractions 5 and 6) to the late polysomal fractions (up to fraction 9) was observed at day 5 compared with day 1 after T cell activation, indicating that RFLAT-1 expression is translationally regulated.
Figure 1.
RFLAT-1 5′-UTR inhibits translation in vitro and in vivo. (a) Absorbance profile (254 nm) of sucrose gradients from lysates of resting (D0) or PHA-activated (D1 and D5) PBLs. (b) Nine fractions were collected from each gradient, and equal volumes of each fraction were separated on an agarose gel and analyzed by Northern hybridization with an RFLAT-1 probe. 18S and 28S rRNA in each fraction were visualized by ethidium bromide staining. (c) RFLAT-1 expression constructs. (full-length [RF] or lacking the 5′-UTR [ΔU]) were subjected to in vitro transcription-translation in the presence of 35S methionine. The products were detected by autoradiography. (d–g) pcDNA3.1 Luc (Luc) or pcDNA 3.1 5′-UTR Luc (5′-UTR Luc) were subjected to in vitro transcription-translation (d) or transiently transfected into Jurkat T cells (e and g) or HEK293 cells (f) and assayed for luciferase activity. The data are presented as relative luciferase activity where the activity of the 5′-UTR Luc construct is arbitrarily set to 1. *P < 0.05 (d, e, and f). Decreasing amounts of lysates from transfected Jurkat cells were subjected to an RPA (g) using luciferase (upper panel) and actin riboprobes (bottom panel). Lanes 2 and 5, 30 μl; 3 and 6, 15 μl; 4 and 7, 7.5 μl lysate.
The RFLAT-1 5′-UTR mediates translational regulation both in vitro and in vivo.
Translational efficiency can be modulated by the 5′ and 3′ UTRs of the message (23). The 5′-UTR of RFLAT-1 is unusually long (360 nucleotides) and over 80% GC rich. Its deletion resulted in over 25-fold increase in RFLAT-1 protein compared with the full-length cDNA (Figure 1c), demonstrating a major role for the 5′-UTR in RFLAT-1 translational regulation.
To further analyze the role of the 5′-UTR, a hybrid construct containing the RFLAT-1 5′-UTR fused to the luciferase ORF was created. This construct (5′-UTR Luc), along with a control plasmid containing the luciferase gene (Luc), was used in an in vitro transcription-translation assay. The presence of the RFLAT-1 5′-UTR inhibited luciferase activity over tenfold (Figure 1d). For in vivo studies, the same constructs were transfected into different cell lines, and translational efficiency was measured based on luciferase activity. In the T cell lines Jurkat (Figure 1e) and Hut78 (data not shown), the presence of the 5′-UTR inhibited expression approximately threefold, while in the fibroblast cell line 3T3 (data not shown) and in the renal cell line HEK293 (Figure 1f), the effect was marginal. To verify that the differences in luciferase activity are not at the level of luciferase mRNA, we conducted RPAs. Both luciferase constructs were transfected into Jurkat T cells, the cells were lysed 72 hours after transfection, and serial dilutions of the lysates were hybridized to an antisense luciferase riboprobe. As shown in Figure 1g, similar amounts of luciferase transcripts were produced from the Luc and 5′-UTR Luc expression vectors, confirming that the observed inhibition of luciferase activity is due to a translational mechanism. Taken together, these data show that the 5′-UTR of RFLAT-1 is involved in translational regulation of its gene expression in vivo and that this effect is specific for T cells.
The role of the RFLAT-1 uORFs in translational regulation.
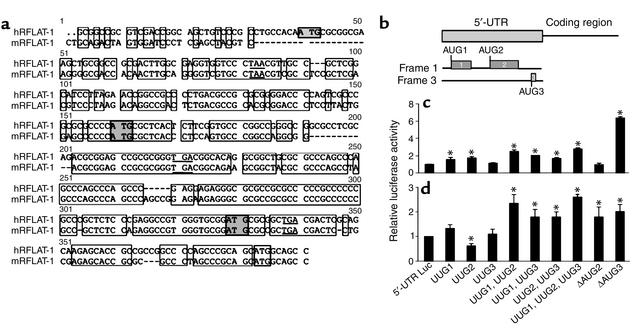
The uAUGs and uORFs can inhibit translation, presumably by interfering with ribosome scanning to the translational start site (29–31). The RFLAT-1 5′-UTR is highly conserved between human and mouse (76% identity). The human sequence contains three uORFs (designated uORF1, 2, and 3), each with a corresponding start and stop codon (Figure 2, a and b).
Figure 2.
Effect of uORFs on RFLAT-1 translation efficiency. (a) Sequence alignment of the 5′-UTR of human and mouse RFLAT-1. The positions of the uAUGs and the first methionine are depicted (shaded box). Underlined are the stop codons for each of the three uORFs. Clear boxes indicate identical nucleotides; dashed lines, missing nucleotides. (b) RFLAT-1 5′-UTR uORF’s, designated 1, 2, and 3, are schematically represented. (c and d) The 5′-UTR Luc (wild-type or mutant) was subjected to in vitro translation (c) or transiently transfected into Jurkat T cells (d) as in Figure 1.
To investigate the importance of the uORFs of RFLAT-1, each uAUG was individually mutated to UUG, a codon that is not recognized by the ribosome as a site of translation initiation (Figure 2b). The three mutations are designated UUG1, UUG2, and UUG3 respectively. Combinations of mutations (UUG1,2; UUG1,3; UUG2,3; and UUG1,2,3) were also created to estimate the importance of each of the uORFs (Figure 2b). The mutant constructs were subjected to in vitro transcription/translation as above (Figure 2c) or tested in vivo by transfection in Jurkat T cells (Figure 2d). In vitro, mutation of either uAUG1 (UUG1) or uAUG2 (UUG2) caused approximately twofold increase in luciferase activity, indicating the equal importance of both uORFs in the regulation of translation (Figure 2c). In vivo, the UUG1 mutation caused a slight increase in luciferase activity, while the UUG2 mutation caused a further inhibition of translation (Figure 2d). However, combinations of mutations had additive effects in relieving translational repression, suggesting that both uORFs are involved in translational regulation of RFLAT-1 expression (Figure 2, c and d). Point mutations in all three uORFs did not completely abolish translational repression in vitro, suggesting that in vitro the secondary structure of the 5′-UTR also plays an important role. Moreover, a deletion of the 5′-UTR preceding AUG2 (ΔAUG2) did not relieve translational repression, while a deletion of the 306 nucleotides preceding uORF3 (ΔAUG3), which removes the first two uORFs and the majority of the 5′-UTR secondary structure, caused sixfold increase in luciferase activity (Figure 2c). This enhancement is significantly higher compared with the UUG1,2 mutant, in which the extensive secondary structure is preserved while uORF1 and uORF2 are absent. The ΔAUG2 and ΔAUG3 mutations did not have an additional effect in vivo (Figure 2d), supporting the above conclusion. We used RPA as above to determine the levels of luciferase transcripts produced by each of the mutant constructs and found that they were similar to that produced by the transfected Luc vector (data not shown). Taken together, these data indicate that translational repression of RFLAT-1 expression is mediated through its 5′-UTR and in vivo is due predominantly to the first two uORFs. In vitro uORF1 and uORF2 seem to be equally important, and additional repression is caused by the secondary structure of the 5′-UTR.
Overexpression of eIF4E increases RFLAT-1 protein levels.
In mammals, most translational regulation occurs at the level of translation initiation where the eIF4F complex mediates recruitment of ribosomes to mRNA (25). The least abundant of all initiation factors, eIF4E is considered to be rate limiting in the binding of ribosomes to mRNA and a major target for regulation (32, 33). The eIF4E protein levels increase during the first 24 hours of T cell activation (34). We observed approximately twofold increase in eIF4E protein levels late in T cell activation preceding or concurrent with RFLAT-1 appearance in the cell (Figures 3, a and b). This suggested that eIF4E might be important for inducing RFLAT-1 protein expression. Since translational repression of many messages with highly structured 5′-UTRs is relieved by overexpression of eIF4E (35, 36), we used the previously described tetracycline-inducible cell line S2-6-4E (37) to test the effect of eIF4E on RFLAT-1 expression. Expression vectors for full-length RFLAT-1 (designated RF) and for truncated RFLAT-1 cDNA, which lacks the 5′-UTR (designated ΔU), were transfected into S2-6-4E and its parental line S2-6. The eIF4E overexpression was induced by removal of tetracycline (37), and RFLAT-1 expression was measured 24 hours later. Overexpression of eIF4E resulted in an approximately threefold increase in the amount of RFLAT-1 produced from the full-length cDNA (Figures 3, c and d). In contrast, the effect on translation of the ΔU construct was minimal, in agreement with cap-dependent translational regulation.
Figure 3.
The eIF4E overexpression increases RFLAT-1 protein levels. (a and b) PHA-activated PBLs were lysed at days 0, 1, 2, 3, and 5. Western blot analysis with anti–RFLAT-1 and anti–eIF4E Ab was performed. Relative amount of eIF4E protein was determined by densitometry followed by normalizing to the amount of α-actinin in the lane. Results are representative of three independent experiments (b). (c and d) S2-6 and S2-6-4E cells were transiently transfected with 1 μg of RFLAT-1 expression constructs in the presence of 1 μg pCMV β-gal. eIF4E was induced 48 hours after transfection, and the cells were harvested 24 hours later. Samples were subjected to Western blot analysis with anti–RFLAT-1 and anti-eIF4E Ab. C, no DNA transfection control. The amount of RFLAT-1 protein was determined by densitometry and normalized to Hsc70 in the lane and to the β-galactosidase activity of the sample (d). Results are representative of two independent experiments. *P < 0.05.
RFLAT-1 expression depends on 4E-BP phosphorylation.
eIF4E is regulated at multiple levels (38). Its activity is inhibited by binding to a family of translational repressor proteins (4E-BPs). The 4E-BPs are inactivated by phosphorylation through a signaling cascade that is phosphatidylinositol 3-kinase (PI-3K) dependent and involves Akt/PKB and FRAP/mTOR (38). To explore the upstream events of RFLAT-1 translational regulation, we examined the effects of the immunosuppressant rapamycin on RFLAT-1 protein levels, which inhibit phosphorylation of 4E-BP by binding to its target FRAP/mTOR (38). A CD8+ IL-2–dependent human cytotoxic T cell line was pretreated with 100 nM rapamycin followed by stimulation with 500 U/ml rIL-2. As shown in Figure 4, a and b, rapamycin treatment resulted in 1.5- to 2-fold decrease in the amounts of RFLAT-1, suggesting that 4E-BP phosphorylation is important in RFLAT-1 translational regulation during late T cell activation.
Figure 4.
Rapamycin inhibits RFLAT-1 protein expression. (a and b) A CD8+ cell line was stimulated with rIL-2 and cultured with or without rapamycin for 0, 2, 4, and 6 hours. Lysates were subjected to Western blot analysis with anti–RFLAT-1 Ab, anti–phospho-4EBP-1 as control for the effect of rapamycin, and anti-Hsc70. The relative amount of RFLAT-1 protein in each sample was determined as above. Results are representative of two independent experiments. *P < 0.05. Rap, rapamycin.
RFLAT-1 translational regulation depends on MAP kinase signaling.
eIF4E is phosphorylated in response to growth factors, mitogens, and hormones (23). Phosphorylated eIF4E has higher binding affinity for the cap (39) and forms a more stable eIF4F complex (40), resulting in enhanced translation. Mnk1, the physiological kinase that acts upon eIF4E, is under the control of both p38 MAPK and ERK-1/2 signal transduction pathways (41). Recently, Mnk1 has been implicated in regulation of cap-dependent translation in HEK293 (42). To investigate whether Mnk1 is involved in RFLAT-1 translational regulation in T cells, we used inhibitors of p38 MAPK, SB203580 (SB), and ERK-1/2, PD98058 (PD). SB202474, a related compound that does not affect p38 MAPK, was used as a control. Human PBLs were activated with mitogen and cultured for 3–5 days in the presence or absence of inhibitors. As shown in Figure 5a, PD caused a threefold decrease in RFLAT-1 protein, while SB had no effect. However, in the presence of both inhibitors the amount of RFLAT-1 protein decreased over 15-fold (SB+PD). This synergistic effect indicates that both the p38 MAPK and ERK-1/2 pathways are involved in the regulation of RFLAT-1 gene expression, consistent with the signal transduction pathways leading to Mnk1 phosphorylation and further activation of eIF4E.
Figure 5.
RFLAT-1 expression is regulated by MAP kinases and Mnk1. (a) PHA-activated PBLs were treated with medium (C), 4 μM SB203580 (SB), 4 μM SB202474 (SB-C), 10 μM PD98058 (PD), or both SB203580 and PD98058 (SB+PD). Western blots with anti–RFLAT-1, anti–ERK-1/2, and phospho-specific anti–ERK-1/2 Ab were performed. (b and c) Jurkat cells were electroporated with 5 μg of the RFLAT-1 expression vector alone or in combination with 10 μg of constructs expressing Mnk1. Western blots were probed with anti–RFLAT-1, anti–FLAG M2, and anti-Hsc70, and the amount of RFLAT-1 in the samples was determined as above. C, no DNA control. Results are representative of two independent experiments. *P < 0.05.
Mnk1 is a positive regulator of RFLAT-1 expression.
To confirm the direct involvement of Mnk1 in RFLAT-1 translational regulation, the RFLAT-1 expression vector was transfected with or without constructs expressing wild-type Mnk1, a constitutively active form of the kinase (T344E) and a dominant negative Mnk1 mutant (TAA) (43, 44) into Jurkat T cells (Figure 5, b and c). RFLAT-1 expression increased 40% in the presence of the constitutively active Mnk1 (T344E), while the dominant negative form (TAA) significantly inhibited (threefold) RFLAT-1 production, reducing the protein to its basal levels (Figure 5, b and c). Taken together, our data indicate that Mnk1 is a positive regulator of RFLAT-1 expression in T cells.
IL-2 induces RFLAT-1 protein expression.
Since growth factors, mitogens, and cytokines can activate the ERK-1/2 signal transduction cascade, we hypothesized that treatment of T cells with IL-2 would rapidly increase eIF4E phosphorylation and activity, induce RFLAT-1 protein production, and thus, increase RANTES expression. To test this hypothesis we used an IL-2–activated T cell line (28). An increase in expression of RANTES, as measured by FACS, was observed as early as 2 hours after stimulation and peaked at 4 hours (Figure 6a). By 6 hours, intracellular RANTES had returned to the basal level. We also examined the amounts of RFLAT-1 and eIF4E proteins at the corresponding time points and found that there was an increase of both proteins concomitant with increased RANTES expression (Figure 6, b and c). Interestingly, the eIF4E blot showed an increase mostly of the slower migrating and presumably phosphorylated form. Thus, RANTES expression in T cells is tightly regulated by changes in the cellular environment through activation of the basic translation machinery.
Figure 6.
IL-2 induces RANTES and RFLAT-1 expression. (a) IL-2–dependent T cells were stimulated with 1,000 U/ml rIL-2, and intracellular RANTES was measured by FACS. Dotted line, no stimulation; solid line, 4 hours after stimulation. (b and c) Aliquots of the cells were lysed and subjected to Western blot analysis using anti–RFLAT-1, anti-eIF4E, and anti-Hsc70 Ab. Results are representative of two independent experiments. *P < 0.05.
Discussion
RFLAT-1 is the major transcription factor that positively regulates expression of RANTES in activated T cells. RFLAT-1 protein expression is developmentally regulated, appearing late after activation of T cells (20). In contrast, its message is ubiquitously present and its steady-state levels do not change during T cell activation. Here we have investigated the molecular mechanism underlying the posttranscriptional control of RFLAT-1 gene expression.
Regulation of RFLAT-1 expression is mediated through its 5′-UTR.
The RFLAT-1 5′-UTR is very GC rich and contains three alternative start sites for short uORFs. Deletion of the 5′-UTR resulted in a significant increase in translation efficiency in an in vitro transcription-translation assay. Furthermore, the fusion of the RFLAT-1 5′-UTR to the luciferase coding region inhibited luciferase activity, both in transfected mammalian cells and in in vitro translation assays. This indicates that the RFLAT-1 5′-UTR is not only necessary, but also sufficient to mediate translational repression. Interestingly, the effect of the 5′-UTR was more pronounced in T cells, suggesting that RFLAT-1 regulation is T cell specific.
Translation may be regulated by various cis-acting elements within the 5′-UTR, that is, start site context, uORFs, binding sites for regulatory proteins, polypyrimidine tracts, and internal ribosome entry sites (45, 46). The uAUGs are known to inhibit translation initiation, presumably by interfering with initiation at the bona fide start codon (23). Mutational analysis of the uAUGs of RFLAT-1 demonstrated that uAUG1 and uAUG2 are both important for translational repression of RFLAT-1, since point mutations of the two uAUGs completely eliminated the inhibitory effect of the 5′-UTR. Interestingly, although mutation of the first AUG caused slight increase in translation efficiency, mutation of the second start site further inhibited translation. The reasons for this effect are unclear, but the absence of the second uORF may interfere with reinitiation of the ribosome at the bona fide translation initiation start site.
Stable secondary structures in the 5′-UTR can interfere with ribosome scanning to the translation start site (24). The extremely high GC content of the RFLAT-1 5′-UTR allows formation of multiple internal hairpin loops, as demonstrated by computer predictions of its secondary structure (21). The RFLAT-1 5′-UTR secondary structure appears to be important mostly in in vitro translation assays, while in vivo most of the translational inhibition could be attributed to the uAUGs in the sequence. This observation does not exclude a role for the extensive secondary structure in repression of translation in the cell. The multiple internal hairpins may be important for facilitation of the inhibitory effect of the uAUGs by promoting positioning of the ribosomes at these sites.
At this time, we cannot exclude a role of RNA-binding proteins in RFLAT-1 translational repression. Nevertheless, it is plausible that the general translation machinery is responsible for regulation of RFLAT-1 expression, since different transcripts have different requirements for translation factors and can thus be regulated in a gene-specific manner (47, 48).
Translation of RFLAT-1 expression is cap dependent.
eIF4E is the rate-limiting factor in translation initiation (32, 33), and overexpression of eIF4E can relieve translational repression of messages with highly structured 5′-UTRs (35, 36). We show here that eIF4E overexpression results in higher levels of RFLAT-1 protein translated from the full-length message but not from the mRNA lacking the 5′-UTR, indicating that RFLAT-1 translation is cap dependent. Furthermore, eIF4E protein levels increase during late T cell activation coincident with RFLAT-1 appearance, supporting the role of the basic translation machinery in regulation of RFLAT-1 expression.
The eIF4E activity is regulated at multiple levels, including interaction with inhibitory proteins (4E-BP) and phosphorylation. Phosphorylation of 4E-BP, which results in activation of eIF4E, is mediated by a PI-3K–dependent pathway and is inhibited by the immunosuppressant rapamycin. Treatment of PBLs with rapamycin resulted in decreased production of RFLAT-1, further implicating eIF4E in regulation of RFLAT-1 expression. The eIF4E kinase Mnk1 is controlled by both p38 MAPK and ERK-1/2 signal transduction pathways (41). RFLAT-1 levels were decreased in PBLs treated with inhibitors of both pathways, suggesting that Mnk1 is involved in RFLAT-1 translational regulation. Expression of a constitutively active Mnk1 mutant increased RFLAT-1 protein levels, while a dominant negative Mnk1 mutant inhibited RFLAT-1 expression. These results indicate that Mnk1 acts as a positive regulator of RFLAT-1 expression, presumably through phosphorylation of eIF4E and promoting translation initiation. Taken together, these data support our conclusion that regulation of RFLAT-1 translation is cap dependent.
Translational rheostat for RFLAT-1/RANTES expression.
Translational regulation is particularly important during cell growth and differentiation (22, 25). Examples of translational regulation in the immune system have been described recently. The report that IFN-γ mRNA expression is regulated through PKR (49) suggests that effector mRNA levels in different immune cells may be regulated by diverse mechanisms. Translational control allows cells to respond rapidly to changes in the environment without de novo synthesis or degradation of specific mRNAs. Since the function of eIF4E can be regulated by multiple physiologic stimuli, including growth factors, mitogens, and cytokines (22), it is conceivable that RFLAT-1, and thus RANTES expression, can undergo dynamic regulation in T lymphocytes. Changes in the abundance and/or phosphorylation state of eIF4E or 4E-BPs in response to these stimuli could modulate the inhibitory effect of the 5′-UTR. Consistent with this idea is our observation that IL-2 can regulate RFLAT-1 and RANTES expression via enhanced eIF4E protein expression and phosphorylation.
In addition to eIF4E, the eIF4F initiation complex consists of other factors, including eIF4A (an RNA-dependent ATPase and helicase) and the scaffold protein eIF4G, which contains binding sites for eIF4E, eIF4A, eIF3, Mnk1, and the poly A-binding protein (23). Thus, RFLAT-1 translation may also be modulated by regulation of other components of translation initiation complex. For example, changes in the RNA helicase activity of eIF4A could be a possible target of regulation, given the highly structured 5′-UTR of RFLAT-1.
Translational regulation of RANTES may be critical in vivo at sites of inflammation. Immune cells use a myriad of receptors to constantly monitor their environment as they move through extracellular spaces to the sites of injury to incite and maintain inflammation. In this context, the “late” (3–5 days after activation) expression of RANTES in T lymphocytes is particularly interesting, suggesting that this chemokine may play a special role in regulating T lymphocyte–mediated inflammation in time and space. Here we show that translational regulation of RANTES expression permits a rapid response to cytokines and growth factors (i.e., IL-2) in mature effector T cells. Subtle changes in chemokine gradients provide a precise rheostat for chemoattraction and events related to immune cell activation.
Acknowledgments
We thank Shu-Chen Lyu, Xiaoli Qin, and Kianoush Khaleghpour for technical assistance and Peter Sarnow for generously sharing his laboratory equipment. This work was supported by NIH grant DK-35008 and in part by grant PF-77419 from the Elizabeth Glaser Pediatric AIDS Foundation and the Satellite Dialysis Centers Young Investigator Grant of the National Kidney Foundation.
Footnotes
Tania Nikolcheva’s present address is: IVDU, Roche Bioscience, Palo Alto, California, USA.
Stephane Pyronnet’s present address is: INSERM U531, Institut Louis Bugnard, Toulouse, France.
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: RANTES factor of late-activated T lymphocytes-1 (RFLAT-1); macrophage inflammatory protein 1α (MIP-1α); Krüppel-like factor 13 (KLF13); basic transcription element-binding protein 1 (BTEB); untranslated regions (UTRs); upstream open reading frames (uORFs); upstream AUGs (uAUGs); phytohemagglutin-P (PHA-P); luciferase (Luc).
References
- 1.Schall TJ, et al. A human T cell-specific molecule is a member of a new gene family. J Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 2.Schall TJ, Bacon K, Toy KJ, Goeddel DV. Selective attraction of monocytes and T lymphocytes of the memory phenotype by cytokine RANTES. Nature. 1990;347:669–671. doi: 10.1038/347669a0. [DOI] [PubMed] [Google Scholar]
- 3.Kameyoshi Y, Dorschner A, Mallet AI, Christophers E, Schroder JM. Cytokine RANTES released by thrombin-stimulated platelets is a potent attractant for human eosinophils. J Exp Med. 1992;176:587–592. doi: 10.1084/jem.176.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rot A, et al. RANTES and macrophage inflammatory protein 1 alpha induce the migration and activation of normal human eosinophil granulocytes. J Exp Med. 1992;176:1489–1495. doi: 10.1084/jem.176.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahinden CA, et al. Monocyte chemotactic protein 3 is a most effective basophil- and eosinophil-activating chemokine. J Exp Med. 1994;179:751–756. doi: 10.1084/jem.179.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taub DD, Sayers TJ, Carter CR, Ortaldo JR. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol. 1995;155:3877–3888. [PubMed] [Google Scholar]
- 7.Kuna P, et al. RANTES, a monocyte and T lymphocyte chemotactic cytokine releases histamine from human basophils. J Immunol. 1992;149:636–642. [PubMed] [Google Scholar]
- 8.Alam R, et al. RANTES is a chemotactic and activating factor for human eosinophils. J Immunol. 1993;150:3442–3448. [PubMed] [Google Scholar]
- 9.Bacon KB, Premack BA, Gardner P, Schall TJ. Activation of dual T cell signaling pathways by the chemokine RANTES. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 10.Cocchi F, et al. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 11.Alkhatib G, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 12.Choe H, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Doranz BJ, et al. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 14.Dragic T, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Nelson PJ, Krensky AM. Chemokines, lymphocytes and viruses: what goes around, comes around. Curr Opin Immunol. 1998;10:265–270. doi: 10.1016/s0952-7915(98)80164-7. [DOI] [PubMed] [Google Scholar]
- 16.Nelson PJ, Kim HT, Manning WC, Goralski TJ, Krensky AM. Genomic organization and transcriptional regulation of the RANTES chemokine gene. J Immunol. 1993;151:2601–2612. [PubMed] [Google Scholar]
- 17.Nelson EL, et al. Tumor-specific, cytotoxic T-lymphocyte response after idiotype vaccination for B-cell, non-Hodgkin’s lymphoma. Blood. 1996;88:580–589. [PubMed] [Google Scholar]
- 18.von Luettichau I, et al. RANTES chemokine expression in diseased and normal human tissues. Cytokine. 1996;8:89–98. doi: 10.1006/cyto.1996.0012. [DOI] [PubMed] [Google Scholar]
- 19.Ortiz, B.D., Nelson, P.J., and Krensky, A.M. 1995. The RANTES gene and the regulation of its expression. In Biology of the chemokine RANTES. A.M. Krensky, editor. Springer-Verlag. New York, New York, USA. 87–99.
- 20.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 21.Song A, Nikolcheva T, Krensky AM. Transcriptional regulation of RANTES expression in T lymphocytes. Immunol Rev. 2000;177:236–245. doi: 10.1034/j.1600-065x.2000.17610.x. [DOI] [PubMed] [Google Scholar]
- 22.Wickens, M., Goodwin, E.B., Kimble, J., Strickland, S., and Hentze, M. 2000. Translational control of developmental decisions. In Translational control of gene expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 295–370.
- 23.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 24.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5’ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 25.Raught, B., Gingras, A.-C., and Sonenberg, N. 2000. Regulation of ribosomal recruitment in eukaryotes. In Translational control of gene expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 245–293.
- 26.Clemens MJ, Bommer UA. Translational control: the cancer connection. Int J Biochem Cell Biol. 1999;31:1–23. doi: 10.1016/s1357-2725(98)00127-7. [DOI] [PubMed] [Google Scholar]
- 27.Imataka H, et al. Cell-specific translational control of transcription factor BTEB expression. The role of an upstream AUG in the 5’-untranslated region. J Biol Chem. 1994;269:20668–20673. [PubMed] [Google Scholar]
- 28.Boytim ML, et al. A human class II MHC-derived peptide antagonizes phosphatidylinositol 3-kinase to block IL-2 signaling. J Clin Invest. 2000;105:1447–1453. doi: 10.1172/JCI8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregorieff A, Pyronnet S, Sonenberg N, Veillette A. Regulation of SOCS-1 expression by translational repression. J Biol Chem. 2000;275:21596–21604. doi: 10.1074/jbc.M910087199. [DOI] [PubMed] [Google Scholar]
- 30.Liu CC, Simonsen CC, Levinson AD. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984;309:82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- 31.Marth JD, Overell RW, Meier KE, Krebs EG, Perlmutter RM. Translational activation of the lck proto-oncogene. Nature. 1988;332:171–173. doi: 10.1038/332171a0. [DOI] [PubMed] [Google Scholar]
- 32.Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 33.Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. 1985;260:7843–7849. [PubMed] [Google Scholar]
- 34.Boal TR, et al. Regulation of eukaryotic translation initiation factor expression during T-cell activation. Biochim Biophys Acta. 1993; 1176:257–264. doi: 10.1016/0167-4889(93)90053-r. [DOI] [PubMed] [Google Scholar]
- 35.Hoover DS, Wingett DG, Zhang J, Reeves R, Magnuson NS. Pim-1 protein expression is regulated by its 5’-untranslated region and translation initiation factor elF-4E. Cell Growth Differ. 1997;8:1371–1380. [PubMed] [Google Scholar]
- 36.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5’ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khaleghpour K, Pyronnet S, Gingras AC, Sonenberg N. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol Cell Biol. 1999;19:4302–4310. doi: 10.1128/mcb.19.6.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raught B, Gingras AC. eIF4E activity is regulated at multiple levels. Int J Biochem Cell Biol. 1999;31:43–57. doi: 10.1016/s1357-2725(98)00131-9. [DOI] [PubMed] [Google Scholar]
- 39.Minich WB, Balasta ML, Goss DJ, Rhoads RE. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA. 1994;91:7668–7672. doi: 10.1073/pnas.91.16.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bu X, Haas DW, Hagedorn CH. Novel phosphorylation sites of eukaryotic initiation factor-4F and evidence that phosphorylation stabilizes interactions of the p25 and p220 subunits. J Biol Chem. 1993;268:4975–4978. [PubMed] [Google Scholar]
- 41.Pyronnet S, et al. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21:5500–5511. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyronnet S. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem Pharmacol. 2000;60:1237–1243. doi: 10.1016/s0006-2952(00)00429-9. [DOI] [PubMed] [Google Scholar]
- 45.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 47.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 49.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]