Abstract
The Polycomb-group (PcG) gene Rae28 is a mammalian homologue of the Drosophila gene polyhomeotic. PcG genes are known to maintain transcription states, once initiated, probably by regulating chromatin structure. Since homozygous Rae28-deficient (Rae28–/–) mice displayed cardiac anomalies similar to congenital heart diseases in humans, we examined the role of Rae28 in cardiac morphogenesis at the molecular level. In Rae28–/– embryos, expression of the cardiac selector gene Nkx2.5/Csx (Nkx2.5) was initiated properly but was not sufficiently sustained later in development. This impaired expression of Nkx2.5 in the maintenance phase proved to have a crucial effect on cardiac morphogenesis, as demonstrated by the results of a genetic complementation experiment in which the cardiac anomalies were suppressed by overexpression of human NKX2.5/CSX1 in Rae28–/– embryos. Ubiquitous expression of exogenous Rae28 likewise restored the impaired Nkx2.5 expression in Rae28–/– embryos, further supporting the notion that Rae28 sustains Nkx2.5 expression in cardiomyocytes. Thus, our data show that a mammalian PcG gene can play a key role in organogenesis by helping to maintain the expression of a selector gene.
Introduction
During development, differential gene expression governs generation of the cellular specificity and diversity required for the establishment of patterning and organogenesis. In Drosophila, the initial pattern of homeotic gene expression, which governs cellular specificity, is established by maternal effect and segmentation gene products. Since expression of segmentation gene products is transient, the expression patterns of homeotic genes must be relayed by Polycomb-group (PcG) and trithorax-group genes, which are known to maintain repressed and active chromatin structures, respectively (1).
Until recently, less has been known about the maintenance phase of the developmental regulation of transcription in mammals. Isolation of mammalian homologues of Drosophila PcG genes yielded insights into its mechanisms and biological functions. There exist at least two classes of PcG complexes, namely PcG complexes 1 and 2 (2). PcG complex 1 may be a homologue to Drosophila Polycomb repressive complex 1, which may competitively antagonize the ATP-dependent nucleosome remodeling ability of SWI/SNF–family complexes (3) and may also directly interact with the general transcription machinery (4, 5). As we previously reported, the Rae28 gene (Rae28) is a mammalian homologue of the Drosophila polyhomeotic gene, a member of PcG genes, and encodes a constituent of PcG complex 1 (2).
Targeted replacement has been used to create mutant mice lacking each member of PcG complex 1 (6–11). All these mutations were found to cause abnormal anteroposterior patterning, similar to PcG mutations in flies, which resulted from altered Hox expression. We generated Rae28-deficient (Rae28–/–) mice and clearly demonstrated that mammalian PcG genes have a highly conserved role in the maintenance of Hox expression but not in its initiation; i.e., Hoxb3 expression was not affected in the initiation phase but its expression domain became progressively enhanced along the anteroposterior axis in the later maintenance phase in the rhombomeres of Rae28–/– embryos (10, 12).
Interestingly, these PcG gene–deficient mice further revealed that mammalian PcG genes have additional roles in hematopoiesis, sex determination, and cerebellar and heart development (6–10). Since Rae28–/– embryos revealed cardiac anomalies compatible with tetralogy of Fallot (TOF) and double outlet of the right ventricle (DORV) (10), Rae28–/– mice provided a clue to clarifying the role of PcG genes in heart development.
The cardiac precursors are generated in the anterior lateral plate mesoderm and migrate ventromedially to form the linear heart tube, which possesses a highly regionalized structure (13). Complex remodeling of the heart tube and immigration of the neural crest cells occur during looping to complete morphogenesis of the heart. The genetic pathways regulating regionalization and these complex processes were recently identified (13). The cardiac homeobox gene Nkx2.5/Csx (Nkx2.5), a mammalian homologue of the Drosophila tinman (14, 15), is presumed to be a cardiac selector gene essential for either cardiogenesis or cardiac morphogenesis (16). This hypothesis is based on the fact that Nkx2.5-deficient embryos display defects in looping morphogenesis of the heart and cardiac gene expression of Hand1, atrial natriuretic factor (ANF), and myosin light chain 2v (MLC2v) (17–19), and that exogenous expression of dominant negative Nkx2.5 into early Xenopus blastomeres abrogates heart formation (20, 21). Several mutations in NKX2.5/CSX1 (NKX2.5) have been reported in patients with a variety of cardiac anomalies, including atrioventricular (AV) conduction delays, atrial septal defects, TOF, and DORV (22–24), further underscoring the importance of Nkx2.5 in cardiac development.
Studies have recently been reported on the cis-regulatory elements required for initiation of Nkx2.5 expression. These elements, however, were unable to sustain the expression later in the development of the heart in the transgenic animals (25, 26). Thus, very little is known about the maintenance phase of cardiac Nkx2.5 expression in mammals.
In the present study, we performed a detailed examination of Rae28–/– embryos to determine the role of Rae28 in the genetic programs governing heart development. First, we propose the presence of a maintenance phase in Nkx2.5 expression in cardiomyocytes, which is sustained by Rae28. Secondly, the maintenance phase of cardiac Nkx2.5 expression plays a crucial role in maturing cardiac morphogenesis. Based on this evidence, we argue the case for biological implication of the PcG gene–mediated transcription maintenance system in mammalian heart development.
Methods
Generation of Rae28–/– embryos and those complemented by NKX2.5 overexpression or exogenous Rae28 expression.
Mice deficient in Rae28 were generated and maintained by heterozygous intercrossing as described previously (10). Transgenic mouse lines overexpressing human NKX2.5/CSX1 cDNA were generated by using pCAGGS vector (27), and those overexpressing the full-length Rae28 cDNA (Rae2819) (28) by using either chicken β-actin promoter with Rous sarcoma-virus enhancer (RSV-Rae28) or β-myosin heavy chain promoter (βMHC-Rae28) (29). These transgenic mice were backcrossed to C57BL/6 to minimize the effects of genetic background before being used in this study. The resulting progeny were mated with heterozygous Rae28-deficient mice (Rae28+/–), and their genotypes were examined by means of PCR. Embryos obtained by cesarean section were examined histologically and subjected to in situ hybridization analyses.
Whole-mount and section in situ hybridization.
Whole-mount in situ hybridization was performed essentially as previously described (12). Briefly, embryos were fixed in PBS containing 4% paraformaldehyde, bleached, and treated with 10 μg/ml proteinase K (Sigma-Aldrich, St. Louis, Missouri, USA). After additional fixation in 0.2% glutaraldehyde and 4% paraformaldehyde in PBS, the embryos were soaked in prewarmed prehybridization buffer for 1 hour at 70°C, and hybridized overnight with a digoxigenin-labeled riboprobe. Hybridization was detected by treatment of embryos with preabsorbed alkaline phosphatase–conjugated anti-digoxigenin antibodies (Roche Diagnostics GmbH, Mannheim, Germany), followed by reaction with 4-nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (Roche Diagnostics GmbH). Section in situ hybridization with αMHC, cardiac α-actin, and cardiac Ca2+-dependent ATPase probes was done as described previously (10).
RT-PCR assay.
Total RNA was prepared from the hearts, which were dissected from embryos at 9.5 days postcoitus (dpc), by means of ISOGENE (Nippon Gene, Tokyo, Japan), treated with RNase-free DNase I (Roche Diagnostics GmbH), and suspended in 20 μl of distilled water. The first-strand cDNA synthesis was performed with Superscript II reverse transcriptase (Invitrogen, Carlsbad, California, USA) and an oligo(dT20) primer according to the manufacturer’s instructions. PCR amplification was performed with a GeneAmp PCR System 2400 (Applied Biosystems, Foster City, California, USA) using the previously described oligonucleotide primers specific for MLC2a, MLC2v, αMHC, ANF, Hand1, Nkx2.5, and β-actin (17, 30–33). PCR cycles were as follows: 95°C for 5 minutes, followed by 25–50 cycles of 95°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute. The PCR products were electrophoresed on 1% agarose gels and stained with ethidium bromide.
Results
Morphology of the embryonic hearts.
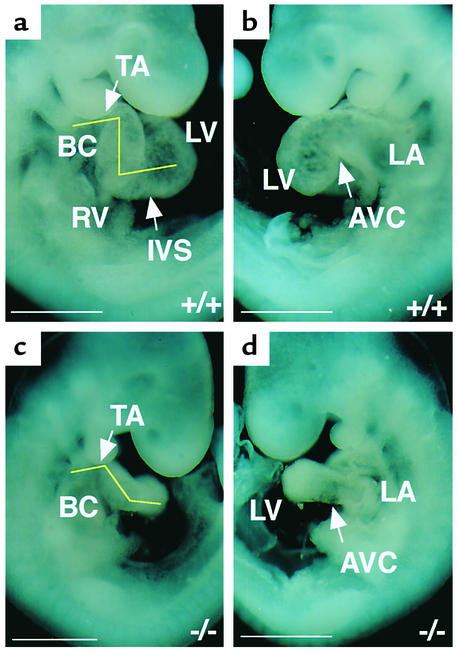
Wild-type and Rae28–/– hearts were microscopically indistinguishable at the linear heart tube stage (8.0 dpc). At 8.5 dpc, the heart tube initiated dextro-looping, which orients the atrial and ventricular chambers and aligns the outflow tracts with the vasculature. At 9.5 dpc, 17 out of 20 Rae28–/– hearts showed apparent positional immaturity and/or deformation, as demonstrated by incomplete looping, deformation of the wall, and/or dilatation of the heart tube (Figure 1). The right frontal view shows that interventricular sulcus formation was incomplete and that the bulbus cordis and truncus arteriosus were extremely deformed (Figure 1c). Angulation between the ventricles, and between the bulbus cordis and truncus arteriosus, was reduced in comparison with that of the wild-type (Figure 1c). The left frontal view shows that the AV canal was neither narrowed nor elongated sufficiently (Figure 1d). This may be the result of impaired development of the AV cushion, which may be also responsible for positional immaturity and mislooping in the mutant heart. Because cardiac looping is an important morphologic event that is critical for the proper alignment and orientation of the atrial, ventricular, and vascular compartments of the heart (13), it is probable that positional and morphological changes in the looping heart caused the abnormal cardiac morphogenesis in Rae28–/– embryos.
Figure 1.
Cardiac looping morphogenesis in 9.5-dpc embryos. (a and c) Right and (b and d) left frontal views of whole embryos are shown. Cardiac looping was morphologically affected in the 9.5-dpc Rae28–/– embryo (c and d). IVS, interventricular sulcus; BC, bulbus cordis; TA, truncus arteriosus; AVC, AV canal; LV, left ventricle; LA, left atrium; RV, right ventricle. +/+, a wild-type embryo; –/–, a Rae28–/– embryo. Scale bar: 500 μm.
In situ hybridization analysis of gene expression.
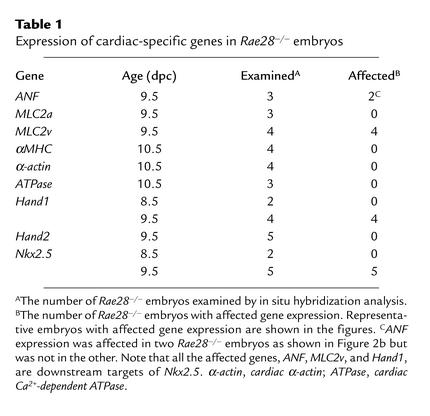
To examine the molecular effect of Rae28 mutation on cardiac segments, expression of the cardiac-specific genes in 9.5-dpc Rae28–/– embryos (13, 30, 34) was observed with the aid of whole-mount in situ hybridization. The domain of ANF expression in the presumptive right ventricle had almost disappeared in Rae28–/– embryos (Figure 2b). Although MLC2v expression is normally restricted to the ventricles of the developing heart (30), MLC2v expression in Rae28–/– embryos had extended to part of the atrium beyond the AV canal (Figure 2d). Although there are wild-type embryos in which MLC2v expression extends to the atrium to some extent, the extension was more pronounced in Rae28–/– embryos. These altered expression domains of ANF and MLC2v suggest that segmental specification is abnormal in the mutant hearts. Other cardiac-specific genes, including MLC2a (Figure 2f), αMHC, cardiac α-actin, and cardiac Ca2+-dependent ATPase, were expressed normally in the mutant hearts (35, 36) (Table 1), suggesting that the Rae28 mutation does not have an overall effect on the expression of heart-specific genes.
Figure 2.
Whole-mount in situ hybridization. Expression of ANF (a and b), MLC2v (c and d), MLC2a (e and f), Hand1 (g–n), Hand2 (o and p), and Nkx2.5 (q–t) was detected. (a–l and o–r) 9.5 dpc; (m, n, s, and t) 8.5 dpc; (a and b) Right lateral views; (c–i, k, q, and r) left lateral views; (j, l–p, s, and t) frontal views. ANF expression in the right ventricle is reduced in the Rae28–/– embryo (b), while MLC2v expression extends into the right atrium (d). Expression of Hand1 was markedly reduced in the hearts of the Rae28–/– embryos (h, k, and l). Since AVC and IVS formations were incomplete in the Rae28–/– embryos, the portions corresponding to AVC and IVS are indicated by an arrowhead and a dotted line, respectively (k and l). Note that Nkx2.5 and Hand1 expressions were markedly reduced in the Rae28–/– embryos at 9.5 dpc (k, l, and r) but were unaffected at 8.5 dpc (n and t). RA, right atrium; LM, lateral mesoderm; AVS, AV sulcus; AT, atrium; P, pharyngeal arch; H, heart. Scale bar: 200 μm.
Table 1.
Expression of cardiac-specific genes in Rae28–/– embryos
Recently, several genes have been implicated in cardiac segmentation and chamber specification. The basic helix-loop-helix genes Hand1 and Hand2 are expressed in complementary patterns in the developing heart and are involved in cardiac looping and chamber specification (37–41). Expression of Hand1 was markedly reduced in the hearts of 9.5-dpc Rae28–/– embryos (Figure 2, h, k, and l) but was unaffected in other tissues in these embryos, such as the pharyngeal arches and lateral mesoderm (Figure 2h). This finding indicates that misregulation of this gene in the mutant embryos is heart-specific. However, Hand2 expression was not altered in the mutant embryos (Figure 2p), which is consistent with the specific, rather than general, misregulation of genes required for heart development.
The genes affected by the Rae28 mutation, ANF, MLC2v, and Hand1, are downstream targets of Nkx2.5 (17–19), which is essential for cardiac looping and morphogenesis (17–19). We therefore examined the expression of Nkx2.5 in 9.5-dpc Rae28–/– embryos. Nkx2.5 expression had almost disappeared in the ventricles but was detectable at a very low level in the common atrium (Figure 2r). Interestingly, however, we could not find any significant difference in expression of Nkx2.5 in the initial stage of cardiac looping (8.5 dpc) (Figure 2t); this was also confirmed by the finding that expression of Hand1, a downstream target of Nkx2.5, was normally initiated (Figure 2n). This finding is consistent with what has been observed in PcG regulation of homeotic genes (1).
The data from in situ hybridization analyses of Rae28–/– embryos are summarized in Table 1. Representative findings are shown in the figures as described above. Although the severity of misexpression somehow varied in each of the embryos, all Rae28–/– embryos showed apparent misexpression of MLC2v, Hand1, and Nkx2.5. ANF expression domain was altered in two out of three Rae28–/– embryos as shown in Figure 2b, but not in the third.
RT-PCR analysis of gene expression.
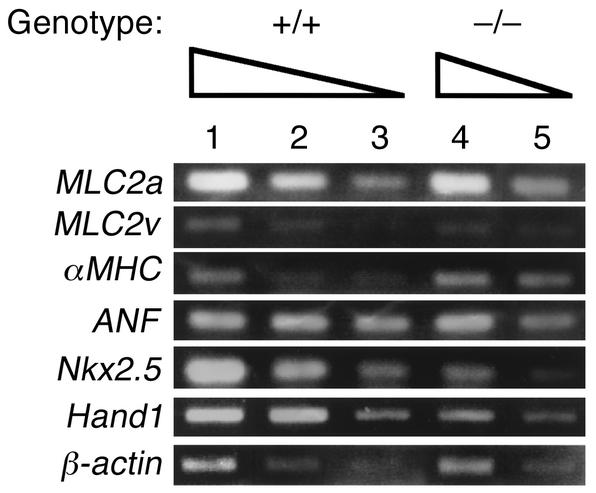
Cardiac gene expression was further examined by semiquantitative RT-PCR analysis. cDNA was generated from total cellular RNAs extracted from the hearts, which were derived from five stage-matched Rae28–/– or wild-type embryos to minimize interembryo variability. A quantitative RT-PCR assay (17) confirmed that the transcripts of Nkx2.5 and Hand1 were downregulated by approximately a factor of four in the mutant hearts, and those of MLC2v by a factor of two (Figure 3). Although the expression domain of ANF in the right ventricle had almost disappeared in two out of three Rae28–/– embryos (Table 1 and Figure 2b), the RT-PCR assay could not detect any significant change in the amount of ANF mRNA in the mutant hearts (Figure 3), probably because ANF expression had not necessarily been altered in all the mutant embryos and because a larger amount of ANF mRNA present in the left ventricle may overcome the difference. The consistency of the changes was confirmed in RNAs extracted from another five embryonic hearts with each of the genotypes (data not shown). The specificity of these bands was also confirmed by Southern blot analysis with radiolabeled gene-specific probes (data not shown).
Figure 3.
Quantitative RT-PCR analysis of gene expression in the hearts. Total cellular RNAs were extracted from the hearts dissected from five wild-type and five Rae28–/– embryos at 9.5 dpc. As indicated by the wedges, twofold serial dilutions of cDNAs were used as templates for PCR amplification with primers specific for MLC2a, MLC2v, αMHC, ANF, Nkx2.5, Hand1, and β-actin. Signals for β-actin mRNA were used as controls to adjust the amount of cDNAs. Equal amounts of cDNAs were subjected to the PCR assays in lanes 1 and 4 as well as in lanes 2 and 5. Although the expression domain of ANF in RV was affected in Rae28–/– (Figure 2b), RT-PCR analysis did not detect a significant difference in the amount of ANF mRNA.
Complementation analysis of Rae28–/– embryos by exogenous human NKX2.5.
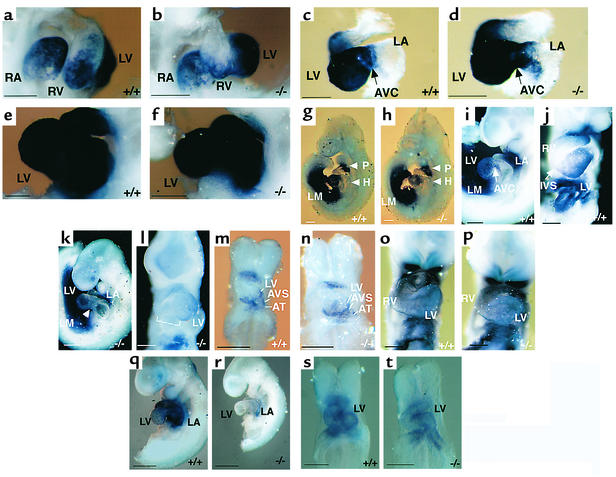
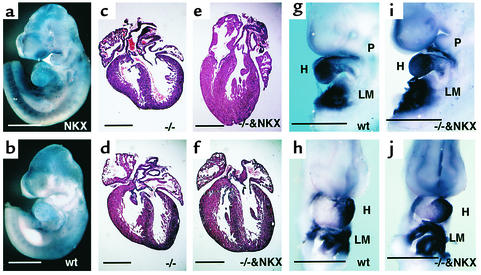
We next wanted to provide functional evidence of the diminished expression of Nkx2.5 in Rae28–/– embryos, and to determine in vivo to what extent downregulation of Nkx2.5 is responsible for the cardiac phenotype. A genetic complementation experiment was performed by crossing Rae28-deficient animals with transgenic mice overexpressing NKX2.5/CSX1 (NKX2.5) (27) (Figure 4a). Although the pCAGGS vector, including the chicken β-actin promoter with a cytomegalovirus enhancer, was used to generate the transgenic mice, NKX2.5 expression was not ubiquitous but detected mainly in the heart and somites, as reported previously (Figure 4a). Northern blot analysis was used to confirm the persistent expression of the NKX2.5 transgene even in the neonatal stage (data not shown). We examined 12 Rae28–/– neonates including eight carrying the NKX2.5 transgene (Rae28–/–/NKX2.5). Four Rae28–/– embryos without the NKX2.5 transgene showed cardiac anomalies, including a large membranous ventricular septal defect straddled on the aorta (Figure 4c), which is compatible with TOF, and a large atrial septal defect with bizarre tricuspid valves (Figure 4d). These abnormal morphological findings were essentially similar to those reported previously in Rae28–/– mice (10). On the other hand, complete reversion of the cardiac phenotype at the morphologic level was found in each of the seven Rae28–/–/NKX2.5 embryos (Figure 4, e and f) except for one, which displayed a muscular-type ventricular septal defect (data not shown). Thus, overexpression of NKX2.5 suppressed the majority of the heart phenotype observed in Rae28–/– mice. Not only Hand1 (Figure 4, i and j), ANF, and MLC2v expressions (data not shown) but also cardiac looping improved remarkably in 9.5-dpc Rae28–/–/NKX2.5 embryos (Figure 4, i and j). Although weak NKX2.5 expression was detected in the regions outside the heart in the NKX2.5 transgenic embryos (Figure 4a), no significant effect of the exogenous NKX2.5 expression was detected on the mutant phenotypes outside the heart, such as abnormal facies with ophthalmic hypoplasia, cleft palate, and parathyroid and thymic hypoplasia (data not shown).
Figure 4.
Suppression of cardiac phenotypes in Rae28–/– embryos by overexpression of NKX2.5. (a and b) Whole-mount in situ hybridization of 9.5-dpc embryos with a NKX2.5 probe. Since we used a human NKX2.5 probe, the signal in the wild-type embryo was weaker, while that from the transgene was detected clearly. (c–f) Histological sections of the mutant hearts. The offspring were generated by intercrossing of Rae28+/– mice carrying the NKX2.5 transgene, and the hearts derived from Rae28–/– embryos with or without the transgene were examined histologically. (g–j) Whole-mount in situ hybridization of 9.5-dpc embryos with a Hand1 probe. The findings were reproducible in the other embryo with the same genotype (data not shown). WT, wild-type embryos; –/–, Rae28–/– embryos; NKX, NKX2.5 transgenic embryos; –/–&NKX, Rae28–/– embryos carrying the NKX2.5 transgene; LM, lateral mesoderm. Scale bar: 500 μm.
Effect of exogenous Rae28 expression on Nkx2.5 expression.
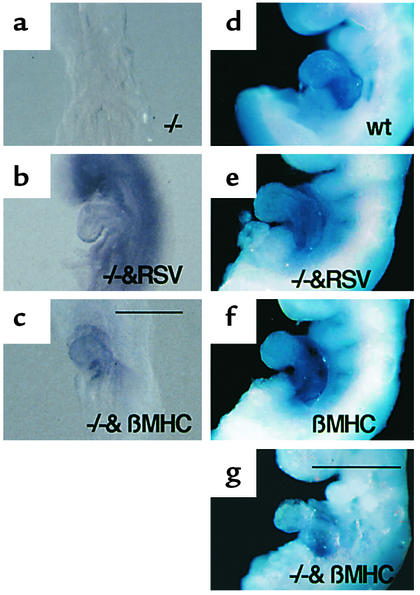
To address the question of how Rae28 regulates Nkx2.5 expression in cardiomyocytes, we generated transgenic mice overexpressing Rae28, using either the chicken β-actin promoter with a Rous sarcoma-virus enhancer (RSV-Rae28) or the β-myosin heavy chain promoter (βMHC-Rae28) (29). Further, we used genetic intercrossing to generate Rae28–/– embryos carrying either RSV-Rae28 (Rae28–/–/RSV-Rae28) or βMHC-Rae28 transgene (Rae28–/–/βMHC-Rae28). Exogenous Rae28 expression was detected at 8.5 dpc (Figure 5, b and c). The Rae28 expression was ubiquitous in Rae28–/–/RSV-Rae28 embryos but was cardiomyocyte-specific in Rae28–/–/βMHC-Rae28 embryos (Figure 5, b and c). The similar expression patterns were also found at 9. 5 dpc (29). Thus the exogenous Rae28 expression was considered to occur prior to or during the time that Nkx2.5 expression was altered in Rae28–/– embryos. The expression of exogenous Rae28 protein was confirmed in both of these transgenic hearts using Western blot analysis (29). All the hearts from Rae28–/–/RSV-Rae28 embryos displayed complete reversion of cardiac anomalies, while those from Rae28–/–/βMHC-Rae28 embryos showed no alteration in the cardiac phenotype (29). Thus, whole-mount in situ hybridization was used to examine Nkx2.5 expression in Rae28–/–/RSV-Rae28 or Rae28–/–/βMHC-Rae28 embryos at 9.5 dpc. Reduced Nkx2.5 expression in Rae28–/– had reverted to the wild-type level in Rae28–/–/RSV-Rae28 embryos (Figure 5e), but not in Rae28–/–/βMHC-Rae28 embryos (Figure 5g); this was highly compatible with phenotypic alterations in these mutant embryos, as mentioned above (29). Thus, βMHC promoter–mediated cardiomyocyte-specific Rae28 expression was unable to revert the reduced Nkx2.5 expression in the Rae28–/– hearts. We further confirmed that overexpression of Rae28 in cardiomyocytes neither induced nor reduced Nkx2.5 expression either at 9.5 dpc (Figure 5f) or in the neonatal period (data not shown).
Figure 5.
Effect of exogenous Rae28 on Nkx2.5 expression in the heart. Expression of the exogenous Rae28 genes and their effect on Nkx2.5 expression were examined by whole-mount in situ hybridization with Rae28 (a–c) and Nkx2.5 probes (d–g). Ubiquitous and cardiomyocyte-specific Rae28 expression was detected at 8.5 dpc in Rae28–/–/RSV-Rae28 and Rae28–/–/βMHC-Rae28 embryos, respectively (b and c), either of which possessed a genetic background of Rae28–/– where Rae28 expression was undetectable (a). Ubiquitous Rae28 expression efficiently reverted reduced expression of Nkx2.5 in the Rae28–/– embryo (e), while cardiomyocyte-specific expression of Rae28 neither augmented nor reduced Nkx2.5 expression in the wild-type heart (f) and did not exert any effect on reduced expression of Nkx2.5 in the Rae28–/– embryo (g). We further confirmed these findings in other embryos (data not shown). WT, wild-type embryo; βMHC, βMHC-Rae28; –/–&RSV, Rae28–/–/RSV-Rae28; –/–&βMHC, Rae28–/–/βMHC-Rae28. Scale bar: 200 μm (a–c), 500 μm (d–g).
Discussion
In general, little has been known about the molecular mechanisms that maintain gene expression after the initial transcriptional decision has been made during mammalian development. Recently, more than 14 mammalian homologues of Drosophila PcG genes have been identified, and genetic evidence is accumulating that mammalian PcG genes are involved in wider biological functions than Drosophila PcG genes (2). Rae28–/– mice not only showed affected anteroposterior patterning in the paraxial mesoderm and rhombomeres but also displayed abnormal cardiac morphogenesis (10). In the study reported here, we revealed that expression of Nkx2.5 and its downstream targets was systematically affected during the later stage of heart development in Rae28–/– embryos. The impaired expression of Nkx2.5 proved to be mainly responsible for cardiac dysmorphogenesis, because complementary expression of human NKX2.5 effectively reverted cardiac anomalies in Rae28–/– embryos. Interestingly, we could not find any significant reduction in Nkx2.5 expression and that of its downstream target Hand1 in the initial stage of cardiac looping at 8.5 dpc, while these expressions were affected in the later stage (9.5 dpc). This finding is consistent with what has been observed for PcG regulation of homeotic genes in Drosophila and in mammals (1, 10, 12). Normal initiation but alteration in Nkx2.5 and Hand1 expression in the maintenance phase may be the reason why cardiac development was not completely eliminated in the looping stage as observed in Nkx2.5- and Hand1-deficient mice (17–19, 39, 42). Instead, these alterations may have generated cardiac anomalies such as TOF and/or DORV, indicating the important role of the maintenance phase of Nkx2.5 expression in maturing cardiac morphogenesis. Ubi-quitous expression of exogenous Rae28 reverted the impaired Nkx2.5 expression in Rae28–/– embryos, further confirming that Rae28 is required for sustaining Nkx2.5 expression in cardiomyocytes.
Since Nkx2.5 is highly conserved and plays an essential role in heart development (43), analysis of the transcription regulation is crucial for understanding the molecular mechanisms of heart development and the pathogenesis of congenital heart diseases in humans. Nkx2.5 transcription was shown to be regulated by complex modular cis-regulatory elements with distinct regional specificity (16), suggesting that the heart is presumed to evolve as a modular organ in higher organisms (43). Although these elements recapitulate the initial expression pattern of Nkx2.5 relatively well, they were unable to sustain Nkx2.5 expression in the maintenance phase after the initiation (25, 26). In Drosophila, it has been suggested that tinman sustains its own transcription through a positive autoregulatory feedback loop in combination with decapentaplegic and wingless signals (44, 45). In mammals, however, it remains controversial whether Nkx2.5 forms a positive feedback loop for sustaining the proper expression. A combination of Nkx2.5 and GATA4 was reported to show a transactivation effect on the Nkx2.5 promoter (25), while a negative feedback loop of Nkx2.5 was also suggested by transgenic experiments (26). Thus, molecular mechanisms for sustaining Nkx2.5 expression had been largely unknown. Since we demonstrated in this study that Rae28 is required for sustaining Nkx2.5 expression in the entire cardiac region later in heart development, it is possible to speculate that Rae28 has a role in systematically governing each module for maturing cardiac morphogenesis. This system might also help to sustain proper expression of the selector gene, once initiated, even in the presence of various internal and external agents affecting the transcription, because a PcG gene is implicated in negatively defining accessibility to transcriptional regulatory elements such as retinoic acid response elements (8).
An interesting aspect of these results is that, whereas PcG genes maintain genes in a repressed state (1), Rae28 is required for sustaining Nkx2.5 expression in cardiomyocytes. A need for PcG genes in the activation of gene activity has been observed in Drosophila (46–48), so activation of Nkx2.5 by Rae28 is not unprecedented. Recently it was reported that, in Drosophila, binding of the ATP-dependent chromatin remodeling factors SWI and SNF to the nucleosomal template is competed for by Polycomb repressive complex 1, including Polycomb, Posterior sex combs, sex combs on midleg, and polyhomeotic, a Drosophila counterpart of Rae28 (3). Furthermore, the SWI/SNF complexes can produce a nucleosome structure both conducive to transcription at some promoters and repressive at others (49). It is possible that Rae28 positively regulates Nkx2.5 expression through regulation of the SWI/SNF–mediated nucleosome remodeling, but notwithstanding the arguments presented above, it remains a possibility that Rae28-mediated regulation of Nkx2.5 is indirect.
We then attempted to examine whether Rae28 regulates Nkx2.5 expression in cardiomyocytes directly or indirectly. Although ubiquitous expression of Rae28 driven by the RSV enhancer reverted the reduced expression of Nkx2.5 in Rae28–/– embryos at 9.5 dpc, cardiomyocyte-specific expression of Rae28 driven by the βMHC promoter was unable to sustain Nkx2.5 expression. The possibility exists that Rae28 could be required at an earlier developmental stage and that Rae28 expression derived from the βMHC-Rae28 transgene did not occur early enough to sustain proper Nkx2.5 expression. However, we detected no significant difference in Nkx2.5 expression in Rae28–/– embryos as early as 8.5 dpc, at which stage the βMHC promoter had already started the transcription (Figure 5c). Thus, it is unlikely that the exogenous expression of Rae28 did not occur early enough to correct reduced Nkx2.5 expression in Rae28–/– embryos. Furthermore, we detected Rae28 protein in the hearts of βMHC-Rae28 embryos, and βMHC-Rae28 embryos carried the same full-length cDNA for Rae28 as was used to generate RSV-Rae28 mice, in which exogenous Rae28 revealed a capacity to suppress all the phenotypes and also reduced Nkx2.5 expression observed in Rae28–/– mice (29). It is therefore also unlikely that the Rae28 protein derived from the βMHC-Rae28 transgene encoded the nonfunctional proteins in the cardiomyocytes. These findings support the hypothesis that Rae28 has a role in sustaining Nkx2.5 expression in cardiomyocytes through a non–cell-autonomous pathway. This assumption might be consistent with the fact that, in general, PcG genes are known to maintain repressed states of transcription (1). Further analysis is, however, required to elucidate the mechanisms by which Rae28 regulates Nkx2.5 expression in cardiomyocytes.
Acknowledgments
We thank Hugh W. Brock, Yoshiyuki Niho, Makoto Nakazawa, Kiyomasa Nishii, and Yoichiro Shibata for their encouragement; Ichiro Shiojima, Akihisa Sawada, Ju Yoo Kim, and Hiroko Endou for their discussion; Eric Olson, Peter Cserjesi, Richard Harvey, and Tadashi Kaname for kindly providing Hand1, Hand2, and Nkx2.5 probes, respectively; and Rie Hasegawa for secretarial help. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan and grants from the Ministry of Health and Welfare of Japan.
Footnotes
Manabu Shirai and Tomoaki Osugi contributed equally to this work.
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: Polycomb-group (PcG); tetralogy of Fallot (TOF); double outlet of the right ventricle (DORV); Nkx2.5/Csx (Nkx2.5); NKX2.5/CSX1 (NKX2.5); atrial natriuretic factor (ANF); myosin light chain 2v (MLC2v); atrioventricular (AV); days postcoitus (dpc); Nkx2.5/Csx (Nkx2.5).
References
- 1.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 2.Takihara Y, Hara J. The Polycomb-group genes and hematopoiesis. Int J Hematol. 2000;72:165–172. [PubMed] [Google Scholar]
- 3.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 4.Breiling A, Turner BM, Bianchi M, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- 5.Saurin AJ, et al. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 6.van der Lugt MT, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1proto-oncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 7.Akasaka T, et al. A role for mel-18, a Polycomb group related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- 8.Coré N, et al. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- 9.Katoh-Fukui Y, et al. Male-to-female sex reversal in M33mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- 10.Takihara Y, et al. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development. 1997;124:3673–3682. doi: 10.1242/dev.124.19.3673. [DOI] [PubMed] [Google Scholar]
- 11.del Mar Lorente M, et al. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- 12.Tomotsune D, Shirai M, Takihara Y, Shimada K. Regulation of Hoxb3 expression in the hindbrain and pharyngeal arches by Rae28, a member of the mammalian Polycomb group of genes. Mech Dev. 2000;98:165–169. doi: 10.1016/s0925-4773(00)00457-3. [DOI] [PubMed] [Google Scholar]
- 13.Olson EN, Srivastava D. Molecular pathways controlling heart development. Science. 1996;272:671–676. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- 14.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 15.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz RJ, Olson EN. Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- 17.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 18.Biben C, Harvey RP. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997;11:1357–1369. doi: 10.1101/gad.11.11.1357. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- 21.Grow MW, Krieg PA. Tinman function is essential for vertebrate heart development: elimination of cardiac differentiation by dominant inhibitory mutants of the tinman-related genes, XNkx2-3 and XNkx2-5. Dev Biol. 1998;204:187–196. doi: 10.1006/dbio.1998.9080. [DOI] [PubMed] [Google Scholar]
- 22.Schott JJ, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 23.Benson DW, et al. Mutations in the cardiac transcription factor NKX2.5affect diverse cardiac developmental pathways. J Clin Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosoda T, et al. Familial atrial septal defect and atrioventricular conduction disturbance associated with a point mutation in the cardiac homeobox gene CSX/NKX2-5 in a Japanese patient. Jpn Circ J. 1999;63:425–426. doi: 10.1253/jcj.63.425. [DOI] [PubMed] [Google Scholar]
- 25.Reecy JM, et al. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2.5. Development. 1999;126:839–849. doi: 10.1242/dev.126.4.839. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M, et al. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development. 1999;126:1439–1450. doi: 10.1242/dev.126.7.1439. [DOI] [PubMed] [Google Scholar]
- 27.Takimoto E, et al. Up-regulation of natriuretic peptides in the ventricle of Csx/Nkx2-5 transgenic mice. Biochem Biophys Res Commun. 2000;270:1074–1079. doi: 10.1006/bbrc.2000.2561. [DOI] [PubMed] [Google Scholar]
- 28.Nomura M, Takihara Y, Shimada K. Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: one of the early inducible clones encodes a novel protein sharing several highly homologous regions with a Drosophila polyhomeotic protein. Differentiation. 1994;57:39–50. doi: 10.1046/j.1432-0436.1994.5710039.x. [DOI] [PubMed] [Google Scholar]
- 29.Koga H, et al. Overexpression of Polycomb-group gene Rae28 in cardiomyocytes does not complement abnormal cardiac morphogenesis in mice lacking Rae28, but causes dilated cardiomyopathy. Lab Invest. 2002;82:375–385. doi: 10.1038/labinvest.3780432. [DOI] [PubMed] [Google Scholar]
- 30.Kubalak SW, Miller-Hance WC, O’Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- 31.Miller-Hance WC, et al. In vitro chamber specification during embryonic stem cell cardiogenesis. Expression of the ventricular myosin light chain-2 gene is independent of heart tube formation. J Biol Chem. 1993;268:25244–25252. [PubMed] [Google Scholar]
- 32.Xu C, Liguori G, Adamson ED, Persico MG. Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev Biol. 1998;196:237–247. doi: 10.1006/dbio.1998.8862. [DOI] [PubMed] [Google Scholar]
- 33.Conway SJ, Henderson DJ, Copp AJ. Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development. 1997;124:505–514. doi: 10.1242/dev.124.2.505. [DOI] [PubMed] [Google Scholar]
- 34.Zeller R, Bloch KD, Williams BS, Arceci RJ, Seidman CE. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev. 1987;1:693–698. doi: 10.1101/gad.1.7.693. [DOI] [PubMed] [Google Scholar]
- 35.Sassoon DA, Garner I, Buckingham M. Transcripts of α-cardiac and α-skeletal actins are early markers for myogenesis in the mouse embryo. Development. 1988;104:155–164. doi: 10.1242/dev.104.1.155. [DOI] [PubMed] [Google Scholar]
- 36.Lompre AM, de la Bastie D, Boheler KR, Schwartz K. Characterization and expression of the rat heart sarcoplasmic reticulum Ca2+-ATPase mRNA. FEBS Lett. 1989;249:35–41. doi: 10.1016/0014-5793(89)80010-9. [DOI] [PubMed] [Google Scholar]
- 37.Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava D, et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 39.Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- 40.Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 41.Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- 42.Rindt H, Gulick J, Knotts S, Neumann J, Robbins J. In vivo analysis of the murine beta-myosin heavy chain gene promoter. J Biol Chem. 1993;268:5332–5338. [PubMed] [Google Scholar]
- 43.Fishman MC, Olson EN. Parsing the heart: genetic modules for organ assembly. Cell. 1997;91:153–156. doi: 10.1016/s0092-8674(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 44.Yin Z, Xu XL, Frasch M. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development. 1997;124:4971–4982. doi: 10.1242/dev.124.24.4971. [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Yin Y, Hudson JB, Ferguson EL, Frash M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fauvarque MO, Zuber V, Dura JM. Regulation of polyhomeotic transcription may involve local changes in chromatin activity in Drosophila. Mech Dev. 1995;52:343–355. doi: 10.1016/0925-4773(95)00412-t. [DOI] [PubMed] [Google Scholar]
- 47.LaJeunesse D, Shearn A. E(z): a polycomb group gene or a trithorax group gene? Development. 1996;122:2189–2197. doi: 10.1242/dev.122.7.2189. [DOI] [PubMed] [Google Scholar]
- 48.Milne TA, Sinclair DA, Brock HW. The Additional sex combs gene of Drosophila is required for activation and repression of homeotic loci, and interacts specifically with Polycomb and super sex combs. Mol Gen Genet. 1999;261:753–761. doi: 10.1007/s004380050018. [DOI] [PubMed] [Google Scholar]
- 49.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]