Respiratory syncytial virus (RSV) and the human parainfluenza viruses (PIVs) are enveloped nonsegmented negative-strand RNA viruses of the paramyxovirus family. RSV is the leading cause of severe viral respiratory disease in infants and children, and in the United States RSV is responsible for 73,000 to 126,000 annual hospitalizations of infants younger than 1 year of age (1). Two antigenic subgroups of RSV, designated A and B, have been distinguished on the basis of antigenic and sequence dimorphism that is most pronounced for the ectodomain of the G protein, one of the two major protective antigens. Although there is substantial cross-reactivity between the two subgroups, they are sufficiently distinct that an effective RSV vaccine likely will need to represent both.
The human PIV serotypes 1, 2, and 3 also cause severe respiratory tract disease that leads to hospitalization of infants and young children (2). PIV1, PIV2, and PIV3 are distinct serotypes that do not induce significant cross-neutralization or cross-protection. In a long term study of infants and children over a 20-year period, PIV1, PIV2, and PIV3 were identified as etiologic agents responsible for 6.0%, 3.2%, and 11.5%, respectively, of hospitalizations for pediatric respiratory tract disease (1). In total, the three PIVs accounted for only slightly less than the 23% of hospitalizations for pediatric respiratory tract disease caused by RSV. Therefore, there is a need for a vaccine to protect against these three serotypes of PIV and the two antigenic subgroups of RSV. An additional pediatric vaccine will likely be needed for the newly described but related human respiratory tract pathogen human metapneumovirus (3).
The molecular virology of RSV and the PIVs
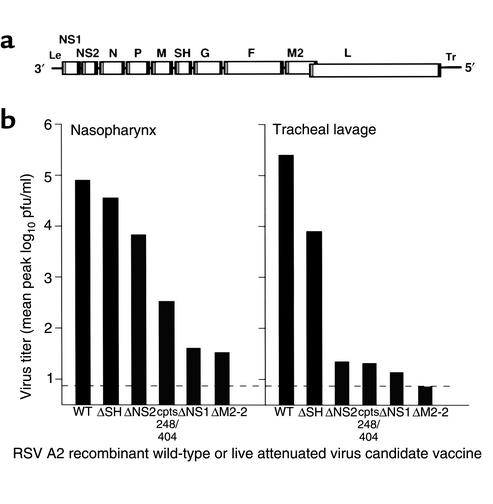
The molecular virology of RSV and the PIVs is relatively well understood and forms a strong foundation on which to build a vaccine development program using reverse genetics. The genome of RSV (Figure 1a) is a single-stranded negative-sense RNA of 15.2 kb that encodes ten subgenomic mRNAs. Transcription of the viral genes is directed by short, conserved gene-start and gene-end cis-acting signals that flank each gene and are separated by intergenic regions of varying length. These mRNAs are translated into 11 known proteins: four nucleocapsid proteins, namely, nucleocapsid N protein, phosphoprotein P, large polymerase subunit L, and transcription elongation factor M2-1; three transmembrane envelope glycoproteins, namely, fusion F protein, attachment G protein, and small hydrophobic SH protein; two nonstructural proteins, NS1 and NS2; a matrix M protein; and an RNA regulatory factor, M2-2. The M2-1 and M2-2 proteins are expressed from separate open reading frames (ORFs) in the M2 mRNA. The G and F glycoproteins are the major protective antigens and induce RSV-neutralizing antibodies and resistance to infection.
Figure 1.
Map of the RSV genome and spectrum of attenuation exhibited by gene-deletion viruses. (a) Map of the RSV genome, a single negative-sense RNA of 15.2 kb. Each viral gene is represented by a box, and the gene-start and gene-end transcription signals that flank each gene are indicated by gray and black bars, respectively. The 3′ and 5′ ends of the genome consist of the extragenic leader (Le) and trailer (Tr) regions, respectively. The genes are separated by short intergenic regions except in the case of the M2 and L genes, which overlap but are nonetheless transcribed into separate mRNAs, as are all of the other genes (1). The diagram is only approximately to scale. (b) Mean peak virus titers in the upper (nasopharynx) and lower (tracheal lavage) respiratory tract of chimpanzees that had been inoculated by the intranasal and intratracheal routes simultaneously with the indicated gene-deletion virus or, as controls, with wild-type RSV or with the rRSVcpts248/404 virus. This last virus serves as a reference point for a virus with mild residual virulence for seronegative infants (4).
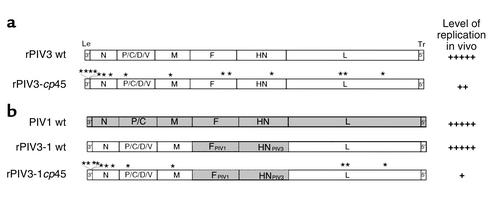
The genetic maps of the PIVs (shown for PIV3 and PIV1 in Figure 2, a and b, respectively) have some similarity to that of RSV, encoding N, P, M, F, and L proteins that are distantly related functional counterparts to the same proteins in RSV. On the other hand, the human PIVs lack NS1, NS2, SH, and M2 proteins, and the PIV P gene gives rise to additional, accessory proteins (designated C, C′, Y1, Y2, V, D and X) that vary in occurrence among the different PIVs (1, 2). The two protective antigens of the PIVs are the HN (hemagglutinin-neuraminidase) attachment protein and the F protein.
Figure 2.
Structure of the PIV genome in wild-type, attenuated, and chimeric viruses. (a) rPIV3-cp45 is a version of wild-type recombinant PIV3 (rPIV3 wt) that contains 15 point mutations (asterisks) that confer the ts and attenuation phenotypes of the biologically derived PIV3-cp45 vaccine candidate virus. (b) The HN and F ORFs of PIV1 wt virus were substituted for their counterparts in PIV3 wt, creating the chimeric rPIV3-1 virus. This was then attenuated by the further introduction of 12 of the 15 mutations of PIV3-cp45, resulting in rPIV3-1cp45, a live-attenuated vaccine candidate for PIV1.
We have focused our efforts on developing live-attenuated intranasally administered pediatric RSV and PIV vaccines. This strategy, which mimics natural infection, has the major advantage of inducing a balanced immune response that includes serum and mucosal virus-neutralizing antibodies as well as protective and regulatory components of cellular and innate immunity. In addition, mucosally administered vaccines partially escape the immunosuppressive effect of maternally derived RSV- and PIV-specific serum antibodies present in the young infant. Another important consideration is that vaccines based on inactivated virus or purified proteins have been associated with disease enhancement upon subsequent RSV infection, whereas live-attenuated RSV vaccines or natural infection induces protective immunity without disease enhancement (4). The attenuated viruses are developed using reverse genetics, whereby infectious virus is produced entirely from cDNA, and predetermined changes in the nucleotide sequence can thus be introduced into infectious virus via the cDNA intermediate. The work described here employed reverse genetics systems based on the RSV subgroup A strain A2 (5), PIV3 virus strain JS (6), and bovine PIV3 (BPIV3) (7). More recently, human PIV1 (8) and PIV2 (9) have also been recovered from cDNA and are now available for attenuation to yield new candidate vaccine strains.
The genetic basis of attenuation in biologically derived, live-attenuated virus vaccine candidates
It is very advantageous to know the specific mutations within a live-attenuated vaccine that confer the attenuation phenotype, as well as to know their relative contributions to that phenotype. Knowledge of the genetic basis of attenuation allows one to monitor the stability of the relevant mutations during all phases of vaccine manufacture and usage in humans. Knowledge of the number of attenuating mutations and their nature (for instance, whether they are point mutations or deletion mutations) also helps in interpreting data on the stability of the attenuation phenotype following growth in vitro and in vivo. Furthermore, once attenuating mutations have been identified, they can be introduced into cDNA-derived viruses to generate improved candidate vaccine viruses.
Reverse genetics provides a powerful means to define the genetic basis of attenuation of existing biologically derived RSV and PIV vaccine candidates that had been attenuated by conventional methods, such as cold-passage or chemical mutagenesis (10, 11). As a first step, the complete consensus nucleotide sequence must be determined for each virus in question and compared with that of its wild-type parent. The identified mutations or sets of mutations must then be inserted into wild-type recombinant virus via reverse genetics to determine whether they confer an attenuation phenotype (10, 11). For example, a cold-passaged (cp) PIV3 (PIV3-cp45) that is a promising vaccine candidate has been found to contain 15 potentially significant nucleotide substitutions (Figure 2). The reintroduction of these 15 mutations into wild-type recombinant virus by reverse genetics showed that only six mutations, including both temperature-sensitive (ts) and non-ts types, contribute substantially to the attenuation phenotype. The presence of multiple ts and non-ts attenuating mutations provides a likely explanation for the high level of phenotypic stability previously observed for PIV3-cp45 following its replication in vivo (12), since experience with live-attenuated influenza virus vaccines indicates that ts mutations are much less likely to revert in the presence of non-ts attenuating mutations (13).
In the case of RSV, the most promising live-attenuated viruses are based on a virus called cpRSV, which was generated by extensive passage of RSV at suboptimal temperatures. cpRSV is only moderately attenuated in chimpanzees and humans and has been found to contain five non-ts amino acid substitutions in three proteins (N, F, and L) that, as a set, confer the attenuation phenotype. cpRSV was subjected to chemical mutagenesis, and two promising ts mutants (RSVcpts248 and RSVcpts530) were identified and subjected to a second round of mutagenesis, yielding several mutants (including RSVcpts248/404 and RSVcpts530/1030) with a lower shut-off temperature. Interestingly, each chemical mutagenesis step introduced a single attenuating ts mutation (10). All but one of these attenuating ts mutations involved single nucleotide substitutions, resulting in single amino acid substitutions in the L polymerase protein. The exception was a strongly attenuating ts mutation in RSVcpts248/404 involving a single nucleotide substitution in the noncoding, cis-acting gene-start signal of the M2 gene. None of these biologically derived RSV vaccine candidates proved to be sufficiently attenuated for use as a pediatric vaccine, but they represented a starting point for further attenuation by reverse genetics. Furthermore, in a number of cases, the amino acid substitution that conferred the attenuation phenotype could be designed to involve two nucleotide changes relative to the wild-type assignment, rather than the single nucleotide substitution found in the original biological mutants, which should greatly reduce the frequency of reversion and thus increase phenotypic stability.
Reverse genetics has also been used to determine the genetic basis of attenuation of a Jennerian PIV3 vaccine candidate, that is, a BPIV3 wild-type virus that is antigenically related to human PIV3 and is highly attenuated and phenotypically stable in humans due to a natural host range restriction (14, 15). Bailly et al. (16) constructed a chimeric recombinant human-bovine PIV3 virus (rPIV3-NB) in which the nucleoprotein (N) ORF of the human PIV3 was replaced by its counterpart from BPIV3. The replication of rPIV3-NB in rhesus monkeys was restricted to an extent similar to that of its BPIV3 parent, showing that the BPIV3 N protein helps determine the host range restriction (that is, the attenuation phenotype) of BPIV3 in primates (16). There are 79 differences out of a total of 515 amino acids between the N proteins of human PIV3 and BPIV3, many of which probably contribute to the host range attenuation phenotype of rPIV3-NB. Therefore, this strain should be stable even following extensive replication in vivo. rPIV3-NB, which combined the antigenic determinants of PIV3 with the host range restriction and attenuation phenotype of BPIV3, induced a level of resistance to PIV3 challenge in monkeys that was indistinguishable from that conferred by immunization with human PIV3. Thus, the process of analyzing the genetic basis of attenuation of BPIV3 for primates led to the development of the promising vaccine candidate rPIV3-NB and identified chimerization of human and bovine PIV3 viruses as a novel method to generate an attenuated PIV3 vaccine. In summary, reverse genetics has allowed us to explore the genetic basis of attenuation of at least ten biologically derived RSV and PIV3 vaccine candidates and has provided insight into the genetic basis of stability of the attenuation phenotype following replication of these vaccine viruses in vivo.
Developing a menu of attenuating viral mutations
Practical considerations dictate that a live-attenuated vaccine candidate must be not only satisfactorily attenuated but also capable of efficient replication in cultured cells suitable for large scale production and use in humans. This is particularly important because attenuated viruses often exhibit decreased infectivity for the host, necessitating the administration of a relatively high dose of the vaccine (17). Attenuating mutations with desired properties can be assembled into a large menu of mutations that can be combined to derive a vaccine strain that possesses the required balance between attenuation and immunogenicity.
In our studies, three major sources of attenuating mutations were used to compile menus for RSV and PIV3. The first source was existing, biologically derived attenuated vaccine candidate viruses, including multiply passaged viruses (such as PIV3-cp45 and cpRSV), mutants derived by chemical mutagenesis (such as the cpts RSV mutants), and host range–restricted viruses (such as BPIV3). These viruses replicate efficiently in cell culture and have been confirmed to be safe and attenuated in preclinical and clinical studies; hence they represent a valuable resource. In addition, the attenuating host range sequences in the BPIV3 N protein exhibit a high level of phenotypic stability. Attenuating ts mutations are desirable because they can be grown efficiently in vitro at permissive temperature and because different mutations can specify different levels of attenuation (18). For respiratory viruses such as RSV and the PIVs, attenuating ts mutations also are desirable because they specify restricted replication preferentially in the warmer lower respiratory tract, the site of the most severe disease. Six RSV and three PIV attenuating ts mutations have been identified by sequence analysis of biologically derived vaccine candidates and by reverse genetics (10, 11).
The second source of attenuating mutations is the generation of novel mutations not found among existing virus mutants or vaccine candidates. RSV and the PIVs encode several nonessential proteins whose major function is to facilitate virus replication in vivo, for instance, by antagonizing host defense factors. Deleting or silencing such accessory genes or ORFs is attractive because the resulting virus frequently retains replicative efficiency in cell culture. If these viral accessory factors enhance pathogenesis or downregulate host immune responses, their ablation might improve the safety and efficacy of the resulting vaccine (10). Such mutations can be made by deleting a complete transcriptional unit encoding an mRNA and protein, such as has been done for the NS1, NS2, and SH genes of RSV, or by deleting or silencing a single ORF contained within a complex mRNA encoding more than one protein, as has been done to eliminate expression of the M2-2 protein of RSV and the C, D, and V proteins of PIV3 (19, 20). Deletion of a complete gene is a desirable mutation because it would be expected to be more genetically and phenotypically stable than point mutations.
The ΔSH, ΔNS1, ΔNS2, and ΔM2-2 deletion mutants of RSV exhibit a spectrum of attenuation for the respiratory tract of chimpanzees (Figure 1b) (21, 22) and other experimental animals (23–25). In particular, the deletion mutant RSVΔNS1 is slightly more attenuated in the chimpanzee than is RSVcpts248/404, a biologically derived vaccine candidate that retains some mild residual virulence for the upper respiratory tract of the 2-month-old human infant (4, 22). Despite its highly restricted replication in chimpanzees, RSVΔNS1 induces a high level of resistance to RSV challenge and represents a very promising vaccine candidate.
Deletion mutants of the D and V ORFs of PIV3 appear to be useful additions to the menu of attenuating mutations, whereas deletion of the PIV3 C ORF produced a virus that appears to be overattenuated (19, 20). Reverse genetics also could be used to develop new point mutations not previously observed in biologically derived strains. For example, mutation could be introduced via the cDNA intermediate into individual proteins either randomly or by replacing single or paired charged residues with alanine residues. In this case, each mutation would have to be characterized with regard to viability, replication efficiency, and ts and attenuation phenotype of the recovered virus. Other novel attenuating mutations that can be generated using reverse genetics include large nucleotide insertions or changes in the gene order. However, these mutations typically reduce the efficiency of virus replication in vitro and thus would complicate the manufacture of vaccine virus (26, 27).
A third source of attenuating mutations involves existing attenuated mutants of other paramyxoviruses. Using existing sequence relatedness among paramyxoviruses as a guide, mutations that involve conserved residues or domains can be “transferred” from one paramyxovirus to another. For example, a point mutation (F521L) in the L polymerase of RSVcpts530 (a biologically derived live-attenuated RSV vaccine candidate mentioned above) has been transferred using reverse genetics to the homologous position (residue 456) of the distantly related PIV3-cp45 (10). The acquisition of the F456L mutation by PIV3-cp45 resulted in a small, incremental increase in its level of attenuation in rhesus monkeys and chimpanzees (28). This virus is now available for evaluation should the ongoing clinical evaluation of PIV3-cp45 indicate that a small increase in attenuation would be desirable. In a second example, a point mutation (F170S) in the C protein of a multiply passaged attenuated mutant of Sendai virus (29) (a PIV1 of murine origin) has been introduced into the homologous position (residue 164) of the C protein of human PIV3 (19). The resulting recombinant PIV3, designated rF164S, replicates very efficiently in vitro but is attenuated in monkeys, especially for the upper respiratory tract. On this basis, the F164S mutation has been added to the menu of attenuating PIV3 mutations. The ability to import attenuating mutations from one virus to another greatly expands the menu of attenuating mutations that can be used to produce candidate vaccines.
An important purpose of compiling a list of attenuating mutations is that mutations from this list can be added to insufficiently attenuated viruses to augment both their level of attenuation and their phenotypic stability in vivo. This principle is illustrated by RSV vaccine candidates presently in clinical trials (30). The RSV 1030 attenuating ts mutation (Y1321N) in the L protein of RSVcpts530/1030 has been introduced into the insufficiently attenuated RSVcpts248/404 vaccine candidate (4) to yield rRSVA2cpts248/404/1030, which is more ts and more attenuated than its RSVcpts248/404 parent and represents a promising new RSV vaccine candidate (Figure 3). A further modification, namely deletion of the SH gene, created rA2cpts248/404/1030ΔSH, which is now in clinical trials in infants and children and appears to be more attenuated than RSVcpts248/404 (31). Thus, rRSVA2cpts248/404/1030ΔSH has mutations gathered using reverse genetics from five different RSV mutants: cpRSV, RSVcpts248, RSVcpts248/404, RSVcpts530/1030, and RSVΔSH. This illustrates the power of combining mutations from a menu of attenuating mutations into a single mutant in response to information gathered from ongoing clinical trials, and it shows that knowledge of the individual properties of the mutations can guide the choice of which mutations to include in a new vaccine candidate.
Figure 3.
Use of reverse genetics to introduce new combinations of mutations into rRSV A2 of subgroup A and to expedite the development of a live-attenuated vaccine virus specific to RSV subgroup B. (a) Recombinant wild-type RSV (rRSV A2 wt) was used as a backbone for the introduction of the five point mutations of cpRSV (cp), two attenuating mutations from RSVcpts248/404 (248-L and 404-M2), and one attenuating mutation from RSVcpts530/1030 (1030) to create the new vaccine candidate rRSVA2cpts248/404/1030. (b) The genes encoding the F and G antigenic determinants of the subgroup B RSV B1 wt virus were substituted into the backbone of rRSV A2 wt to create the chimeric rRSV AB wt virus, which was then attenuated by the introduction of three mutations of cpRSV, two mutations from RSVcpts248/404, and one mutation from RSVcpts530/1030. The level of viral replication in the respiratory tract of rodents or chimpanzees is indicated on the right.
Several factors need to be considered in compiling a menu of useful attenuating mutations for respiratory viruses. First, it is preferable to include mutations that yield various levels of attenuation, since it might be necessary to further restrict the level of replication of an incompletely attenuated candidate over a 10- to 1000-fold range. Second, the menu should include non-ts mutations, which can enhance the genetic stability of ts mutations present in a vaccine virus. Third, mutations or sequences that specify a high level of genetic and phenotypic stability should be included, such as deletion mutations or host range determinants (e.g., BPIV3 N ORF). Fourth, the menu should be reasonably large, since not all combinations of attenuating mutations are viable (30) and since the level of attenuation specified by a combination of mutations is not always the sum of the attenuation specified by the individual mutations. For example, the attenuating effect of an individual mutation can be masked when placed in the context of certain other attenuating mutations. Including such mutations in a single strain can be desirable, since it allows a greater number of attenuating mutations to be incorporated into a vaccine candidate, which would restrain phenotypic change should reversion occur at one or more sites. Fifth, as mentioned above, it is desirable, when possible, to generate amino acid point mutations using codons that differ from the wild-type assignment by more than one nucleotide, in order to reduce the frequency of reversion.
Trial and error is needed to identify compatible mutations and to confirm the level of attenuation of any particular combination. It might seem that the potential number of vaccine candidates would be unworkably large, but in practice the design of new viruses is guided and simplified by comparison to previous vaccine candidates that have undergone preclinical and clinical evaluation. This is illustrated in Figure 1a, where RSV deletion mutants are compared with the existing, well-characterized RSVcpts248/404 virus. Using these principles, a menu of attenuating mutations can be assembled and employed in the design of a vaccine virus that exhibits the proper balance between attenuation and immunogenicity.
The design and use of antigenic chimeric viruses to create new vaccines
As indicated above, human RSV exists as two antigenic subgroups A and B, both of which should be represented in a vaccine. To expedite the development of a vaccine for subgroup B — for which promising vaccine candidates, a reverse genetics system, and a menu of attenuating mutations were lacking — we have modified the existing recombinant RSV A2 (subgroup A) so that its G and F glycoprotein genes were replaced with their counterparts from the RSV subgroup B strain B1 (Figure 3b) (32). This replacement of the protective antigens was initially done for the wild-type recombinant RSV A2 virus to create an RSV AB wild-type chimeric virus (rRSVAB wt), and then for a series of RSV A2 derivatives that contain various combinations of RSV A2–derived attenuating mutations located in genes other than F and G, one of which is depicted in Figure 3b (32). The rRSVAB wt chimeric virus replicates in cell culture with an efficiency comparable to that of the RSV A2 and B1 wild-type parents. Likewise, the rRSVABcpts248/404/1030 chimeric recombinant containing ts mutations in the RSV A2 background exhibits a level of temperature sensitivity in vitro similar to that of the RSV A2 recombinant bearing the same mutations.
In chimpanzees, the replication of the rRSVAB wt chimera is intermediate between that of the RSV A2 and B1 wild-type viruses (Figure 3b) and is accompanied by moderate rhinorrhea. rRSVABcpts248/404/1030 is highly attenuated in both the upper and lower respiratory tract of chimpanzees and is immunogenic and protective against challenge with rRSVAB wt virus. Therefore, rRSVABcpts248/404/1030 chimeric virus represents a promising vaccine candidate for RSV subgroup B and should be evaluated in humans. Furthermore, these results suggest that additional attenuating mutations derived from RSV A2 can be inserted into the RSV A2 background of rRSVAB as necessary to modify the attenuation phenotype in a reasonably predictable manner to achieve an optimal balance between attenuation and immunogenicity in a virus bearing the major protective antigens of RSV subgroup B.
We took a similar approach to designing a live-attenuated vaccine for PIV1, for which promising vaccine candidates did not exist and which did not have a reverse genetics system available until very recently (8). Taking advantage of the existing PIV3 reverse genetics system, we replaced the HN and F ORFs of PIV3 with their PIV1 counterparts, creating a chimeric virus, rPIV3-1 wt, that replicated as efficiently as a wild-type virus in vitro and in rodents and monkeys (Figure 2b). In the same way, we introduced the PIV1 HN and F ORFs into a recombinant version of the well-characterized PIV3-cp45 vaccine candidate to create a live-attenuated chimeric PIV1 vaccine candidate, termed rPIV3-1cp45, that combines the attenuated backbone of PIV3-cp45 with the neutralization antigens of PIV1 (Figure 2b) (33). Three of the 15 mutations of PIV3-cp45 lie within the HN and F genes and thus were lost by the swap involving these genes. Nonetheless, the chimeric rPIV3-1cp45 virus was found to be somewhat more restricted in replication in hamsters than was rPIV3-cp45, indicating that the introduction of the heterologous PIV1 HN and F proteins into PIV3 had an attenuating effect that was additive to that conferred by the 12 PIV3-cp45 mutations. rPIV3-1cp45 is immunogenic and protective against challenge with wild-type PIV1 in hamsters. This virus shows sufficient promise that it should be evaluated further as a candidate live-attenuated vaccine strain for the prevention of severe lower respiratory tract PIV1 disease in infants and young children.
In another permutation of this strategy, we used reverse genetics to replace the HN and F glycoprotein genes of BPIV3, the Jennerian vaccine candidate for human PIV3 described above, with their human PIV3 counterparts, creating an attenuated chimeric virus called rB/HPIV3. Because this chimeric virus bears the HN and F protective antigens of human PIV3, it induces a higher level of serum antibody to HPIV3 than does the BPIV3 vaccine candidate (7, 34). rB/HPIV3, like the rPIV3-NB chimera described above, combines the antigenic determinants of human PIV3 with the host range restriction and attenuation phenotype of BPIV3, but in the case of rB/HPIV3 all of the genes other than the HN and F are of bovine PIV3 origin. It is very likely that more than one BPIV3 gene is involved in the host range restriction of replication of BPIV3 for primates, in which case rB/HPIV3 would be even more phenotypically stable than rPIV3-NB.
Although antigenic chimeric viruses provide a strategy for the rapid generation of new vaccine candidates, there might be a price to pay for their use. The epidemiology of RSV and PIV1, PIV2, and PIV3 suggests that it would be optimal to administer vaccines in a sequential schedule, and the use of antigenic chimeric viruses as vaccines must be compatible with this sequential schedule. RSV and PIV3 cause significant illness within the first 4 months of life, whereas most of the illness caused by PIV1 and PIV2 occurs after 6 months of age. A desirable immunization sequence employing live-attenuated RSV and PIV vaccines would be administration of RSV and PIV3 vaccines together as a combined vaccine given two or more times, with the first dose administered at or before 1 month of age, followed by a bivalent PIV1 and PIV2 vaccine at 4 and 6 months of age. Should cross-protection between a PIV3 vaccine and a recombinant PIV3-1 vaccine virus occur, it would complicate this type of sequential immunization scheme. We have found that prior infection with PIV3 moderately decreases the replication, immunogenicity, and efficacy of an rPIV3-1 vaccine candidate against PIV1 (35). This interference likely was mediated by resistance conferred by T cell immunity to the shared internal PIV3 proteins of the two viruses. In hamsters, this effect wanes within a period of 4 months. The magnitude and duration of cell-mediated immunity to PIV3 in primates and, in particular, young human vaccinees is unknown. Thus, a sequential immunization protocol using an rPIV3-1 antigenic chimeric vaccine in PIV3-immune persons might pose potential problems for the infectivity and immunogenicity of these vaccines. Importantly, this limitation does not apply to the RSV subgroup A vaccine virus and its A/B antigenic chimeric counterpart, since these two viruses would be given simultaneously. A successful vaccine against PIV1 and PIV2 might require the development of attenuated human PIV1 and PIV2 vaccine viruses using the recently described reverse genetics systems and employing the general principles outlined here.
References
- 1.Collins, P.L., Chanock, R.M., and Murphy, B.R. 2001. Respiratory syncytial virus. In Fields virology. 4th edition. Volume 1. D.M. Knipe et al., editors. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 1443–1486.
- 2.Chanock, R.M., Murphy, B.R., and Collins, P.L. 2001. Parainfluenza viruses. In Fields virology. 4th edition. Volume 1. D.M. Knipe et al., editors. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 1341–1379.
- 3.van den Hoogen BG, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright PF, et al. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis. 2000;182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- 5.Collins PL, et al. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durbin AP, et al. Recovery of infectious human parainfluenza virus type 3 from cDNA. Virology. 1997;235:323–332. doi: 10.1006/viro.1997.8697. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt AC, et al. Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates. J Virol. 2000;74:8922–8929. doi: 10.1128/jvi.74.19.8922-8929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman JT, et al. Sequence analysis of the Washington/1964 strain of human parainfluenza virus type 1 (HPIV1) and recovery and characterization of wild type recombinant HPIV1 produced by reverse genetics. Virus Genes. 2002;24:77–92. doi: 10.1023/a:1014042221888. [DOI] [PubMed] [Google Scholar]
- 9.Kawano M, et al. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology. 2001;284:99–112. doi: 10.1006/viro.2001.0864. [DOI] [PubMed] [Google Scholar]
- 10.Collins PL, et al. Rational design of live-attenuated recombinant vaccine virus for human respiratory syncytial virus by reverse genetics. Adv Virus Res. 1999;54:423–451. doi: 10.1016/s0065-3527(08)60374-7. [DOI] [PubMed] [Google Scholar]
- 11.Skiadopoulos MH, et al. Identification of mutations contributing to the temperature-sensitive, cold-adapted, and attenuation phenotypes of the live-attenuated cold-passage 45 (cp45) human parainfluenza virus 3 candidate vaccine. J Virol. 1999;73:1374–1381. doi: 10.1128/jvi.73.2.1374-1381.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karron RA, et al. A live human parainfluenza type 3 virus vaccine is attenuated and immunogenic in healthy infants and children. J Infect Dis. 1995;172:1445–1450. doi: 10.1093/infdis/172.6.1445. [DOI] [PubMed] [Google Scholar]
- 13.Murphy BR, Park EJ, Gottlieb P, Subbarao K. An influenza A live attenuated reassortant virus possessing three temperature-sensitive mutations in the PB2 polymerase gene rapidly loses temperature sensitivity following replication in hamsters. Vaccine. 1997;15:1372–1378. doi: 10.1016/s0264-410x(97)00031-5. [DOI] [PubMed] [Google Scholar]
- 14.Karron RA, et al. A live attenuated bovine parainfluenza virus type 3 vaccine is safe, infectious, immunogenic, and phenotypically stable in infants and children. J Infect Dis. 1995;171:1107–1114. doi: 10.1093/infdis/171.5.1107. [DOI] [PubMed] [Google Scholar]
- 15.Karron RA, et al. Evaluation of a live attenuated bovine parainfluenza type 3 vaccine in two- to six-month-old infants. Pediatr Infect Dis J. 1996;15:650–654. doi: 10.1097/00006454-199608000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Bailly JE, et al. A recombinant human parainfluenza virus type 3 (PIV3) in which the nucleocapsid N protein has been replaced by that of bovine PIV3 is attenuated in primates. J Virol. 2000;74:3188–3195. doi: 10.1128/jvi.74.7.3188-3195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy BR. Use of live attenuated cold-adapted influenza A reassortant virus vaccines in infants, children, young adults and elderly adults. Infectious Diseases in Clinical Practice. 1993;2:174–181. [Google Scholar]
- 18.Richman DD, Murphy BR. The association of the temperature-sensitive phenotype with viral attenuation in animals and humans: implications for the development and use of live virus vaccines. Rev Infect Dis. 1979;1:413–433. doi: 10.1093/clinids/1.3.413. [DOI] [PubMed] [Google Scholar]
- 19.Durbin AP, McAuliffe JM, Collins PL, Murphy BR. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology. 1999;261:319–330. doi: 10.1006/viro.1999.9878. [DOI] [PubMed] [Google Scholar]
- 20.Nagai Y, Kato A. Paramyxovirus reverse genetics is coming of age. Microbiol Immunol. 1999;43:613–624. doi: 10.1111/j.1348-0421.1999.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 21.Whitehead SS, et al. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng MN, et al. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J Virol. 2000;74:9317–9321. doi: 10.1128/jvi.74.19.9317-9321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin H, et al. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology. 1998;251:206–214. doi: 10.1006/viro.1998.9414. [DOI] [PubMed] [Google Scholar]
- 24.Jin H, Cheng X, Zhou HZ, Li S, Seddiqui A. Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J Virol. 2000;74:74–82. doi: 10.1128/jvi.74.1.74-82.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin H, et al. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology. 2000;273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- 26.Skiadopoulos MH, Surman SR, Durbin AP, Collins PL, Murphy BR. Long nucleotide insertions between the HN and L protein coding regions of human parainfluenza virus type 3 yield viruses with temperature-sensitive and attenuation phenotypes. Virology. 2000;272:225–234. doi: 10.1006/viro.2000.0372. [DOI] [PubMed] [Google Scholar]
- 27.Wertz GW, Perepelitsa VP, Ball LA. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc Natl Acad Sci USA. 1998;95:3501–3506. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skiadopoulos MH, et al. Attenuation of the recombinant human parainfluenza virus type 3 cp45 candidate vaccine virus is augmented by importation of the respiratory syncytial virus cpts530 L polymerase mutation. Virology. 1999;260:125–135. doi: 10.1006/viro.1999.9802. [DOI] [PubMed] [Google Scholar]
- 29.Itoh M, Isegawa Y, Hotta H, Homma M. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC- MK2 cells. J Gen Virol. 1997;78:3207–3215. doi: 10.1099/0022-1317-78-12-3207. [DOI] [PubMed] [Google Scholar]
- 30.Whitehead SS, et al. Addition of a missense mutation present in the L gene of respiratory syncytial virus (RSV) cpts530/1030 to RSV vaccine candidate cpts248/404 increases its attenuation and temperature sensitivity. J Virol. 1999;73:871–877. doi: 10.1128/jvi.73.2.871-877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karron, R.A., et al. 2001. Evaluation of live rRSV A2 vaccines in infants and children. In Respiratory syncytial viruses after 45 years. Instituto de Salud Carlos, Ministerio de Sanidad y Consumo. Segovia, Spain. p. 151. (Abstr.)
- 32.Whitehead SS, et al. Replacement of the F and G proteins of respiratory syncytial virus (RSV) subgroup A with those of subgroup B generates chimeric live attenuated RSV subgroup B vaccine candidates. J Virol. 1999;73:9773–9780. doi: 10.1128/jvi.73.12.9773-9780.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skiadopoulos MH, Tao T, Surman SR, Collins PL, Murphy BR. Generation of a parainfluenza virus type 1 vaccine candidate by replacing the HN and F glycoproteins of the live-attenuated PIV3 cp45 vaccine virus with their PIV1 counterparts. Vaccine. 1999;18:503–510. doi: 10.1016/s0264-410x(99)00227-3. [DOI] [PubMed] [Google Scholar]
- 34.Haller AA, Miller T, Mitiku M, Coelingh K. Expression of the surface glycoproteins of human parainfluenza virus type 3 by bovine parainfluenza virus type 3, a novel attenuated virus vaccine vector. J Virol. 2000;74:11626–11635. doi: 10.1128/jvi.74.24.11626-11635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao T, et al. A live attenuated recombinant chimeric parainfluenza virus (PIV) candidate vaccine containing the hemagglutinin-neuraminidase and fusion glycoproteins of PIV1 and the remaining proteins from PIV3 induces resistance to PIV1 even in animals immune to PIV3. Vaccine. 2000;18:1359–1366. doi: 10.1016/s0264-410x(99)00406-5. [DOI] [PubMed] [Google Scholar]