Abstract
Corepressors N-CoR and SMRT participate in diverse repression pathways and exist in large protein complexes including HDAC3, TBL1 and TBLR1. However, the roles of these proteins in SMRT–N-CoR complex function are largely unknown. Here we report the purification and functional characterization of the human N-CoR complex. The purified N-CoR complex contains 10–12 associated proteins, including previously identified components and a novel actin-binding protein IR10. We show that TBL1/TBLR1 associates with N-CoR through two independent interactions: the N-terminal region and the C-terminal WD-40 repeats interact with the N-CoR RD1 and RD4 region, respectively. In vitro, TBL1/TBLR1 bind histones H2B and H4, and, importantly, repression by TBL1/TBLR1 correlates with their interaction with histones. By using specific small interference RNAs (siRNAs), we demonstrate that HDAC3 is essential, whereas TBL1 and TBLR1 are functionally redundant but essential for repression by unliganded thyroid hormone receptor. Together, our data reveal the roles of HDAC3 and TBL/TBLR1 and provide evidence for the functional importance of histone interaction in repression mediated by SMRT–N-CoR complexes.
Keywords: HDAC3/histone interaction/N-CoR/TBL1/TBLR1
Introduction
The organization of chromatin is of fundamental significance in transcriptional control in eukaryotic cells (Wolffe, 1998). The effect of chromatin structure on transcription is determined, at least in part, by the post-translational modifications of histones such as acetylation and methylation (Kouzarides, 1999; Jenuwein and Allis, 2001; Roth et al., 2001). The acetylation of lysine residues in histone N-terminal tails is catalyzed by histone acetyltransferases (HATs) and is reversed by the activity of histone deacetylases (HDACs). The fact that many important transcriptional cofactors possess HAT activities and that the transcriptional corepressors are often associated with HDACs underscores the importance of this dynamic modification in transcriptional regulation. Acetylation is believed to facilitate transcription by destabilizing nucleosomes and/or high order chromatin structure and serving as docking sites (histone code) for regulatory proteins, whereas deacetylation reverses such effects and thus reinforces the repressive effect of chromatin.
The nuclear receptors belonging to the steroid/thyroid/retinoid receptor superfamily are ligand-inducible transcription factors (Mangelsdorf et al., 1995). Nuclear receptors such as thyroid hormone receptors (TRs) and retinoic acid receptors (RARs) have the capacity to make use of chromatin to repress or activate transcription alternatively dependent on the absence or presence of ligands (Glass et al., 1989; Baniahmad et al., 1990; Wong et al., 1995). In the absence of ligands, TR and RAR are believed to interact with corepressor proteins such as nuclear receptor corepressor (N-CoR) (Horlein et al., 1995) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) (Chen and Evans, 1995), two highly related proteins (Ordentlich et al., 1999; Park et al., 1999). Although identified initially as corepressors for nuclear receptors, N-CoR and SMRT have also been implicated in repression by many other transcription factors including Mad/Mxi, BCL6/LAZ3, ETO and CBF (for a review, see Glass and Rosenfeld, 2000). Recent studies with N-CoR knockout mice underscore the functional importance of N-CoR in early embryonic development and demonstrate that N-CoR is required for both short-term repression by unliganded nuclear receptors and long-term repression events mediated by REST/NRSF (Jepsen et al., 2000).
SMRT and N-CoR are believed to mediate repression at least in part through their ability to recruit HDACs to generate repressive chromatin. Early studies provided evidence for the recruitment of the HDAC1/2-containing mSin3A complex as a potential mechanism for repression mediated by SMRT and N-CoR (Heinzel et al., 1997; Nagy et al., 1997). Further studies indicated that a small fraction of N-CoR and SMRT also associates with class II HDACs (Huang et al., 2000; Kao et al., 2000). Recent efforts in biochemical purification and characterization of both SMRT and N-CoR demonstrate that both N-CoR and SMRT proteins exist in large protein complexes and are associated primarily with HDAC3 (Guenther et al., 2000; Li et al., 2000; Wen et al., 2000). In addition to HDAC3, the purified SMRT complex also contained TBL1, a protein with six WD-40 repeats (Guenther et al., 2000; Li et al., 2000). Recent purification of the N-CoR complex by Zhang et al. (2002) identified two additional N-CoR/SMRT-associated proteins, namely GPS2, a protein involved in intracellular signaling, and TBLR1, a six WD-40 repeat protein with extensive sequence homology with TBL1. The presence of TBL1 and TBLR1 in SMRT and N-CoR complexes is reminiscent of RbAp46 and RbAp48, two histone-binding proteins, in the mSin3A and Mi-2–NURD corepressor complexes (Zhang et al., 1997, 1998; Xue et al., 1998; Wade et al., 1999). However, the functional significance of HDAC3, TBL1 and TBLR1 in SMRT and N-CoR complex function in mammalian cells remains obscure.
As SMRT and N-CoR complexes are estimated to have a size of 1.5–2 MDa (Guenther et al., 2000; Li et al., 2000), the components so far identified in SMRT or N-CoR complexes are unlikely to account for all SMRT- or N-CoR-associated proteins. Here we report the purification of the N-CoR complex and functional characterization of HDAC3 and TBL1/TBLR1. The N-CoR complex purified from HeLa nucelar extracts contains 10–12 proteins. By mass spectrometry, we identified, in addition to other previously identified proteins, a novel coronin-like actin-binding protein IR10 as a component of the N-CoR complex. We show that TBL1/TBLR1 interact with N-CoR through two different domains. In addition, we show that TBL1/TBLR1 bind through their N-terminal region preferentially to histones H2B and H4, and that histone interaction correlates with their transcriptional repression function. Finally, by knock-down experiments using small interference RNA (siRNA) techniques, we demonstrate that HDAC3 is essential and TBL1 and TBLR1 are functionally redundant but essential for repression by unliganded TR.
Results
Purification of the N-CoR protein complex from HeLa nuclear extracts
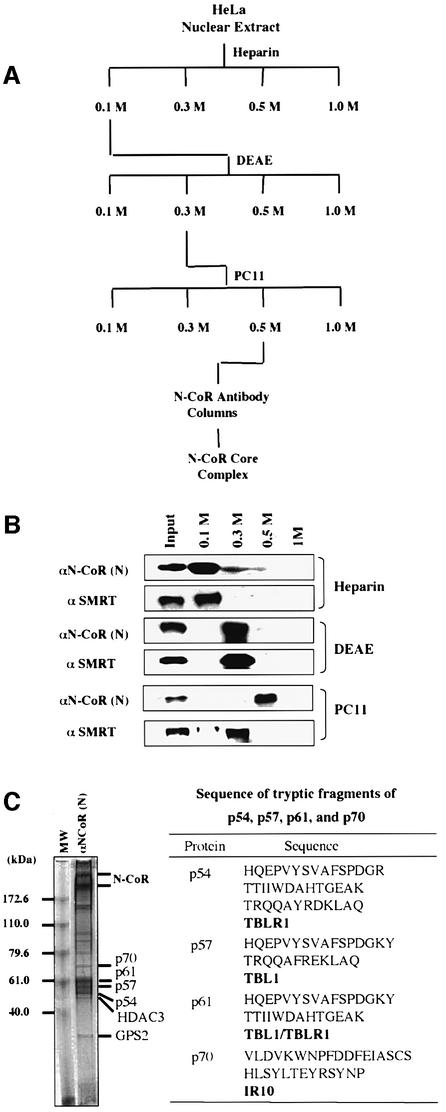
To purify N-CoR complexes from HeLa nuclear extracts, we developed a purification scheme as shown in Figure 1A. Through these three chromatographic steps, N-CoR was found in the phosphocellulose (PC) 0.5 M fraction (Figure 1B) with an estimated 20- to 30-fold enrichment (data not shown). Gel filtration analysis indicated that the N-CoR proteins in the PC 0.5 M fraction co-fractionated with HDAC3 and TBL1, and maintained the same gel filtration profile as the N-CoR proteins in HeLa nuclear extracts (Figure 2B).
Fig. 1. Purification of the N-CoR complex from HeLa nuclear extracts and identification of N-CoR-associated proteins by mass spectrometry. (A) Diagram illustrating the purification scheme of the N-CoR complex. PC, phosphocellulose p11 resins; DEAE, DEAE–Sepharose Fast Flow resins. (B) The differential fractionation of SMRT and N-CoR proteins by various chromatographic steps. The protein samples from each fraction were analyzed by western blotting using the specific antibodies indicated. (C) Immunoaffinity purification and identification of the N-CoR complex. The pooled fractions of immunoaffinity-purified N-CoR complex were resolved using an 8% NuPAGE (Novex) gel and visualized by Coomassie staining. The peptide sequences derived from p54, p57, p61 and p70 determined by mass spectrometry and the identity of these proteins are shown on the right.
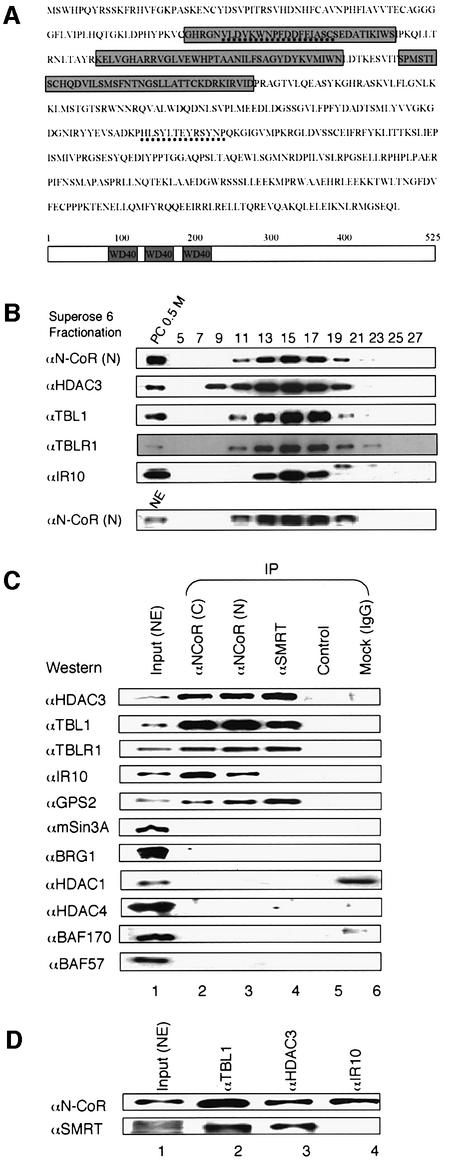
Fig. 2. Characterization of N-CoR-associated proteins. (A) The amino acid sequence of human IR10 protein. The regions identified by mass spectrometry are underlined. The WD-40 repeats are boxed. (B) N-CoR proteins remained in a large protein complex after three chromatographic steps. The PC 0.5 M or HeLa nuclear extract was fractionated with a Superose 6 gel filtration column. Fractions were collected, and analyzed by western blotting using the antibodies indicated on the left. (C) Western blot analyses of N-CoR–SMRT-associated proteins. N-CoR or SMRT complex was immunoprecipitated from HeLa nuclear extracts using N-CoR- or SMRT-specific antibodies, and the presence of various proteins was analyzed by western blotting using antibodies as indicated. Input, 10% HeLa nuclear extract used for immunoprecipitation (IP); control, no antibody; and mock, IP with rabbit anti-mouse IgG. (D) IR10 associates with N-CoR but not SMRT. HeLa nuclear extracts were immunoprecipitated with antibodies against TBL1, HDAC3 and IR10, respectively, and probed by western blotting using N-CoR or SMRT antibody.
The N-CoR complex in the PC 0.5 M fraction was purified subsequently with an N-CoR-specific antibody affinity column. Coomassie staining indicated that immuno purified N-CoR complex contained ∼10–12 protein bands (Figure 1C, left panel). All these bands were probably the subunits of the N-CoR complex, as they were absent in the control experiment using rabbit IgG as affinity resin (data not shown). However, it is unclear from our immunopurification whether all these proteins represent a single N-CoR complex or multiple heterogeneous N-CoR complexes. For simplicity, we refer to them hereafter collectively as the N-CoR complex.
Components of the purified N-CoR complex
Protein bands were excised from the SDS–poly acrylamide gel (Figure 1C, left panel) and their identities were determined by mass spectrometry. The top two bands were identified as N-CoR, and p50 was identified as HDAC3. p54 was identified as TBLR1 and p57 as TBL1, whereas the p61 band contained both TBL1 and TBLR1 (see the table in the right panel of Figure 1C). The identity of these bands was verified by western analysis using HDAC3-, TBL1- and TBLR1-specific antibodies (data not shown, see Figure 2C). Although we failed to identity the p35 protein by mass spectrometry, the size of this protein suggests it to be the GPS2 protein reported by Roeder’s group (Zhang et al., 2002). This assumption was confirmed by western analysis using a GPS2-specific antibody (data not shown, see Figure 2C). Thus, our independent work on purification of the N-CoR complex is in agreement with the results from Roeder’s group (Zhang et al., 2002). In the purified complex, HDAC3, GPS2, TBL1 and TBLR1 appear to be present in approximately stoichiometric amounts to N-CoR, suggesting that these proteins are likely to be the core subunits of the N-CoR complex.
In addition to the above subunits, several proteins present in substoichiometric amounts also co-purified with N-CoR. The p70 band was found to contain HSP70 as well as IR10, a novel coronin-like actin-binding protein (Zaphiropoulos and Toftgard, 1996). Interestingly, IR10 is a protein with three WD-40 repeats (Figure 2A), but shares little sequence similarity with TBL1 and TBLR1. The association of IR10 with N-CoR was supported by the findings that IR10 was co-immunoprecipitated with two different N-CoR antibodies (Figure 2C) and that IR10 co-fractionated with N-CoR complex in gel filtration analysis (Figure 2B). In addition, like antibodies against HDAC3 and TBL1, an IR10-specific antibody also co-immunoprecipitated N-CoR (Figure 2D). Interestingly, IR10 appears to associate only with N-CoR, but not with SMRT, since IR10 was not co-immunoprecipitated with SMRT, and SMRT was not detected in immunoprecipitation of IR10 (Figure 2C and D). The presence of IR10 in the N-CoR complex suggests a possible functional connection of the N-CoR complex with actin. As actin is involved in various cellular processes, the functional significance of this association currently is under investigation.
The identity of several other proteins in the N-CoR complex has yet to be determined due to their relatively low levels (Figure 1C). However, by western analyses, we failed to detect in our purified complex the presence of mSin3A or BRG1 and its associated BAFs, as reported (Underhill et al., 2000; Jones et al., 2001). As shown in Figure 2C, immunoprecipitation of HeLa nuclear extracts by two different N-CoR antibodies and a SMRT-specific antibody significantly enriched HADC3, TBL1, TBLR1 and GPS2. In contrast, neither Sin3A, HDAC1, BRG1, BAF170 nor BAF57 were detected. In addition, we also failed to detect the presence of N-CoR and SMRT in reciprocal immunoprecipitations using BRG1- or BAF-specific antibodies (data not shown). Taken together, we conclude that our purified N-CoR complex contains GPS2, HDAC3, TBL1, TBLR1, IR10 and several unidentified proteins. However, those unidentified proteins are unlikely to be the proteins found in Sin3A or BRG1–BAF complexes.
TBL1 and TBLR1 interact with two different regions in N-CoR
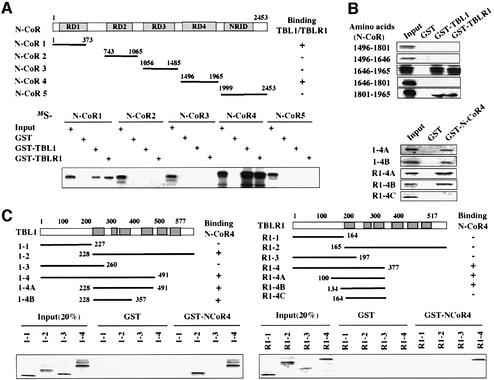
The identification of TBL1 and TBLR1 in SMRT and N-CoR complexes indicates that the HDAC3-containing SMRT and N-CoR complexes also contain two WD-40 repeat core subunits. We next wished to elucidate the structural and functional roles of these two proteins. First, we confirmed by in vitro pull-down assay the published results that TBL1/TBLR1 interact with SMRT and N-CoR, but not HDAC3 (Guenther et al., 2000; Zhang et al., 2002; data not shown). Although TBL1 was shown previously to interact with repression domain 1 (RD1) of N-CoR and SMRT (Guenther et al., 2000; Zhang et al., 2002), the identification of TBLR1 raised the question of whether both TBL1 and TBLR1 bind to the same region. We thus carried out a systematic interaction assay to define the interaction between N-CoR and TBL1/TBLR1. The results in Figure 3A showed that the N-CoR(1–373) fragment containing RD1 interacted with both TBL1 and TBLR1, in agreement with previous results (Guenther et al., 2000; Zhang et al., 2002). However, the N-CoR(1496–1965) fragment also bound strongly to both TBL1 and TBLR1, whereas the rest did not bind. To map more precisely the interaction of TBL1/TBLR1 with N-CoR(1496–1965), we constructed several new expression constructs within N-CoR4(1496–1965) (Figure 3B, left hand side). When tested by in vitro pull-down assays, the region including N-CoR 1646–1965 (N4-3) interacted efficiently with both TBL1 and TBLR1 (Figure 3B). The region between 1801 and 1965 (N4-3B) also interacted, although less efficiently. Thus, a minimal interaction domain was mapped to the region between N-CoR 1801 and 1965. Taken together, we conclude that N-CoR contains two distinct TBL1/TBLR1 interaction domains, one in RD1 and the second mapped to 1801–1965.
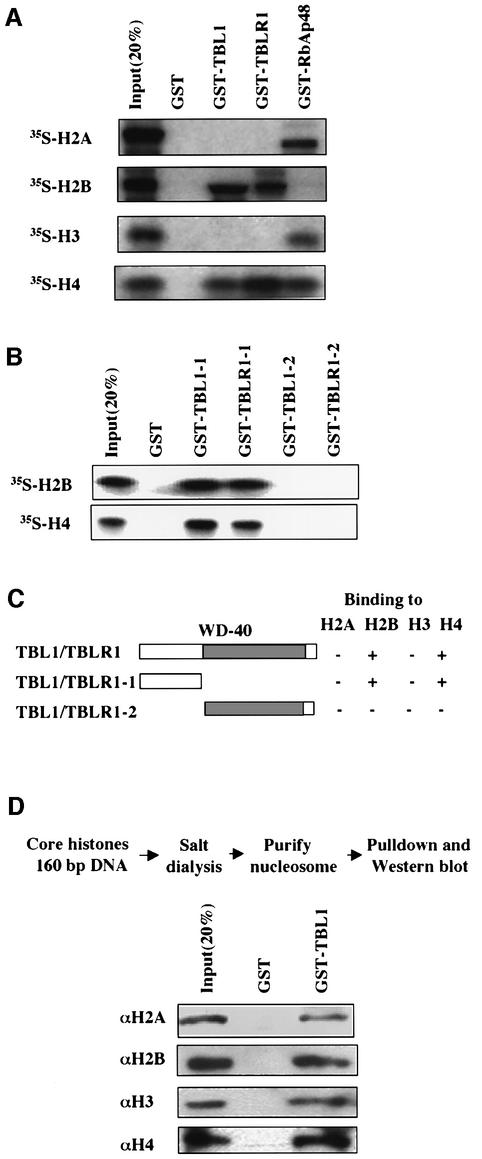
Fig. 3. TBL1/TBLR1 associate with N-CoR through two independent, reciprocal interactions. (A) TBL1 and TBLR1 interact with N-CoR1 and N-CoR4 in vitro. The upper panel shows the structure of N-CoR and deletion constructs used for mapping experiments. Each fragment was in vitro translated, [35S]methionine labeled and used for in vitro pull-down assays using GST–TBL1 and GST–TBLR1 fusion proteins. The result of the pull-down assay is summarized in the top right panel. Input was 20% of the sample used for pull-down assay. (B) The TBL1/TBLR1 interaction region in N-CoR4 was mapped to N-CoR 1801–1965. The left hand side shows the N-CoR4 deletion mutants, and experiments were performed as in (A). (C) The interaction with N-CoR4 was mapped to the WD-40 repeat region of TBL1/TBLR1. The deletion constructs of TBL1 and TBLR1 were as indicated. The results of pull-down assays are summarized on the right side of each construct.
Two distinct regions in TBL1/TBLR1 interact with N-CoR
We next wished to determine the N-CoR interaction region(s) in TBL1 and TBLR1. By in vitro pull-down assay, we first confirmed that the N-CoR1 fragment containing the RD1 interacts only with the N-terminal, but not the C-terminal region of TBL1/TBLR1 (Guenther et al., 2000; Zhang et al., 2002; data not shown). In contrast, using immobilized GST–N-CoR4, we found that the WD-40 repeat region (TBL1-2) interacted specifically with the N-CoR4 region. Further mapping showed that the region containing the first three WD-40 repeats (TBL1-4B) was sufficient for this interaction. Inter estingly, the first three WD-40 repeats of TBLR1 (TBLR1-4C) by themselves failed to bind to GST– N-CoR4, and a short sequence in the N-terminal region is essential for binding (Figure 3C). Nevertheless, the WD-40 repeat region of TBLR1 is essential for binding, as no binding was detected for the TBLR1-3 fragment.
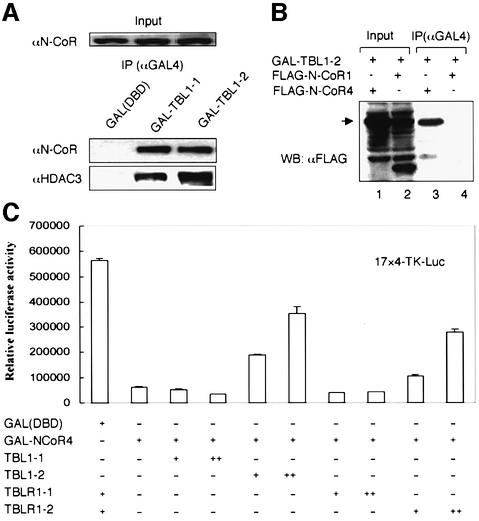
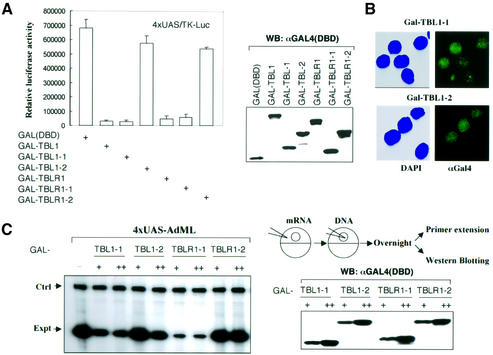
To substantiate the in vitro interaction results described above, we co-transfected an expression construct encoding either the TBL1-1 or TBL1-2 region fused to the GAL4 DNA-binding domain (DBD) into HeLa cells. Subsequent immunoprecipitation experiments revealed the association of endogenous N-CoR and HDAC3 with both TBL1-1 and TBL1-2, but not with the control GAL4(DBD) (Figure 4A). To substantiate further our in vitro results that the WD-40 repeats of TBL1/TBLR1 interact with N-CoR4 fragment, we co-transfected GAL4-TBL1-2 with Flag-tagged N-CoR1 or N-CoR4 into HeLa cells. The result in Figure 4B indicates that N-CoR4 but not the N-CoR1 fragment co-immunoprecipitated with GAL4-TBL1-2. Similar results were obtained when TBLR1-1 and TBLR1-2 were used (data not shown).
Fig. 4. Both N-CoR interaction domains in TBL1 are sufficient for interaction with endogenous N-CoR. (A) HeLa cells were transfected with a GAL4(DBD), GAL4-TBL1-1 or GAL4-TBL1-2 expression construct and, after 24 h, cell extracts were prepared and immunoprecipitated with a GAL4(DBD)-specific antibody. The association with N-CoR and HDAC3 was revealed by western analysis. (B) The WD-40 repeat domain of TBL1 interacts specifically with the N-CoR4 region in HeLa cells. The GAL4-TBL1-2 construct was co-transfected with either a Flag-tagged N-CoR1 or N-CoR4 expression plasmid into HeLa cells. The interaction was assayed by immunoprecipitation using a GAL(DBD)-specific antibody followed by western analysis using a FLAG-specific antibody (M2, Sigma). (C) The N-CoR4 region repressed transcription when tethered to DNA, and this repression could be partially blocked by TBL1/TBLR1 WD-40 repeats. The amount of plasmids used for transfection: 4xUAS/TK-Luc reporter vector, 0.2 µg, and expression vectors, +, 0.3 µg; ++, 1.0 µg. The luciferase data, expressed as RLU, are the mean ± SD of three independent transfection experiments.
Taken together, we conclude that two distinct domains of TBL1/TBLR1 interact with two different regions in N-CoR. The N-terminal domain of TBL1/TBLR1 interacts with N-CoR RD1, whereas the first three WD-40 repeats in the C-terminal domain interact with N-CoR residues 1801–1965.
The identification of a fourth repression domain in N-CoR
Previous studies have identified three autonomous repression domains in N-CoR (RD1–3, Figure 3A) (Horlein et al., 1995; Ordentlich et al., 1999; Park et al., 1999). The ability of N-CoR4 fragment to interact with TBL1/TBLR1 prompted us to test if this region also exhibits repression activity when tethered to DNA. As shown in Figure 4C, a GAL4(DBD) fusion of the N-CoR4 fragment could effectively repress a TK-luciferase reporter bearing four GAL4-binding sites in transient transfection assays. We thus designated this region as the RD4 of N-CoR. The repression activity of RD4 appears to correlate with interaction with TBL1/TBLR1, because co-expression of TBL1/TBLR1 WD-40 repeat domains (dominant-negative) could partially block the repression activity, whereas co-expression of the N-terminal regions which do not interact with the N-CoR4 fragment failed to do so (see Supplementary figure 1 available at The EMBO Journal Online).
TBL1/TBLR1 bind histones H2B and H4 through the N-terminal domain
We next analyzed the interaction of TBL1/TBLR1 with histones. Histone proteins were individually in vitro translated and labeled with [35S]methionine, and their binding to TBL1 and TBLR1 was compared with that of RbAp48 using in vitro pull-down assays. The results (Figure 5A) showed that H2A and H3 exhibited considerable binding to GST–RbAp48 but not to GST–TBL1/TBLR1. On the other hand, H2B bound well to GST–TBL1/TBLR1 but not to GST–RbAp48, whereas H4 bound to all three proteins. In all cases, no binding to GST was observed for any histone proteins, indicating that the observed interaction with histones is specific to TBL1/TBLR1. We also determined the domain in TBL1/TBLR1 responsible for interaction with H2B and H4. As shown in Figure 5B, both H2B and H4 bound to the N-terminal region of TBL1/TBLR1 (GST–TBL1-1 and GST– TBLR1-1), but not the WD-40 repeat region. The results of histone interaction are summarized in Figure 5C.
Fig. 5. Both TBL1 and TBLR1 interact with core histones H2B and H4. (A) GST pull-down assays with GST–TBL1 or TBLR1 and in vitro translated, [35S]methioine-labeled core histones. Bound core histone was detected by fluorography. GST–RbAp48 was used as positive control. (B) In vitro translated histones H2B or H4 were incubated with the indicated GST fusion proteins and the interaction was determined by GST pull-down. (C) Diagram summarizing the results of GST pull-down assays. (D) TBL1 is capable of binding to in vitro reconstituted nucleosomes. In vitro reconstitution and purification of mononucleosome were essentially as described previously (Wong et al., 1995). The retention of nucleosomes by GST–TBL1 was revealed by western analysis using core histone-specific antibodies.
To test whether TBL1 also binds to histones in chromatin, we reconstituted a mononucleosome in vitro with a 160 bp DNA fragment derived from the Xenopus TRβA promoter and purified core histones using a salt dialysis protocol as described previously (Wong et al., 1995). The resulting nucleosomes were tested for binding to GST–TBL1 by in vitro pull-down assay. The results in Figure 5D showed that all four core histones were retained by GST–TBL1 but not by control GST, indicating that TBL1 is capable of binding to this nucleosome, presumably through its interaction with histones H2B and H4.
Repression by TBL1/TBLR1 may require interaction with histones
As subunits of SMRT and N-CoR complexes, tethering TBLR1 to DNA through GAL4(DBD) would be expected to result in transcription repression, as reported for TBL1 (Guenther et al., 2000). Indeed, as shown in Figure 6A, transfection of GAL4-TBL1 and GAL4-TBLR1 into HeLa cells repressed transcription from a TK-luciferase reporter bearing four GAL4-binding sites. As our interaction assays clearly demonstrate that both the N- and C-terminal regions of TBL1/TBLR1 interact with N-CoR, one would expect that tethering either N- or C-terminal regions to DNA would lead to repression. Surprisingly, we found that while transfection of the N-terminal region fused to GAL4(DBD) (GAL-TBL1-1 and GAL-TBLR1-1) resulted in repression, the C-terminal region fusions (GAL-TBL1-2 and GAL-TBLR1-2) failed to repress transcription from the same reporter. The failure in repression was not due to variation in protein expression, since western analysis using a GAL4(DBD)-specific antibody showed similar expression of all TBL1/TBLR1 derivatives. In addition, as shown in Figure 6B, both GAL-TBL1-1 and GAL-TBL1-2 were localized to nuclei, as revealed by immunostaining using a GAL(DBD)-specific antibody. Furthermore, the same results were observed when the transcription assays were performed in Xenopus oocytes. In this case, in vitro synthesized mRNAs encoding the N- or C-terminal region of TBL1/TBLR1 fused to GAL4(DBD) were injected into Xenopus oocytes, followed by injection of an AdML (adenovirus major later promoter)-based reporter bearing four GAL4-binding sites. While the N-terminal region from both TBL1 and TBLR1 repressed transcription effectively, both C-terminal GAL4 fusion proteins showed little, if any repression activity (Figure 6C). Again, western analysis revealed fairly equal levels of expression for all GAL4 fusion proteins. Given that both N- and C-terminal regions of TBL1/TBLR1 show independent interaction with N-CoR (Figures 3 and 4), these results indicate that interaction with N-CoR alone is not sufficient for repression. Since the N-terminal region of TBL1/TBLR1 also interacts with histones (Figure 5), these results suggest that the interaction with histones may be required for repression, although a possible contribution by another interaction cannot be excluded.
Fig. 6. The repression activity of TBL1/TBLR1 correlates with histone interaction but not with N-CoR interaction. (A) Repression assay in HeLa cells. HeLa cells were transiently transfected with 0.2 µg of 4xUAS/TK-Luc reporter and GAL4(DBD) fusion plasmids as indicated. Whole-cell extracts were used in the luciferase assay and western blotting employing anti-GAL4(DBD) antibody. The luciferase data, expressed as RLU, are the mean ± SD of three independent transfection experiments. (B) Immunostaining showing that both GAL-TBL-1 and GAL-TBL1-2 were nuclear proteins. (C) Repression assay in Xenopus oocytes. Groups of Xenopus oocytes were injected with mRNAs encoding GAL4(DBD) fusion proteins as indicated at two different concentrations (+, 100 ng/µl, 18.4 nl/oocyte; and ++, 300 ng/µl, 18.4 nl/oocyte) and a 4xUAS-AdML reporter (25 ng/µl, 18.4 nl/oocyte). The injected oocytes were incubated overnight, and the levels of transcription were then analyzed by primer extension assay. Ctrl, the primer extension product from the endogenous storage histone H4 mRNA; Expt, the primer extension product from the injected reporter. The expression of fusion proteins was detected by western analysis using a GAL4(DBD)-specific antibody.
Functional analysis of components of SMRT–N-CoR complexes using siRNAs: the importance of HDAC3 and TBL1/TBLR1 in TR repression
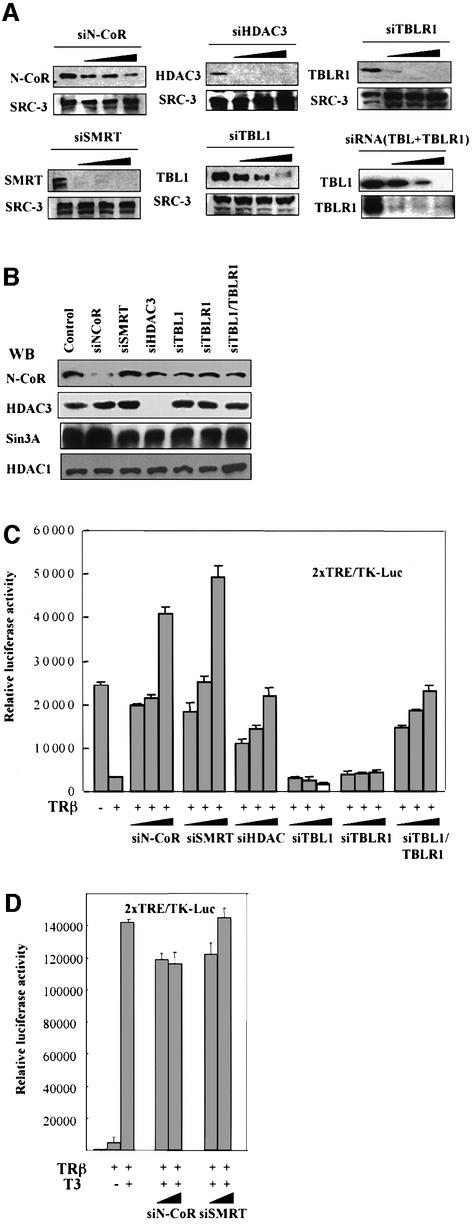
Despite strong biochemical evidence for HDAC3 being the major HDAC associated with SMRT–N-CoR complexes, the roles of HDAC3 in repression mediated by mammalian SMRT–N-CoR is much less clear (Jepsen et al., 2000). Furthermore, as TBL1/TBLR1 are required neither for HDAC3 activity in SMRT–N-CoR complexes nor for interaction with unliganded receptors (Guenther et al., 2000; Zhang et al., 2002), the roles of TBL1/TBLR1 in SMRT–N-CoR complexes, if any, remain to be determined. Recent studies indicate that 21–23 nucleotide double-stranded RNA (siRNA) can act as a guide sequence within a multicomponent nuclease complex to target complementary mRNA for degradation (Elbashir et al., 2002). To address the role of HDAC3 and TBL1/TBLR1 in SMRT–N-CoR complex function, we thus designed siRNA against individual components of SMRT–N-CoR complexes (see Materials and methods). To test the effect and specificity of siRNA, HeLa cells were transfected with siRNA against different components of N-CoR–SMRT complexes. Two days after transfection, HeLa cells were collected, and the effect of siRNA was analyzed by western blotting. siRNA treatment clearly had an effect on the expression of their corresponding target proteins. The siRNAs against SMRT, HDAC3 and TBLR1 worked more efficiently than the siRNAs against N-CoR and TBL1 (Figure 7A). As a control, we measured the levels of SRC-3, a coactivator for nuclear receptors, which showed no significant change after any siRNA treatment. In agreement with the notion that siRNA is highly specific, we have not observed any significant cross-effect by siRNAs used (see Figure 7B).
Fig. 7. The roles of HADC3 and TBL1/TBLR1 in SMRT–N-CoR function. (A) ‘Knock-down’ components of SMRT–N-CoR complexes by siRNAs. HeLa cells were cultured with hormone-free medium and transfected with an increasing amount of siRNA (7.5, 15 and 30 nM) and, 2 days after transfection, cells were harvested and the levels of the corresponding target proteins were analyzed by western blotting. The same blots were reprobed with an SRC-3-specific antibody (negative control). (B) Specificity of siRNAs. Western analyses showed that each siRNA used was highly specific for its target and had little, if any, cross-effect or no specific effect. (C) HDAC3 is essential and TBL1 and TBLR1 are functionally redundant in mediating repression by un liganded TR. HeLa cells were first transfected with siRNA as in (A), and 2 days after were transfected again with 1.0 µg of pSG5-TRβ and 0.2 µg of 28TRE/TK-Luc reporter. Cells were harvested after another 2 days. Whole-cell extracts were used in the luciferase assay. The luciferase data, expressed as RLU, are the mean ± SD of three independent transfection experiments. (D) Treatment with siN-CoR and siSMRT had no effect on transcriptional activation in the presence of T3. The experiment was as in (C) for siN-CoR and siSMRT except that two doses of siRNA (15 and 30 nM) were used and T3 was added 12 h before harvesting cells for luciferase assay.
We next tested the effect of these specific siRNAs on TR repression. HeLa cells were first transfected with siRNA as indicated and, after 48 h, were transfected again with a human TRβ expression construct and a luciferase reporter bearing two TR response elements (2× TRE-TK-luc). As shown in Figure 7C, while transfection of TRβ led to an ∼10-fold repression, treatment with siRNA against N-CoR or SMRT effectively relieved repression. Interestingly, the ‘relief’ of repression was observed under the condition where the N-CoR level was reduced only ∼50% (dosage 1 and 2). This result is somewhat surprising since N-CoR and SMRT would be expected to be functionally redundant. One likely explanation is that unliganded TR could partially activate transcription if not associated with corepressor proteins, as suggested by experiments in yeast (Ohashi et al., 1991). In this case, the effect of siRNA against N-CoR or SMRT on transcription would be the combinatorial effect of ‘relief’ of repression as well as ‘activation’ by corepressor-free unliganded TR. As expected, the effect of siRNAs against N-CoR and SMRT is specific for repression and had no effect on activation induced by liganded TRβ (Figure 7D).
Treatment with HDAC3 siRNA also relieved the repression by unliganded TR, indicating that HDAC3 is probaby the major HADC required for repression mediated by SMRT–N-CoR complexes. With regard to the function of TBL1 and TBLR1 in TR repression, siRNA against either TBL1 or TBLR1 alone had very minimal, if any, effect on TR repression. Importantly, treatment with siRNAs against both TBL1 and TBLR1 could attenuate the repression effectively (Figure 7C). As siRNA against TBL1 and TBLR1 can knock-down the corresponding protein independently under our experimental conditions (Figure 7A), these results demonstrate a redundant but essential role for TBL1 and TBLR1 in mediating repression by unliganded TR.
HDAC3 and TBL1/TBLR1 are also required for repression of an endogenous TR target gene
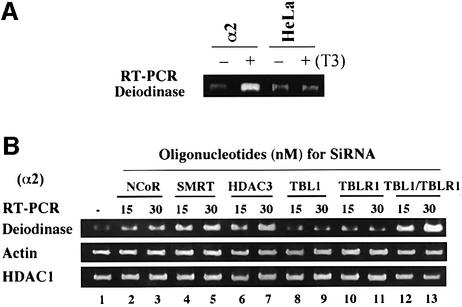
Our previous study demonstrated that in the HeLa α2 cell line, which expresses a stable integrated Flag-tagged TRα, expression of the deiodinase gene is regulated by T3 (Sharma and Fondell, 2002). RT–PCR analysis in Figure 8A confirmed that the deiodinase gene could be induced by T3 treatment (12 h) only in α2 cells but not in the parental cells lacking TR. We thus made use of HeLa α2 cells to test the effect of various siRNAs on deiodinase gene expression. The HeLa α2 cells were transfected with two different concentrations of siRNA (15 and 30 nM), and 2 days after transfection the cells were collected and the levels of transcription from the deiodinase gene were measured by quantitative RT–PCR. The results in Figure 8B showed that treatment with siRNA against N-CoR or SMRT increased the levels of transcription from the deiodinase gene. siRNA against HDAC3 also led to increased deiodinase gene expression, thus further substantiating the importance of HDAC3 in mediating TR repression. To our satisfaction, we found that siRNA against either TBL1 or TBLR1 alone had a minimal effect on deiodinase gene expression. However, siRNAs against TBL1 and TBLR1 together resulted in a clear increase in deiodinase gene expression. In contrast, RT–PCR using the same groups of samples showed no significant changes in levels of actin and HDAC1 mRNAs. These results thus nicely corroborate the results in Figure 7 obtained using transient transfection assays, and further establish an essential, but redundant role for TBL1 and TBLR1 in SMRT–N-CoR complex function.
Fig. 8. HDAC3 and TBL1/TBLR1 are also essential for repression of an endogenous TR target gene, deiodinase. (A) RT–PCR analysis confirmed that the deiodinase gene was regulated by TRα in HeLa α2 cells. (B) HeLa α2 cells were transfected with 15 or 30 nM of siRNA as indicated. Two days after transfection, total RNA was prepared from each sample and used for quantitative RT–PCR to measure the expression of the TR target gene deiodinase. As controls, the levels of actin and HDAC1 mRNA were also measured by quantitative RT–PCR using the same batch of RNA samples.
Discussion
Components of N-CoR and SMRT complexes
We and others have shown previously that corepressors SMRT and N-CoR exist in 1.5–2 MDa large protein complexes containing HDAC3 (Li et al., 2000; Guenther et al., 2001). Recently, Zhang et al. (2002) reported the purification of the N-CoR complex while this manuscript was in preparation. In addition to HDAC3 and TBL1, they also identified TBLR1 and GPS2 as the stoichiometric subunits of N-CoR complexes. By using a combination of conventional and immmunoaffinity chromatography, we purified the N-CoR complex from HeLa nuclear extracts. The purified N-CoR complex contains 10–12 proteins (Figure 1C) and is thus more consistent with the estimated size of 1.5–2 MDa.
In comparison with GPS2, HDAC3, TBL1 and TBLR1, which are likely to be the stoichiometric core subunits (Figure 1C; Zhang et al., 2002), other proteins co-purified with N-CoR appear to be less abundant (substoichometric). It is unclear whether these less abundant proteins only associate with a fraction of the N-CoR complex or if they dissociate during purification. Among these less abundant proteins, we identified a novel N-CoR-associated protein, IR10. IR10 is coronin-like actin-binding protein (Zaphiropoulos and Toftgard, 1996). Interestingly, IR10 was found to associate with N-CoR but not SMRT. Thus, IR10 is the first protein shown to be differentially associated with N-CoR and SMRT complexes. The function of IR10 in the N-CoR complex remains to be determined.
Although we have yet to identify all the proteins in our purified N-CoR complex, western blotting analysis allowed us to exclude the presence of both Sin3A and BRG1–BAF. Furthermore, our extensive reciprocal immunoprecipitation and western analyses provided no evidence for the association of either Sin3A or BRG1–BAF complexes with N-CoR in HeLa nuclear extracts (H.-G.Yoon and J.Wong, data not shown). While these results contradict two reports on purification of N-CoR complexes (Underhill et al., 2000; Jones et al., 2001), our data are in agreement with results from other groups and our own previous results (Guenther et al., 2000; Huang et al., 2000; Li et al., 2000; Rietveld et al., 2002; Zhang et al., 2002). In view of recent reports that SMRT–N-CoR and Sin3A are distinct class I HDAC-containing complexes that can be targeted to gene repression by different transcription factors (Li et al., 2002; Rietveld et al., 2002), we conclude that Sin3A and BRG1–BAF components are unlikely to exist in stable complexes with N-CoR.
Protein–protein interaction network in the N-CoR complex
Among the core subunits of the N-CoR–SMRT complex, N-CoR and SMRT have been reported to interact directly with HDAC3, TBL1 and GPS2 (Guenther et al., 2000; Zhang et al., 2002), whereas GPS2 also binds to TBL1 and TBLR1 (Zhang et al., 2002). The interaction between TBL1 and SMRT–N-CoR was mapped previously to the RD1 of SMRT and N-CoR and the N-terminal non-WD-40 repeat region in TBL1 (Guenther et al., 2000; Zhang et al., 2002). While this interaction was confirmed in this study, we have identified a second interaction between TBL1/TBLR1 and N-CoR that involves the TBL1/TBLR1 WD-40 repeat region and N-CoR RD4 (Figures 3 and 4). This second interaction between TBL1/TBLR and N-CoR is supported by various interaction and co-immunoprecipitation experiments and is mapped to the first three WD-40 repeat regions in TBL1/TBLR1 and the amino acids 1801–1965 of N-CoR. The presence of two independent interactions could either allow the simultaneous binding of both TBL1 and TBLR1 to N-CoR and/or contribute to the stable association of TBL1/TBLR1 with N-CoR. Future work will determine whether such two independent interactions also occur for SMRT.
The role of HDAC3 and TBL1/TBLR1 in repression mediated by SMRT–N-CoR complexes
While our purification data and that of others clearly establish HDAC3 as the major HDAC associated with SMRT–N-CoR complexes, previous studies using antibody injection have failed to show any role for HDAC3 in repression mediated by mammalian SMRT and N-CoR (Jepsen et al., 2000). By using siRNAs that suppress gene expression by targeting specifically the mRNA containing the complementary sequence for degradation (Elbashir et al., 2002), we showed that treatment of HeLa cells with an HDAC3-specific siRNA relieved repression of a transiently transfected TR reporter as well as an endogenous TR target gene (Figures 7 and 8). Thus, in accord with the fact that HDAC3 is the major HDAC associated with SMRT–N-CoR complexes, our result establishes a crucial role for HDAC3 in repression mediated by SMRT–N-CoR complexes.
By using siRNA specific for either TBL1 or TBLR1, we found that ‘knock-down’ of either one had no significant effect on repression by TR of either transfected reporter or an endogenous target gene, deiodinase. However, simultaneous treatment with both TBL1 and TBLR1 siRNAs resulted in loss of repression function of TR. These results indicate that TBL1 and TBLR1 are functionally redundant but essential for SMRT–N-CoR complex functions, at least in mediating repression by unliganded TR. As TBL1 and TBLR1 share 89% sequence identity over the entire sequence, a functional redundancy between TBL1 and TBLR1 is not unexpected.
Interaction of TBL1/TBLR1 with histones and its potential function in repression
In SMRT–N-CoR complexes, previous studies indicate that TBL1 and TBLR1 are required neither for HDAC3 activity nor for binding of SMRT–N-CoR to unliganded TR, since SMRT and N-CoR interact directly with HDAC3 through a deacetylase-activating domain (DAD), and this interaction activates HDAC3 activity (Guenther et al., 2001; Zhang et al., 2002). Why would TBL1/TBLR be required for repression by unliganded TR? The answer to this question may lie in their ability to interact with histones (Figure 5). Like RbAp46 and RbAp48 in Sin3A and Mi-2–NURD complexes, TBL1 and TBLR1 interact with core histones, although with distinct specificity (Figure 5). While RbAp48 binds to H2A, H3 and H4, TBL1 and TBLR1 bind preferentially to H2B and H4 (Figure 5). Such a difference in histone interaction may allow simultaneous binding of SMRT–N-CoR complexes and Sin3A or Mi-2–NURD complexes to chromatin for repression. In vitro, TBL1 is also capable of binding to a reconstituted nucleosome, presumably through its ability to interact with H2B and H4. It is noteworthy that TBL1 previously was reported to interact with histone H3 by a pull-down assay using nuclear extracts as input (Guenther et al., 2000). The discrepancy with our result could be explained by the association of histone H3 with other histones such as H4. The interaction with histones H2B and H4 was mapped to the N-terminal region of TBL1 and TBLR1 (Figure 6). Importantly, although both the N-terminal domain and the C-terminal WD-40 repeat domain are sufficient for interaction with N-CoR, only tethering of the N-terminal domain to DNA through a GAL4 DBD is capable of repressing transcription both in mammalian cells and in Xenopus oocytes (Figure 7). Therefore, these results correlate repression activity by TBL1/TBLR1 directly with histone interaction and provide evidence to suggest that the essential role of TBL1/TBLR1 in repression mediated by SMRT–N-CoR complexes lies in their interaction with histones.
In support of this idea, a recent study has demonstrated elegantly a crucial role for histone interaction in targeting the Tup1–Ssn6 corepressor complex for repression in yeast (Davie et al., 2002). Like TBL1/TBLR1, Tup1 contains six WD-40 repeats and binds histones, and its repression activity correlates with its histone-binding activity (Edmondson et al., 1996). Deletion of the histone H3 tail or mutation of the H4 tail was found to severely reduce the binding of Tup1 to its target genes and compromise its repression function (Davie et al., 2002). Taken together, histone interaction seems to be a conserved function among the WD-40 repeat proteins present in all class I HDAC-containing corepressor complexes. Thus, it is tempting to propose a working model in which chromatin-bound, unliganded TR first interacts with SMRT–N-CoR. This interaction alone, however, is not sufficient for stable targeting of the SMRT–N-CoR complexes to chromatin. The interaction between TBL1/TBLR1 and histones (H2B and H4) presumably is required for stable association of the SMRT–N-CoR complexes with chromatin, which in turn is required for efficient histone deacetylation by HDAC3 and subsequent transcriptional repression.
Materials and methods
Constructs and siRNAs
The GST–TBL1 construct was kindly provided by Dr H.G.Stunnenberg (University of Nijmegen). Full-length TBLR1 cDNA was generated by RT–PCR according to GI:15294576. The various TBL1 and TBLR1 deletions were generated by high fidelity PCR and subcloned into pSG5-HH2M1 (Sigma) for in vitro translation, pGEX4T-1 for GST fusions, pCMV-GAL4(DBD) for expression in mammalian cells as GAL4 fusions, and pSP64poly(A)-GAL4 vector for Xenopus oocyte injection. The N-CoR deletion constructs used for in vitro interaction were as described previously (Li et al., 2000). The siRNAs for N-CoR, SMRT, TBL1, TBLR1 and HDAC3 were chemically synthesized (Dharmacon Research, Lafayette, Co.), de-protected, annealed and transfected according to the manufacturer’s instructions. The siRNA sequences are as following: N-CoR, 2218AAUGCUACUUCUCGAGGAAACA2238; SMRT, 2584AAGGGUAUCAUCACCGCUGUG2604; HDAC3, 859AAU AUCCCUCUACUCGUGCUGA879; TBL1, 151AAGAUGAUCAGCA UAACCAGUGAC171; and TBLR1, 797AAGGCCCUAUAUUUGCAU UAA817.
Purification of N-CoR complex from HeLa nuclear extract
HeLa nuclear extracts were prepared as described previously (Dignam et al., 1983). HeLa nuclear extracts derived from 50 l of HeLa cells (National Cell Culture Center) were applied for chromatography essentially as described previously (Li et al., 2000) except using the scheme shown in Figure 1A. The N-CoR eluted from the PC11 column in the 0.5 M fraction was pooled together for affinity purification using N-CoR (N) antibody affinity resins essentially as described previously (Li et al., 2000). The proteins eluted from the N-CoR affinity resins were separated by 8% SDS–PAGE and identified by mass spectrometry as described previously (Qin et al., 1997).
Cell culture, transfection and RT–PCR
HeLa and α2 cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% charcoal-stripped serum with standard cell culture techniques. For transfection of siRNA, cells were plated at 40–50% confluency and transfected with various amounts of siRNA by using TransIT-TKO transfection reagent (Mirus) as instructed. For the experiments in Figure 7, 1.0 µg of pSG5-TRβ and 28TRE/TK-Luc reporter constructs were transfected using Lipofectamine 2000 (Invitrogen) 2 days after transfection of siRNA. Luciferase activity was quantitated using a luminometer (Dynex Technologies) and is expressed as arbitrary light units (RLU). For RT–PCR, total RNA was isolated from transfected cells using the RNeasy Mini kit (Qiagen), reverse transcribed with random primers by using a StrataScript™ reverse transcriptase kit (Stratagene) and followed by quantitative PCR.
GST pull-down assay, immunoprecipitation and western blotting
All in vitro translation and labeling with [35S]methionine were performed using the indicated expression constructs and a TNT T7 coupled transcription/translation kit from Promega. Various GST fusion proteins were expressed in Escherichia coli BL21 strain and affinity purified using glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech) according to the manufacturer’s instructions. GST pull-down and immunoprecipitation using HeLa nuclear extracts (100 µl) or whole-cell extracts derived from transfected cells were essentially as described (Li et al., 2000). The in vitro reconstitution and purification of mononcleosomes were as described previously (Wong et al., 1995). The antibodies against N-CoR, SMRT and HDAC1 were as described previously (Li et al., 2000). The antibody against TBL1 and TBLR1 was raised against GST–TBL1 and a synthetic peptide CGSAKNGENTANGEEN (Genemed Synthesis, Inc.), respectively. No cross-reaction was detected between TBL1 and TBLR1 antibodies (data not shown). The commercial antibodies used include HDAC3 monoclonal antibody (BD Transduction Laboratories), HDAC4 and core histones (Cell Signaling), FLAG-specific M2 (Sigma), Sin3A and GAL4(DBD) (Santa Cruz Biotechnology).
In vitro mRNA preparation and microinjection of Xenopus oocytes
The SP64poly(A)-GAL4(DBD)s containing various TBL1 and TBLR1 fragments were first linearized with EcoRI, and mRNA synthesis was then carried out using a SP6 Message Machine kit (Ambion, Inc.) as described by the manufacturer. The preparation of stage VI Xenopus oocytes, microinjection, and transcription analysis by primer extension were essentially as described previously (Wong et al., 1998).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank H.G.Stunnenberg for GST–TBL1 plasmid, Weidong Wang for antibodies against BRG1, BAF57 and BAF 170, Robert Roeder for GPS2 antibody and R.Toftgard for IR10 antibody. This work is supported by grants DK56324 and DK58679 to J.W.
References
- Baniahmad A., Steiner,C., Kohne,A.C. and Renkawitz,R. (1990) Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell, 61, 505–514. [DOI] [PubMed] [Google Scholar]
- Chen J.D. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Davie J.K., Trumbly,R.J. and Dent,S.Y. (2002) Histone-dependent association of Tup1–Ssn6 with repressed genes in vivo. Mol. Cell. Biol., 22, 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Weber,K. and Tuschl,T. (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods, 26, 199–213. [DOI] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Glass C.K., Lipkin,S.M., Devary,O.V. and Rosenfeld,M.G. (1989) Positive and negative regulation of gene transcription by a retinoic acid–thyroid hormone receptor heterodimer. Cell, 59, 697–708. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Lane,W.S., Fischle,W., Verdin,E., Lazar,M.A. and Shiekhattar,R. (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev., 14, 1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Barak,O. and Lazar,M.A. (2001) The smrt and n-cor corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol., 21, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Horlein A.J. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Huang E.Y., Zhang,J., Miska,E.A., Guenther,M.G., Kouzarides,T. and Lazar,M.A. (2000) Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev., 14, 45–54. [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jepsen K. et al. (2000) Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell, 102, 753–763. [DOI] [PubMed] [Google Scholar]
- Jones P.L., Sachs,L.M., Rouse,N., Wade,P.A. and Shi,Y.B. (2001) Multiple N-CoR complexes contain distinct histone deacetylases. J. Biol. Chem., 276, 8807–8811. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Downes,M., Ordentlich,P. and Evans,R.M. (2000) Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev., 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- Li J., Wang,J., Nawaz,Z., Liu,J.M., Qin,J. and Wong,J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J., 19, 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lin,Q., Wang,W., Wade,P. and Wong,J. (2002) Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev., 16, 687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L., Kao,H.Y., Chakravarti,D., Lin,R.J., Hassig,C.A., Ayer,D.E., Schreiber,S.L. and Evans,R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Ohashi H., Yang,Y.F. and Walfish,P.G. (1991) Rat liver c-erb Aβ1 thyroid hormone receptor is a constitutive activator in yeast (Saccharomyces cerevisiae): essential role of domains D, E and F in hormone-independent transcription. Biochem. Biophys. Res. Commun., 178, 1167–1175. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Downes,M., Xie,W., Genin,A., Spinner,N.B. and Evans,R.M. (1999) Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc. Natl Acad. Sci. USA, 96, 2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.J., Schroen,D.J., Yang,M., Li,H., Li,L. and Chen,J.D. (1999) SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc. Natl Acad. Sci. USA, 96, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Fenyo,D., Zhao,Y., Hall,W.W., Chao,D.M., Wilson,C.J., Young,R.A. and Chait,B.T. (1997) A strategy for rapid, high-confidence protein identification. Anal. Chem., 69, 3995–4001. [DOI] [PubMed] [Google Scholar]
- Rietveld L.E., Caldenhoven,E. and Stunnenberg,H.G. (2002) In vivo repression of an erythroid-specific gene by distinct corepressor complexes. EMBO J., 21, 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Sharma D. and Fondell,J.D. (2002) Ordered recruitment of histone acetyltransferases and the TRAP/mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl Acad. Sci. USA, 99, 7934–7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill C., Qutob,M.S., Yee,S.P. and Torchia,J. (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem., 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Gegonne,A., Jones,P.L., Ballestar,E., Aubry,F. and Wolffe,A.P. (1999) Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet., 23, 62–66. [DOI] [PubMed] [Google Scholar]
- Wen Y.D., Perissi,V., Staszewski,L.M., Yang,W.M., Krones,A., Glass,C.K., Rosenfeld,M.G. and Seto,E. (2000) The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl Acad. Sci. USA, 97, 7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. (1998) Chromatin: Structure and Function. Academic Press, San Diego, CA, pp. 97–108.
- Wong J., Shi,Y.B. and Wolffe,A.P. (1995) A role for nucleosome assembly in both silencing and activation of the Xenopus TRβA gene by the thyroid hormone receptor. Genes Dev., 9, 2696–711. [DOI] [PubMed] [Google Scholar]
- Wong J., Patterton,D., Imhof,A., Guschin,D., Shi,Y.B. and Wolffe,A.P. (1998) Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J., 17, 520–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos P.G. and Toftgard,R. (1996) cDNA cloning of a novel WD repeat protein mapping to the 9q22.3 chromosomal region. DNA Cell Biol., 15, 1049–1056. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kalkum,M., Chait,B.T. and Roeder,R.G. (2002) The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell, 9, 611–623. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Iratni,R., Erdjument-Bromage,H., Tempst,P. and Reinberg,D. (1997) Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell, 89, 357–364. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]