The impact of influenza on individuals and on society remains underappreciated. Up to 20% of the population may develop influenza in any given year, and epidemics of influenza are responsible — on average — for 20,000 deaths per year in the US. In some epidemics, this number may climb to more than 40,000 deaths, with over 300,000 influenza-related hospitalizations in a single winter season (1). By far the most catastrophic impact of influenza during the past 100 years was the pandemic of 1918, which cost more than 500,000 lives in the United States (2) and lowered the average life expectancy by almost 10 years.
Epidemiology
The epidemiological success of influenza can be largely explained by the fact that the viruses undergo antigenic change. Strains belonging to two different types (A and B) of influenza virus circulate in the population and are associated with most cases of human influenza. Although their genes have less than 30% overall sequence identity, the two types share a common ancestor and both possess genomes consisting of eight RNAs of negative sense polarity. Hemagglutinin (HA) and neuraminidase (NA) decorate the surface of the lipid-containing virus particles, and these proteins are primarily responsible for the antigenic changes observed in influenza viruses.
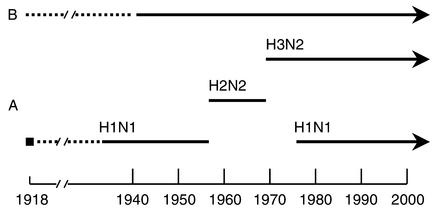
New subtypes emerge, it is believed, when genes from animal influenza viruses are captured by the human virus via reassortment, which occurs when a host has been simultaneously infected by both virus types. Such an antigenic shift appears to present the human immune system with a novel antigenic experience, usually resulting in high morbidity and mortality. As shown in Figure 1, influenza A viruses of the H1N1 (hemagglutinin 1, neuraminidase 1) subtype circulated from 1918 until 1957. They were then replaced by viruses of the H2N2 subtype, which continued to circulate until 1968. Since 1968, H3N2 viruses have been found in the population. Because H1N1 viruses returned in 1977, two influenza A viruses are presently cocirculating. In addition to antigenic shift, slight changes in the surface proteins caused by point mutations (antigenic drift) also allow the viruses to evade the human system, but it may take 3–5 years for a virus of a given subtype to accumulate enough point mutations to cause disease when it reinfects a previously exposed person.
Figure 1.
Epidemiology of human influenza A and B viruses. Three different influenza A virus hemagglutinin subtypes (H1, H2, and H3) and two neuraminidase subtypes (N1 and N2) have been identified in humans over the last century. Although no live 1918 virus (solid square) is available, viral RNAs have been sequenced following RT-PCR from formalin-fixed or frozen tissue samples of 1918 victims (35). Rescue of virus containing reconstructed 1918 genes allows study of virulence characteristics of this long-gone strain (36).
Because type B influenza viruses circulate almost exclusively in humans, these viruses cannot undergo reassortment with animal strains and thus are changed only by antigenic drift. The type C influenza virus represents a distant third in disease-causing potential and is probably of little public health concern.
The present
Inactivated influenza virus vaccines.
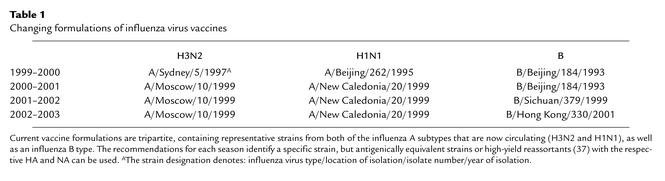
The main option for reducing the impact of influenza is vaccination. In the US, only inactivated influenza virus vaccines are approved at this time. To be effective, current vaccines must contain an H1N1, an H3N2, and a B virus component (see Table 1). Over the past several years, at least one of the components in the formulation had to be changed due to antigenic drift of the strain circulating in the human population. To prepare vaccines, the viral strains are grown in embryonated eggs, and the virus is then purified and made noninfectious by chemical inactivation.
Table 1.
Changing formulations of influenza virus vaccines
The effectiveness of the vaccines that are currently available for influenza depends primarily on the antigenic “match” of the circulating viruses with the strains used for vaccination, as well as on a subject’s age and immune status. If efficacy is defined as preventing illness, studies show that current inactivated vaccines prevent illness in approximately 70–80% of healthy people under the age of 65, although this number may be far lower (30–40%) in the elderly population. On the other hand, if efficacy is defined as preventing death in the vaccinated individual, the current inactivated vaccines may be up to 80% effective, even in such a high-risk segment of the population as the elderly (3). Recommendations at present include vaccination of all people over the age of 50 and of high-risk groups and those who can transmit influenza to high-risk individuals. The potential benefits of vaccination in preventing disease, hospitalization, or even death greatly outweigh the possible risk of side effects or rare adverse reactions associated with the vaccine.
Antiviral agents.
Any discussion of measures against influenza must consider the availability of effective antivirals. Four drugs are approved at present in the US: Amantadine and rimantadine are chemically related inhibitors of the ion-channel M2 protein involved in viral uncoating (4); and zanamivir and oseltamivir are NA inhibitors (5) and prevent the proper release of influenza virus particles from the cytoplasmic membrane. These four drugs are important adjuncts for any medical intervention against influenza (6–8), and three of them can be used in prophylaxis against the virus (zanamivir has not yet been approved). Although these drugs are generally effective, their widespread use is limited by concerns over side effects, patients’ compliance, and the possible emergence of drug-resistant variants. Nevertheless, these agents could be of extraordinary value should a new pandemic strain emerge against which a vaccine has not yet been developed. There is good reason to predict that these drugs would be effective against an emerging virus subtype that threatens to start a new pandemic.
The future of influenza vaccine design
Cold-adapted influenza virus vaccine.
In Russia, live, cold-adapted influenza virus vaccines have been administered to tens of millions of children with protective efficacy and without evidence of deleterious side effects. In addition, there is no sign of the spread of virulent revertants, either within Russia or globally (9). In the US, the development of a cold-adapted influenza virus vaccine for humans has been pursued for more than 20 years, but licensure has not yet been obtained. Maassab and colleagues (10) passaged influenza viruses in chicken kidney cells and in embryonated eggs to adapt them to growth at 25°C. The resulting cold-adapted master strains are temperature-sensitive and are well suited for use as live vaccines because their pathogenicity, in animals as well as humans, is strongly attenuated. The annually updated vaccine formulations could therefore be generated by making “6:2 reassortants” in which the two genes encoding major viral surface antigens (HA and NA) reflect the sequence found in the current strains, whereas the remaining six genes derive from the cold-adapted master strains.
Such live-virus vaccines can be administered by nasal spray — a distinct advantage over the more difficult and costly route of intramuscular injection using needles. Live viruses can also induce local neutralizing immunity and cell-mediated immune responses, which may be associated with a longer-lasting and more cross-protective immunity than is elicited by chemically inactivated virus preparations. Finally, overall protection may be better in certain age groups (children 6 months to 9 years), and there is also evidence of a drastic reduction in secondary bacterial infections causing otitis media, and thus in the need for use of antibiotics (11). The further use of live influenza virus vaccines will shed light on the benefits, potential risks, and economic and logistical consequences of this approach. Continuing surveillance and monitoring will be needed to safeguard against unexpected complications that might arise from the widespread use of cold-adapted vaccines.
Genetically engineered live influenza virus vaccines.
The advent of techniques to engineer site-specific changes in the genomes of negative-strand RNA viruses (12, 13) has made it possible to consider new vaccine approaches. Specifically, it is now possible to tailor-make strains with unique properties that lead to attenuation. For example, exchanging the promoter region of the NA gene of an influenza A virus with that of an influenza B virus gene attenuates that strain in mice (14). Alternatively, engineered changes in the PB2 gene led to a live influenza A virus candidate (15, 16) with interesting biological characteristics.
Live influenza virus vaccine candidates expressing altered NS1 genes.
A further improvement of reverse genetics techniques now allows the rescue of influenza virus vaccine candidates from cells transfected with plasmids (Figure 2) (17, 18). This plasmid-only rescue system makes it possible to engineer deletions in the genomes of influenza viruses for improved stability.
Figure 2.
Plasmid-only rescue of infectious influenza virus. Twelve plasmids are introduced into mammalian cells: four plasmids lead to expression of the viral proteins required for viral RNA replication (PA, PB1, PB2, and NP), and the eight transcription plasmids express precise copies of the eight viral RNA segments (PA, PB1, PB2, HA, NP, NA, M, and NS). The resulting viral RNAs are replicated and transcribed by the reconstituted influenza virus RNA-dependent RNA polymerase. Recombinant infectious influenza virus is generated 48–72 hours after transfection of cells (17).
We have shown that the NS1 protein of influenza viruses has IFN-antagonist activity (19). Following infection by a virus, the host usually mounts an antiviral IFN response. Many, if not all, viruses express an anti-IFN protein or an IFN-antagonist activity. In the case of influenza viruses, changing the NS1 protein can result in an altered virulence characteristic. Specifically, truncated NS1 proteins are responsible for increased attenuation of both influenza A and influenza B viruses in mice (20). In humans, a virus lacking the NS1 gene may be too attenuated to be suitable for vaccines. However, we have found that viruses expressing the N-terminal 99 or 126 amino acids of the NS1 protein possess intermediate IFN-antagonist activity in mice. Such genetically engineered viruses may have optimal phenotypic characteristics for stimulating a robust immune response in humans, while at the same time being safely attenuated because they cannot completely overcome the IFN response by the host. It is thought that humans have redundant IFN genes in order to make the IFN response “fail-safe.” Consequently, humans — even when immune-compromised — may respond effectively to a virus that has a reduced anti-IFN activity, and thus would not become ill. The fact that the IFN response should be higher in humans infected with NS1 mutants than in those infected with wild-type virus may lead to a vigorous (enhanced) humoral immune response. This is based on the finding that type I IFNs can potently enhance the primary antibody response to proteins and can act as adjuvants in mice (21). If this immune-stimulatory effect is also observed in humans inoculated with NS1-mutant viruses, lower amounts of virus may be used for vaccination. If the inoculum size could be reduced by a factor of 100 relative to the dosage needed for the cold-adapted vaccine, protection of large segments of the population in developing as well as developed countries would become feasible.
Use of replication-defective influenza viruses as vaccine candidates.
Another promising approach for the development of live influenza virus vaccines is the construction of virus particles that undergo only a single cycle of replication. For example, infection of cells with a preparation of virus particles lacking the gene for the NEP (NS2) protein will express viral proteins but will not result in the formation of infectious particles (22). Thus, these preparations induce a protective antibody response and also stimulate a strong cell-mediated immune response without allowing the replication of infectious virus. Another way to attenuate the virus and generate a replication-defective strain is by the elimination of the M2 gene. Such a deletion strain grows efficiently in tissue culture substrates but only poorly in mice and thus represents a potential live-virus vaccine candidate (23). Since it is believed that the analogous genes of influenza A and B viruses have identical functions, mutations or deletions found to be effective in influenza A viruses could also be tried in influenza B viruses.
DNA vaccination.
DNA vaccination involves the administration — by injection or by topical application — of plasmid DNA encoding one or more of the influenza virus proteins. To date, reports on DNA vaccination against influenza have been limited to studies in animal models, including nonhuman primates (24), with most of the work being done in mice, chickens, and ferrets (25, 26). Although progress has been impressive, with protection shown against influenza challenge following DNA vaccination, it is likely that vaccination using DNA will be more appropriate for diseases such as AIDS, for which the use of attenuated vaccine strains may be difficult for safety reasons (see Letvin, this Perspective series, ref. 27). On the other hand, further improvements with DNA vaccination techniques may offer a universal approach to generating protective humoral and cell-mediated responses to a variety of foreign antigens, which may result in the development of effective vaccines against a wide variety of pathogens.
Novel adjuvant approaches.
The killed or inactivated influenza virus vaccines presently in use are administered by intramuscular injection. In order to improve their immunogenicity, liposome-like preparations containing cholesterol and viral particles (ISCOMS [immune-stimulating complexes]) have been devised. These preparations are highly effective in mice when delivered by subcutaneous or intranasal administration (28). Whether this approach will be successful in humans awaits further testing.
Another approach involves the use of Escherichia coli heat-labile toxin as adjuvant, complexed with lecithin vesicles to improve the immunogenicity of the trivalent inactivated influenza virus vaccine. Such “virosomal” preparations can be given intranasally and have been found to elicit a protective immune response, including an influenza-specific cell-mediated immunity (29). Unfortunately, this vaccine, which was licensed for use in Switzerland, has been recalled since cases of Bell’s palsy were reported in some vaccinees.
A universal vaccine against influenza?
The possibility of developing a universal influenza vaccine has attracted the attention of many researchers, because the continuing antigenic change of influenza viruses necessitates reformulating the vaccine on a nearly annual basis. Although some components of the virus are more conserved than others, a convincing approach to a universal vaccine based on these conserved parts of the virus has yet to emerge, primarily because the most conserved components of the virus, the minor antigens, are less immunogenic and thus less likely to induce a protective response. Neirynck et al. (30) have attempted to generate a universal vaccine against influenza A virus by fusing the extracellular domain of the conserved M2 protein of an influenza A virus to the hepatitis B virus core protein. They found that intraperitoneal or intranasal administration in mice provided a high degree of protection against viral challenge, but it remains to be seen whether an immune response against a minor surface antigen like the M2 protein will be sufficient to provide full protection against influenza in the human population at large. Vaccines lacking HA and based solely on viral proteins like M2 (30) or NA (31) may not represent an improvement over currently approved vaccines unless the immune responses to these antigens can be strengthened, perhaps by adding appropriate cytokines to the viral protein preparations (32) or by developing acceptable adjuvants for use in humans (33).
As an alternative to targeting only the well-conserved minor antigens, it may be possible to construct a “generic” HA that could be used as an immunogen in vaccines, based on a framework of conserved amino acids. Alternatively, predicting the evolution of human influenza viruses (34) or at least identifying an antigenic trend could be of great benefit in determining the most appropriate strains for use in an influenza virus vaccine. It is even possible that genetic engineering could be used to construct synthetic strains that anticipate evolutionary trends, and that these could be used in future vaccine formulations. Such vaccine candidates may have a better fit with concurrently circulating strains and might not need to be so frequently changed.
Conclusion
Influenza remains a serious disease despite the availability of antivirals and inactivated trivalent vaccines, which are effective for most recipients. Although these modalities of medical intervention are helpful, new approaches are being developed. Specifically, cold-adapted live-virus vaccines, which have been used in millions of people outside the US, are now being considered for approval by the FDA. Other second-generation live-virus vaccines are being designed and tested in animals and are destined to be studied in humans. Major improvements, based on novel adjuvants and recombinant DNA techniques, promise to change the landscape of vaccinology against influenza and many other infectious diseases.
References
- 1.Prevention and control of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP) Morb Mortal Wkly Rep. 2001;50:1–49. [PubMed] [Google Scholar]
- 2.Heilman C, La Montagne J. Influenza: status and prospects for its prevention, therapy and control. Pediatr Clin North Am. 1990;37:669–688. doi: 10.1016/s0031-3955(16)36910-3. [DOI] [PubMed] [Google Scholar]
- 3.Patriarca P, et al. Efficacy of influenza vaccine in nursing homes. Reduction in illness and complications during an influenza A (H3N2) epidemic. JAMA. 1985;253:1136–1139. [PubMed] [Google Scholar]
- 4.Hay A, Wolstenholme A, Skehel J, Smith M. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palese P, Compans RW. Inhibition of influenza virus replication in tissue culture by 2-deoxy- 2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol. 1976;33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 6.Tominack R, Hayden F. Rimantadine hydrochloride and amantadine hydrochloride use in influenza A virus infections. Infect Dis Clin North Am. 1987;1:459–478. [PubMed] [Google Scholar]
- 7.Treanor J, et al. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. JAMA. 2000;283:1016–1024. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 8.Monto A, Moutl A, Sharp S. Effect of zanamivir on duration and resolution of influenza symptoms. Clin Ther. 2000;22:1294–1305. doi: 10.1016/s0149-2918(00)83026-x. [DOI] [PubMed] [Google Scholar]
- 9.Wareing M, Tannock G. Live attenuated vaccines against influenza; an historical review. Vaccine. 2001;19:3320–3330. doi: 10.1016/s0264-410x(01)00045-7. [DOI] [PubMed] [Google Scholar]
- 10.Maassab H, Heilman C, Herlocher M. Cold-adapted influenza viruses for use as live vaccines for man. Adv Biotechnol Processes. 1990;14:203–242. [PubMed] [Google Scholar]
- 11.Nichol K. Live attenuated influenza virus vaccines: new options for the prevention of influenza. Vaccine. 2001;19:4373–4377. doi: 10.1016/s0264-410x(01)00143-8. [DOI] [PubMed] [Google Scholar]
- 12.Enami M, Luytjes W, Krystal M, Palese P. Introduction of site-specific mutations into the genome of influenza virus. Proc Natl Acad Sci USA. 1990;87:3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sastre A. Negative-strand RNA viruses: applications to biotechnology. Trends Biotechnol. 1998;16:230–235. doi: 10.1016/s0167-7799(98)01192-5. [DOI] [PubMed] [Google Scholar]
- 14.Muster T, Subbarao EK, Enami M, Murphy BR, Palese P. An influenza A virus containing influenza B virus 5′ and 3′ noncoding regions on the neuraminidase gene is attenuated in mice. Proc Natl Acad Sci USA. 1991;88:5177–5181. doi: 10.1073/pnas.88.12.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkin N, Chiu P, Coelingh K. Genetically engineered live attenuated influenza A virus vaccine candidates. J Virol. 1997;71:2772–2778. doi: 10.1128/jvi.71.4.2772-2778.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy B, Park E, Gottlieb P, Subbarao K. An influenza A live attenuated reassortant virus possessing three temperature-sensitive mutations in the PB2 polymerase gene rapidly loses temperature sensitivity following replication in hamsters. Vaccine. 1997;15:1372–1378. doi: 10.1016/s0264-410x(97)00031-5. [DOI] [PubMed] [Google Scholar]
- 17.Fodor E, et al. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann G, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 20.Talon J, et al. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc Natl Acad Sci USA. 2000;97:4309–4314. doi: 10.1073/pnas.070525997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBon A, et al. Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–470. doi: 10.1016/s1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T, Watanabe S, Neumann G, Kida H, Kawaoka Y. Immunogenicity and protective efficacy of replication-incompetent influenza virus-like particles. J Virol. 2002;76:767–773. doi: 10.1128/JVI.76.2.767-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Watanabe S, Ito H, Kida H, Kawaoka Y. Influenza A virus can undergo multiple cycles of replication without M2 ion channel activity. J Virol. 2001;75:5656–5662. doi: 10.1128/JVI.75.12.5656-5662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly J, et al. Preclinical efficacy of a prototype DNA vaccine: enhanced protection against antigenic drift in influenza virus. Nat Med. 1995;1:583–587. doi: 10.1038/nm0695-583. [DOI] [PubMed] [Google Scholar]
- 25.Ljungberg K, et al. Effective construction of DNA vaccines against variable influenza genes by homologous recombination. Virology. 2000;268:244–250. doi: 10.1006/viro.2000.0199. [DOI] [PubMed] [Google Scholar]
- 26.Kodihalli S, Kobasa D, Webster R. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine. 2000;18:2592–2599. doi: 10.1016/s0264-410x(99)00485-5. [DOI] [PubMed] [Google Scholar]
- 27.Letvin NL. Strategies for an HIV vaccine. J Clin Invest. 2002;110:15–20. doi: 10.1172/JCI15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambhara S, et al. Heterosubtypic immunity against human influenza A viruses, including recently emerged avian H5 and H9 viruses, induced by FLU-ISCOM vaccine in mice requires both cytotoxic T-lymphocyte and macrophage function. Cell Immunol. 2001;211:143–153. doi: 10.1006/cimm.2001.1835. [DOI] [PubMed] [Google Scholar]
- 29.Glueck R. Review of intranasal influenza vaccine. Adv Drug Deliv Rev. 2001;51:203–211. doi: 10.1016/s0169-409x(01)00174-0. [DOI] [PubMed] [Google Scholar]
- 30.Neirynck S, et al. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med. 1999;5:1157–1163. doi: 10.1038/13484. [DOI] [PubMed] [Google Scholar]
- 31.Kilbourne E, et al. Purified influenza A virus N2 neuraminidase vaccine is immunogenic and non-toxic in humans. Vaccine. 1995;13:1799–1803. doi: 10.1016/0264-410x(95)00127-m. [DOI] [PubMed] [Google Scholar]
- 32.Babai I, et al. A novel liposomal influenza vaccine (INFLUSOME-VAC) containing hemagglutinin-neuraminidase and IL-2 or GM-CSF induces protective anti-neuraminidase antibodies cross-reacting with a wide spectrum of influenza A viral strains. Vaccine. 2001;20:505–515. doi: 10.1016/s0264-410x(01)00326-7. [DOI] [PubMed] [Google Scholar]
- 33.Illum L, Jabbal-Gill I, Hinchcliffe M, Fisher A, Davis S. Chitosan as a novel nasal delivery system for vaccines. Adv Drug Deliv Rev. 2001;51:81–96. doi: 10.1016/s0169-409x(01)00171-5. [DOI] [PubMed] [Google Scholar]
- 34.Bush R, Bender C, Subbarao K, Cox N, Fitch W. Predicting the evolution of human influenza A. Science. 1999;286:1866–1867. doi: 10.1126/science.286.5446.1921. [DOI] [PubMed] [Google Scholar]
- 35.Reid A, Fanning T, Hultin J, Taubenberger J. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basler CF, et al. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci USA. 2001;98:2746–2751. doi: 10.1073/pnas.031575198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilbourne E. Future influenza vaccines and the use of genetic recombinants. Bull World Health Organ. 1969;41:643–645. [PMC free article] [PubMed] [Google Scholar]