Abstract
Recent reports have increased our knowledge of the consecutive steps during 60S ribosome biogenesis substantially, but 40S subunit formation is less well understood. Here, we investigate the maturation of nucleolar 90S pre-ribosomes into cytoplasmic 40S pre-ribosomes. During the transition from 90S to 40S particles, the majority of non-ribosomal proteins (∼30 species) dissociate, and significantly fewer factors associate with 40S pre-ribosomes. Notably, some of these components are part of both early 90S and intermediate 40S pre-particles in the nucleolus (e.g. Enp1p, Dim1p and Rrp12p), whereas others (e.g. Rio2p and Nob1p) are found mainly on late cytoplasmic pre-40S subunits. Finally, temperature-sensitive mutants mapping either in earlier (enp1-1) or later (rio2-1) components exhibit defects in the formation and nuclear export of pre-40S subunits. Our data provide an initial biochemical map of the pre-40S ribosomal subunit on its path from the nucleolus to the cytoplasm. This pathway involves fewer changes in composition than seen during 60S biogenesis.
Keywords: Enp1/nuclear export/ribosome biogenesis/ribosome export/40S subunit
Introduction
The biogenesis of eukaryotic ribosomes is a multistep process that initiates in the nucleolus with the transcription of ribosomal genes and ends in the cytoplasm after export and final maturation of 60S and 40S subunits (Venema and Tollervey, 1999). rDNA is arranged in tandem repeats within the nucleolar chromatin and transcribed by two distinct RNA polymerases, with synthesis of a large 35S primary transcript by RNA polymerase I (Pol I) and pre-5S rRNA synthesis by RNA polymerase III (Pol III). The 35S rRNA precursor is processed to yield the mature 25S, 18S and 5.8S rRNAs. During this process, numerous factors of different function associate with the 35S pre-rRNA and its processing intermediates, including small nucleolar ribonucleoprotein complexes (snoRNPs) and many non-ribosomal proteins. The latter include endo- and exonucleases, which process the pre-rRNA, pseudouridine synthases and methyltransferases, which catalyze covalent nucleotide modifications, and helicases and chaperone-like factors, which probably facilitate RNA folding, RNP remodeling and protein association/dissociation (reviewed in Kressler et al., 1999; Venema and Tollervey, 1999; Warner, 2001). In this way, a large (>2 MDa) complex composed of the U3 snoRNA and associated Utp proteins, termed the ‘terminal knob’ (Miller and Beatty, 1969; Mougey et al., 1993) or small subunit (SSU) processome (Dragon et al., 2002), co-transcriptionally assembles at the 5′ end of the nascent pre-rRNA. Initial work three decades ago identified important pre-ribosomal particles including the 90S pre-ribosome that contains the 35S pre-rRNA, and the derived 66S and 43S particles, the precursors to the 60S and 40S subunits, respectively (Udem and Warner, 1972; Trapman et al., 1975). However, the pathway of ribosome synthesis exhibits more intermediates than these studies initially indicated. In particular, several distinct pre-60S ribosomal particles were identified that differ in their content of associated non-ribosomal proteins and pre-rRNA species on the pathway of 25 rRNA and 5.8S rRNA synthesis (Baßler et al., 2001; Harnpicharnchai et al., 2001; Saveanu et al., 2001; Fatica et al., 2002; Nissan et al., 2002). The 90S pre-ribosome has also been characterized biochemically and shown to contain ∼35 non-ribosomal components including proteins associated with U3 (e.g. Nop56p, Nop58p, Sof1p, Rrp9p, Dhr1p, Imp3p, Imp4p and Mpp10p) and many other factors required for 18S rRNA synthesis (Grandi et al., 2002). Thus, the 40S synthesis machinery is associated predominantly with the 35S pre-rRNA, whereas factors required for 60S formation bind later. Moreover, the Baserga group recently reported the identification of a huge U3 snoRNP particle (>2 MDa), designated the SSU processome, which contains many U3-binding proteins, the U3 snoRNA and 17 additional proteins of unknown function, called Utp1–17 (Dragon et al., 2002). The SSU processome appears to associate co-transcriptionally with the 35S pre-rRNA and could correspond to the ‘terminal balls’ in the electron microscope, because these terminal knobs on the 35S pre-rRNA were lost in mutants which were depleted for U3 snoRNA or U3-specific proteins. Following the assembly of 90S pre-ribosomes, three early endonucleolytic cleavages in the 35S pre-rRNA (at sites A0 to A2) generate the 20S and 27SA2 pre-rRNAs, the precursors to 18S and 25S/5.8S rRNA, respectively. Consequently, these cleavages initiate 40S and 60S subunit formation. A few of the 90S factors (e.g. Enp1p) were shown to remain associated with 20S pre-rRNA (Grandi et al., 2002), whereas a different set of non-ribosomal proteins assembles onto the 27S pre-rRNAs to generate pre-60S subunits (Nissan et al., 2002). Processing of the 27S pre-rRNAs into 25S and 5.8S rRNA particles occurs in several distinct pre-60S intermediates, which pass from the nucleolus via the nucleoplasm to the nuclear periphery (Baßler et al., 2001; Harnpicharnchai et al., 2001; Saveanu et al., 2001; Fatica et al., 2002; Nissan et al., 2002). Finally, nuclear export of pre-60S particles requires Nmd3p, an adaptor protein that carries a nuclear export signal (NES) and serves to couple the large subunit protein Rpl10p to the nuclear export receptor Xpo1p/Crm1/exportin-1 (Ho et al., 2000; Gadal et al., 2001b).
Formation of the 40S subunit is less complicated than 60S biogenesis (Udem and Warner, 1973; Venema and Tollervey, 1999), since a single cleavage reaction converts 20S pre-rRNA into mature 18S rRNA, a process that requires at least Rrp10p (Rio1p) (Vanrobays et al., 2001). The 20S pre-rRNA also undergoes adenine dimethylation, catalyzed by the Dim1p dimethylase, at two positions close to its 3′ end (Lafontaine et al., 1998). Nuclear fractionation experiments indicated that both the modification and 3′ cleavage of the 20S pre-rRNA occur in the cytoplasm in yeast (Stevens et al., 1991), which is also supported by in situ localization of the excised pre-rRNA fragment (Moy and Silver, 1999). Nuclear export of 40S pre-ribosomes requires Xpo1p and nucleoporins of the Nup82p complex (Gleizes et al., 2001; Moy and Silver, 2002), but which pre-40S components carry NES signals is unknown to date.
This study reports an analysis of the composition of 40S pre-ribosomal particles. We have investigated how early 90S pre-ribosomes develop into later pre-40S particles, and defined changes in their protein and RNA composition during transport from the nucleolus to the cytoplasm. Strikingly, nucleolar 40S pre-ribosomes carry only eight major non-ribosomal proteins. Most of these remain associated during export to the cytoplasm, where they are likely to participate in the late steps of small subunit biogenesis.
Results
Biochemical separation of 90S and 40S pre-ribosomal particles
We previously isolated pre-90S ribosomal particles by tandem affinity purification (TAP) of associated components (Grandi et al., 2002). These contained the 35S pre-rRNA, small subunit ribosomal proteins (r-proteins), the U3 snoRNP and many 40S subunit processing factors, but predominantly lacked 60S synthesis factors. One of the co-purifying proteins, Enp1p, was significantly enriched in smaller 40S pre-ribosomes and was associated with the dimethlyated 20S pre-rRNA, strongly indicating that Enp1p is a component of both 90S and 40S pre-ribosomes (Grandi et al., 2002). To unravel the consecutive steps during 40S subunit formation, we concentrated on Enp1p as a non-ribosomal reporter of 40S biogenesis.
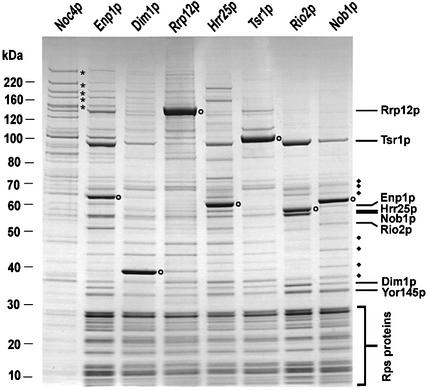
Re-examination of sedimentation on sucrose gradients revealed that Enp1p-TAP is present in two distinct peaks, one at ∼40S, and a second one at ∼90S (Figure 1A; see also Grandi et al., 2002). To affinity-purify Enp1p-TAP from each of these peaks, we scaled up the preparation and fractionated an Enp1p-TAP-containing cell lysate derived from ∼1011 cells on sucrose density gradients. The 40S and 90S peak fractions were pooled and subjected to TAP (Figure 1A). Comparison of these two purifications on the Coomassie Blue-stained SDS–polyacrylamide gel (Figure 1B) revealed the transition from 90S to 40S pre-ribosomes. As previously reported (Grandi et al., 2002), Enp1p-TAP purified from a non-fractionated whole-cell lysate (WCL) is associated with >30 non-ribosomal proteins that form the core of 90S pre-ribosomes (Figure 1B, WCL). As expected, many of these processing and assembly factors were co-enriched within the Enp1p-TAP preparation purified from the 90S pool (Figure 1B; Table I; e.g. Utp20p, Rrp5p, Utp10p, Ecm16p and Utp22p; marked by asterisks). A few bands that are superstoichiometric in the Enp1p-TAP preparation derived from the total lysate were not enriched in the Enp1p-TAP purification from the 90S pool (Figure 1B, marked by open circles). In contrast, the numerous components of the 90S particle were absent when Enp1p-TAP was isolated from the 40S pool, whereas the superstoichiometric bands were still co-enriched (Figure 1B). These major bands, which were identified as Enp1p, Dim1p, Nob1p, Hrr25p, Rrp12p, Rio2p, Trs1p and Yor145p, therefore define a pre-40S particle or family of particles. The 40S Enp1p particle also co-precipitated a few less strongly stained bands, which correspond to Nop14p/Noc5p, Ltv1p and Asc1p (Table I). These may be substoichiometric or transient components of the same pre-40S particle.
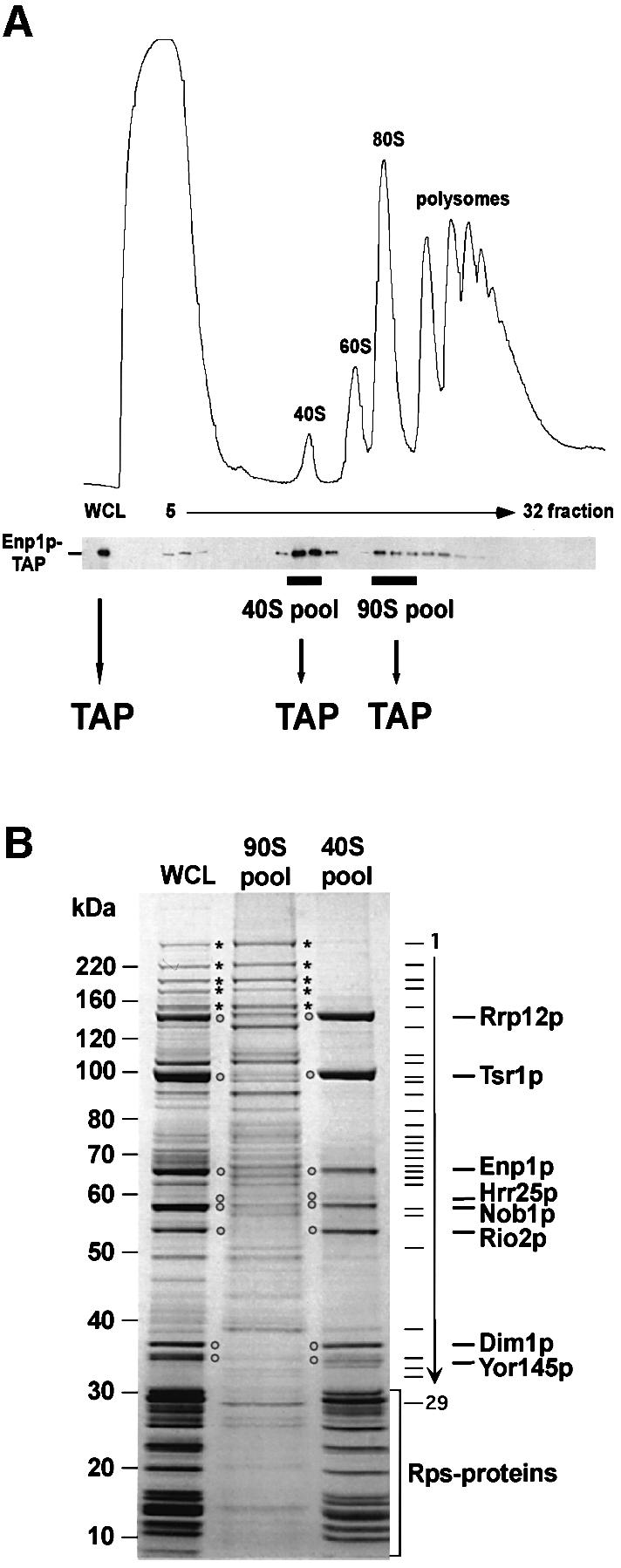
Fig. 1. Enp1p is associated with 90S and 40S pre-ribosomes. (A) The sedimentation behavior of TAP-tagged Enp1p was analyzed by sucrose density gradient centrifugation. A whole-cell lysate (WCL) and fractions 5–32 from the sucrose gradient were analyzed by OD254nm measurement (upper graph) and western blotting using anti-protein A antibodies to reveal the position of TAP-tagged Enp1p (lower panel). A WCL and the pooled 40S and 90S fractions were subjected to tandem affinity purification (TAP). (B) TAP-tagged Enp1p was isolated from an unfractionated yeast WCL, from the 90S pool and the 40S pool by the TAP method. Co-purifying proteins were separated on a 4–12% SDS–polyacrylamide gradient gel, stained with Coomassie Blue and identified by mass spectrometry (see also Table I). Major non-ribosomal proteins of the 40S pre-ribosomes are marked by open circles, high molecular weight factors of the 90S pre-ribosomes by asterisks. Bands 1–29 are 90S factors identified by MS and listed in Table I. Proteins in the low molecular weight range of the gel are predominantly ribosomal Rps proteins (see also Table I).
Table I. Identification of Enp1p-associated proteins from TAP purifications of whole-cell lysate (WCL), 90S ribosomal precursors (90S pool) and pre-40S particles (40S pool, see Figure 1B).
| Mol. wt (kDa) | WCE | 90S pool | Band | 40S pool | |||
|---|---|---|---|---|---|---|---|
| 287.4 | Utp20p | YBL004W | Utp20p | YBL004W | 1 | ||
| 193.0 | Rrp5p | YMR229C | Rrp5p | YMR229C | 2 | ||
| 199.9 | Utp10p | YJL109C | Utp10p | YJL109C | 3 | ||
| 144.8 | Ecm16p | YMR128W | Ecm16p | YMR128W | 4 | ||
| 140.4 | Utp22p | YGR090W | Utp22p | YGR090W | 5 | ||
| 135.6 | Bms1p | YPL217C | Bms1p | YPL217C | 5 | ||
| 137.5 | Rrp12p | YPL012W | Rrp12p | YPL012W | Rrp12p | YPL012W | |
| 119.2 | Kre33p | YNL132W | Kre33p | YNL132W | 6 | ||
| 106.2 | Dip2p | YLR129W | Dip2p | YLR129W | 7 | ||
| 103.9 | Pwp2p | YCR057C | Pwp2p | YCR057C | 7 | ||
| 104.7 | Utp21p | YLR409C | Utp21p | YLR409C | 8 | ||
| 94.2 | Nop14p | YDL148C | Nop14p | YDL148C | 8 | Nop14p | YDL148C |
| 90.6 | Tsr1p | YDL060W | Tsr1p | YDL060W | |||
| 101.1 | Nan1p | YPL126W | 9 | ||||
| 91.0 | Utp13p | YLR222C | Utp13p | YLR222C | 10 | ||
| 81.6 | Enp2p | YGR145W | 11 | ||||
| 87.8 | Utp4p | YDR324C | Utp4p | YDR324C | 11 | ||
| 66.8 | Mpp10p | YJR002W | Mpp10p | YJR002W | 12 | ||
| 80.0 | Utp8p | YGR128C | Utp8p | YGR128C | 13 | ||
| 61.2 | Bfr2p | YDR299W | 14 | ||||
| 72.0 | Utp5p | YDR398W | Utp5p | YDR398W | 14 | ||
| 64.9 | Rrp9p | YPR137W | 15 | ||||
| 65.4 | Ded1p | YOR204W | 16 | ||||
| 65.1 | Utp9p | YHR196W | Utp9p | YHR196w | 17 | ||
| 63.7 | Rok1p | YGL171W | 18 | ||||
| 62.2 | Utp7p | YER082C | Utp7p | YER082C | 18 | ||
| 55.0 | Enp1p | YBR247C | Enp1p | YBR247C | Enp1p | YBR247C | |
| 53.2 | Ltv1p | YKL143W | 19 | Ltv1p | YKL143W | ||
| 56.8 | Nop58p | YOR310C | Nop58p | YOR310C | 20 | ||
| 63.5 | Utp19p | YPR144C | Utp19p | YPR144C | 21 | ||
| 56.7 | Sik1p | YLR197W | Sik1p | YLR197W | 21 | ||
| 57.4 | Hrr25p | YPL204W | Hrr25p | YPL204W | |||
| 51.6 | Nob1p | YOR056C | Nob1p | YOR056C | |||
| 56.8 | Sof1p | YLL011W | 22 | ||||
| 57.6 | Utp15p | YMR093W | 23 | ||||
| 49.1 | Rio2p | YNL207W | Rio2p | YNL207W | |||
| 52.3 | Utp6p | YDR449C | Utp6p | YDR449C | 24 | ||
| 33.5 | Imp4p | YNL075W | Imp4p | YNL075W | 25 | ||
| 35.8 | Dim1p | YPL266W | Dim1p | YPL266W | |||
| 34.7 | Asc1p | YMR116C | Asc1p | YMR166C | 26 | Asc1p | YMR166C |
| 30.2 | YOR145C | YOR145C | YOR145C | ||||
| 30.6 | Bud23p | YCR047C | 27 | Bud23p | YCR047C | ||
| 31.5 | YKR060W | 28 | |||||
| 30.2 | Utp11p | YKL099C | 29 |
Ribosomal proteins S0, S1, S2, S5, S6, S7, S8, S9, S11, S13, S14, S16, S18, S19, S22, S23, S24, S25, L3, L16 and L17 were also found in the WCL. Co-purifying proteins were identified by mass spectrometry; both ORF and SGD standard names are listed. Bands 1–29 correspond to identified proteins in the 90S pool. Major co-purifying proteins of the 40S pool are in bold. Contaminants (e.g. heat shock proteins) are not depicted.
Some of the superstoichiometric bands (e.g. Rio2p and Tsr1p) were absent from the purified 90S pre-ribosomes (Figure 1, compare lanes ‘90S’ and ‘40S’), suggesting that they join the developing 40S particle during or shortly after 90S to 40S transition. All three Enp1p-TAP preparations, isolated from either the 90S pool, 40S pool or non-fractionated lysate, contained small subunit r-proteins in the low molecular weight range of the SDS–polyacryl amide gel (Figure 1B; Table I). These were characterized only from the cell lysate but were clearly more numerous in the 40S fractions than in the 90S fractions.
We conclude that Enp1p is associated with the early 90S pre-ribosomes, together with ∼30 other non-ribosomal processing and assembly factors, and with subsequent 40S pre-ribosomes that contain substantially fewer non-ribosomal proteins but more small subunit r-proteins.
Sedimentation and intracellular location of non-ribosomal proteins associated with 40S pre-ribosomes
We next determined the sedimentation behavior of the major proteins predicted to be associated with 40S pre-ribosomes (Figure 2). Sucrose density gradients were prepared from cells treated with cycloheximide to allow for visualization of 40S and 60S subunits, 80S ribosomes and polysomes (Figure 2, upper panels). The sedimentation of the different TAP-tagged proteins was revealed by western analysis (Figure 2, lower panels). Each of the tagged proteins tested, Enp1p, Dim1p, Nob1p, Hrr25p, Rrp12p, Rio2p, Trs1p and Yor145p, sedimented at positions appropriate for association with 40S subunits or 43S pre-ribosomes (Figure 2). Three proteins tested, Rrp12p, Dim1p and Enp1p, were also present in larger particles that correspond to 90S pre-ribosomes. In contrast, Nob1p, Hrr25p and Yor145p exhibited broad sedimentation peaks below 80S, possibly reflecting association with polysomes. When cycloheximide, which stabilizes polysomes, was omitted from the protocol, the sedimentation of Nob1p-TAP largely coincided with 80S ribosomes, whereas the sedimentation of Enp1p-TAP was not altered (data not shown).
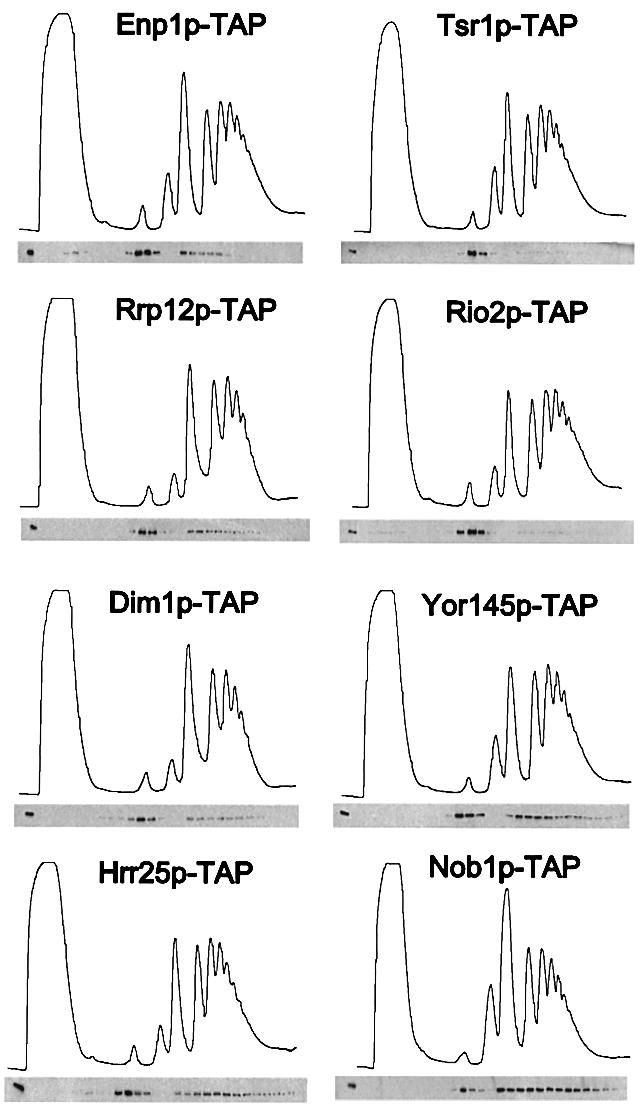
Fig. 2. Sedimentation behavior of TAP-tagged proteins associated with 40S pre-ribosomes on sucrose density gradients. Ribosomal fractions (40S, 60S, 80S and polysomes) were determined by OD254nm measurement of the gradient fractions (upper graph), and the indicated TAP-tagged protein baits were visualized by western blot analysis of these gradient fractions using anti-protein A antibodies (lower panel).
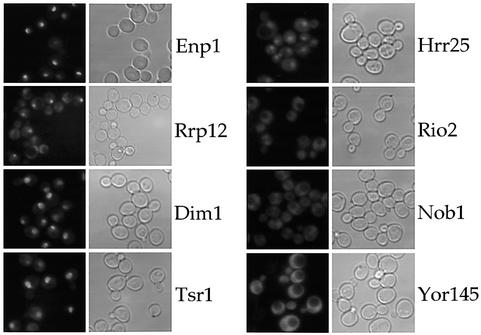
To determine the intracellular distribution of the factors associated with 40S pre-ribosomes, strains expressing green fluorescent protein (GFP)-tagged Enp1p, Rrp12p, Dim1p, Tsr1p, Hrr25p, Rio2p, Nob1p and Yor145p were examined by fluorescence microscopy (Figure 3, and Supplementary figure 1, available at The EMBO Journal Online). These analyses revealed that Enp1p is concentrated in the nucleolus with only a weak cytoplasmic signal (Figure 3). Rrp12p, Dim1p and Tsr1p are enriched in the nucleolus but also show substantial presence in the cytoplasm. In contrast, Hrr25p appears to have a location throughout the nucleus (i.e. nucleolus and nucleoplasm) together with a significant cytoplasmic signal. Finally, GFP-tagged Rio2p, Nob1p and Yor145p show a predominant cytoplasmic location (Figure 3). Thus, the steady-state locations of factors associated with 40S pre-ribosomes varied significantly, providing prima facie evidence for the spatial organization of 40S subunit maturation.
Fig. 3. Subcellular localization of GFP-tagged protein baits associated with 40S pre-ribosomes. The in vivo location of the indicated tagged proteins, associated with different 40S pre-ribosomes, was analyzed in the fluorescence microscope. For microscopic inspection, cells were grown to mid-log phase, mounted on a microscopic slide and photographed with identical exposure times. Strains were also transformed with pRS314-DsRed-NOP1 as a marker for the nucleolus and stained with DAPI to visualize DNA (see Supplementary figure 1).
Isolation of late cytoplasmic 40S pre-ribosomes
Sedimentation analysis in combination with intracellular location studies indicated that Rio2p is associated predominantly with cytoplasmic pre-40S subunits. We therefore wanted to characterize Rio2p further as a marker for late 40S pre-subunits. TAP of Rio2p-TAP from a WCL revealed a considerably simpler pattern of associated non-ribosomal proteins in the SDS–polyacrylamide gel than observed for Enp1p-TAP (Figure 4). The major Rio2p-associated bands were identified by mass spectrometry as Tsr1p, Enp1p, Hrr25p, Nob1p, Dim1p and Yor145p, as well as many small subunit Rps proteins (Figure 4, see also Table II). A similar pattern of co-enriched bands was found for TAP-purified Nob1p (Figure 4). In contrast, purifications of Dim1p-TAP and Rrp12p-TAP resulted in a highly complex pattern of associated proteins reminiscent of early 90S particles (Figure 4; see TAP-purified Noc4p for a marker of 90S particles). Finally, the protein pattern co-purifying with Hrr25p-TAP and Tsr1p-TAP was of intermediate complexity. Hrr25p-associated bands were identified as Gcn2p (a protein kinase phosphorylating initiation factor eIF2-α), Iki3p and Elp2p (both components of the Pol II transcription elongation complex), Scp160p, Rpm2p, Tsr1p, Ded1p, Ltv1p, Enp1p, Utp7p, Rok1p (an RNA helicase involved in rRNA processing), Rio2p, Dim1p and Yor145p (Table II). Thus, Hrr25 also associates with proteins that at first sight do not appear to play a direct role in 40S formation. Tsr1p-associated proteins were Rrp12p, Spt5p, Tif4631p, Def1p, Tsr1p, Ded1p, Enp1p, Nob1p, Rio2p, Ymr315p, Dim1p and Yor145p. Unfortunately, Yor145p-TAP could not be purified (data not shown). These analyses indicate that Rio2p and Nob1p are associated predominantly with related, late cytoplasmic pre-40S particles, while Hrr25p and Tsr1p apparently associate with intermediate pre-40S particles.
Fig. 4. Isolation of late cytoplasmic 40S pre-ribosomes. The indicated TAP-tagged protein baits were isolated from yeast lysates by the TAP method, and eluates were separated on a 4–12% SDS–polyacrylamide gradient gel followed by Coomassie Blue staining. Bait proteins are indicated by open circles. Prominent co-purifying proteins were identified by mass spectrometry (see also Table II). Non-ribosomal factors of the pre-40S particels are labeled. As a marker for 90S pre-ribosomes, TAP-purified Noc4p was also loaded on the gel (see also Grandi et al., 2002). High molecular weight 90S factors are marked by asterisks. Putative contaminants in the preparations (indicated by diamonds) are (from top to bottom): Ssa1/2p, Ssb1/2p, Pab1p, eEF-1α, Rpl3p, Rpl4p and Escherichia coli matrix porin f.
Table II. Identification of non-ribosomal proteins associated with late 40S pre-ribosomes.
| Mol. wt (kDa) | Hrr25p | Tsr1p | Rio2p | Nob1p | ||||
|---|---|---|---|---|---|---|---|---|
| 190.0 | Gcn2p | YDR283C | ||||||
| 152.8 | Iki3p | YLR384C | ||||||
| 137.5 | Rrp12p | YPL012W | ||||||
| 134.7 | Scp160p | YJL080C | ||||||
| 125.2 | Rpm2p | YML091C | ||||||
| 115.5 | Spt5p | YML010W | ||||||
| 107.0 | Tif4631p | YGR162W | ||||||
| 128.6 | YIL151C | |||||||
| 83.8 | Def1p | YKL054C | ||||||
| 90.6 | Tsr1p | YDL060W | Tsr1p | YDL060W | Tsr1p | YDL060W | Tsr1p | YDL060W |
| 89.3 | Elp2p | YGR200C | ||||||
| 65.4 | Ded1p | YOR204W | Ded1p | YOR204W | Ded1p | YOR204W | Ded1p | YOR204W |
| 53.2 | Ltv1p | YKL143W | Ltv1p | YKL143W | Ltv1p | YKL143W | ||
| 60.9 | Dbp2p | YNL112W | ||||||
| 55.0 | Enp1p | YBR247C | Enp1p | YBR247C | Enp1p | YBR247C | Enp1p | YBR247C |
| 57.4 | Hrr25p | YPL204W | Hrr25p | YPL204W | Hrr25p | YPL204W | ||
| 51.6 | Nob1p | YOR056C | Nob1p | YOR056C | Nob1p | YOR056C | ||
| 62.2 | Utp7p | YER082C | ||||||
| 63.7 | Rok1p | YGL171W | ||||||
| 49.1 | Rio2p | YNL207W | Rio2p | YNL207W | Rio2p | YNL207W | Rio2p | YNL207W |
| 38.1 | YMR315W | |||||||
| 35.8 | Dim1p | YPL266W | Dim1p | YPL266W | Dim1p | YPL266W | Dim1p | YPL266W |
| 30.2 | YOR145C | YOR145C | YOR145C | YOR145C |
TAP-tagged protein baits are indicated in the top row. Co-purifying proteins were identified by mass spectrometry; both ORF and SGD standard names are listed. The major pre-ribosomal components are in bold. Contaminants (e.g. heat shock proteins) are not depicted.
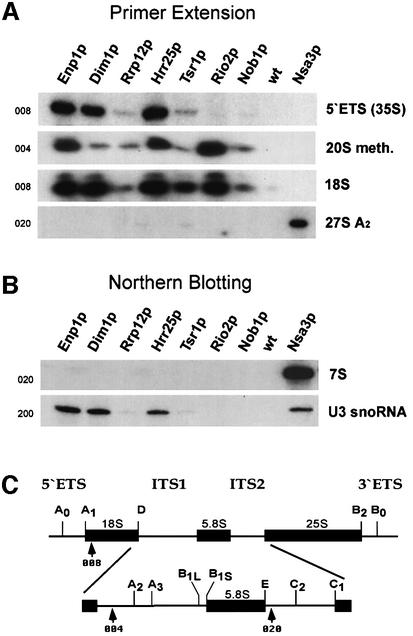
To obtain information about the rRNA composition of the different 40S pre-ribosomes, we TAP-purified each of the eight tagged proteins under standardized conditions and determined their RNA content by primer extension (Figure 5A) and northern blot hybridization (Figure 5B). Apparently, only rRNA precursors to the small ribosomal subunit (i.e. 35S, 20Smeth. and 18S) were found to co-enrich with the bait proteins, while pre-rRNAs of the large subunit (i.e. 27SA2 and 7S) were not detected and only co-purified with Nsa3p. Nsa3 was used as control since it previously was shown to be associated with U3 snoRNA and 27SA2 rRNA, but not with 35S, 20S and 18S rRNA (Nissan et al., 2002). 35S pre-rRNA and U3 snoRNA were detected mainly in TAP preparations of Enp1p, Dim1p and Hrr25p, suggesting that their associated complexes are precursors to later stages represented by Tsr1p, Rio2p and Nob1p baits. Despite a generally lower rRNA content, Rrp12p did not significantly co-enrich U3 sno-RNA, although this protein was identified previously as a component of 90S ribosomal precursors (Grandi et al., 2002).
Fig. 5. RNA analysis of the different pre-40S particles. (A) Primer extension to detect 35S, 20Smeth., 18S and 27SA2 rRNA, and (B) northern hybridization to detect 7S rRNA and U3 snoRNA were performed with RNA extracted from affinity-purified TAP-tagged Enp1p, Dim1p, Rrp12p, Hrr25p, Tsr1p, Rio2p, Nob1p, Nsa3p, and a non-tagged wild-type strain (negative control). Oligonucleotides used are indicated to the left of each corresponding panel, and their annealing position in the pre-rRNAs is depicted in (C).
Rio2p–GFP accumulates in the nucleus in the leptomycin B-sensitive xpo1 mutant
Rio2p is predominantly cytoplasmic at steady state, but this does not exclude a possible transient interaction with nucleolar 40S pre-ribosomes. To assess whether Rio2p is able to shuttle between the nucleus and the cytoplasm, a leptomycin B (LMB)-sensitive xpo1 mutant was used. In this strain, addition of LMB rapidly inhibits nuclear export of NES-containing cargos, as well as pre-60S and pre-40S subunits (Neville and Rosbash, 1999; Gadal et al., 2001b; Moy and Silver, 2002). As shown in Figure 6, Rio2p–GFP is cytoplasmic in the LMB-sensitive xpo1 strain in the absence of LMB, but accumulates throughout the nucleus in the presence of LMB. Thus, Rio2p is likely first to be imported into the nucleus before export to the cytoplasm in association with 40S pre-ribosomes. Similarly, the cytoplasmically located Yor145p becomes concentrated in the nucleus in the LMB-sensitive xpo1 mutant (Figure 6). Moreover, Tsr1p–GFP strongly accumulates in the nucleus and completely loses its cytoplasmic signal upon LMB treatment of cells (Figure 6). In contrast, Nob1p–GFP does not accumulate in the nucleus of the LMB-sensitive mutant within the first 20 min of LMB treatment, but only after extended incubation times (>1 h) (Figure 6). Taken together, these data suggest that Rio2p, Yor145c and also Tsr1p shuttle between the nucleus and cytoplasm. The delayed shuttling behavior of Nob1p, however, remains unclear.
Fig. 6. Rio2p, Yor145p, Tsr1p and Nob1p accumulate in the nucleus of a leptomycin-sensitive xpo1 mutant. In vivo location of Rio2p–GFP, Yor145p–GFP, Tsr1p–GFP and Nob1p–GFP, all chromosomally integrated at the 3′ end of their respective gene locus in the LMB-sensitive xpo1 mutant, after addition of LMB to the culture medium. The fluorescence signals of the GFP fusion proteins were observed in a fluorescence microscope. Note that Rio2p–GFP, Yor145p–GFP and Tsr1p–GFP accumulate in the nucleus already 20 min after addition of LMB, whereas Nob1p–GFP requires 1–2 h.
ENP1, RIO2 and TSR1 are necessary for synthesis and nuclear export of 40S subunits
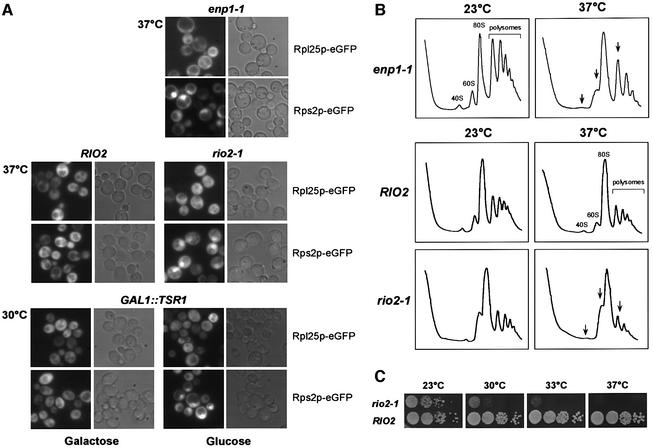
To assess the involvement of factors associated with 40S pre-ribosomes in export to the cytoplasm, we made use of a fusion between the 40S subunit r-protein Rps2p and eGFP (Grandi et al., 2002; Milkereit et al., 2003). We previously employed this reporter construct to demonstrate that kre33 and krr1 ts mutants, both mapping in components of the 90S pre-ribosome, are defective in 40S biogenesis and export (Grandi et al., 2002). Strikingly, an enp1-1 ts mutant (Chen at al., 2003) exhibited strong accumulation of Rps2p–eGFP in the nucleolus and the nucleoplasm following a shift to 37°C (Figure 7A). In contrast, nuclear export of 60S subunits, as indicated by the Rpl25p–eGFF construct, was not impaired in the enp1-1 ts strain. Moreover, the amount of 40S relative to 60S subunits was significantly decreased on polysomal gradients, when the enp1-1 strain was shifted to 37°C prior to the preparation of cell lysates (Figure 7B).
Fig. 7. enp1 and rio2 ts mutants as well as a GAL1::TSR1 depletion strain are defective in synthesis and nuclear export of 40S subunits. (A) In vivo assay to analyze ribosomal export of 40S and 60S subunits. Ts mutants were shifted for 4 h to 37°C before nuclear accumulation of Rps2p–eGFP (40S subunit reporter) and Rpl25p–eGFP (60S subunit reporter) was determined by fluorescence microscopy. The GAL1::TSR1 depletion stain was shifted from galactose- to glucose-containing medium for 12 h at 30°C. (B) Analysis of ribosomal and polysomal fractions isolated by sucrose density gradient centrifugation from enp1-1 and rio2-1 ts strains, grown at 23°C or shifted for 4 h to 37°C. The UV profiles (OD254nm) of the sucrose gradient are depicted. (C) Growth properties of RIO2 and rio2-1 strains. Serial dilutions were spotted on YPD plates and incubated either for 3 days at 23°C or 4 days at 30, 33 and 37°C.
Rio2p is a late marker of 40S biogenesis and likely to be exported from the nucleus to the cytoplasm with the maturing pre-40S particles (see above). To assess its function in 40S subunit formation and export, we generated ts alleles of RIO2 by PCR-mediated mutagenesis (Figure 7C; see Materials and methods). The mutations in the ts allele rio2-1 have been mapped and are Y(73)>H, I(186)>V, Y(210)>H, E(364)>D, D(371)>G and D(417)>G. As anticipated, the TAP-tagged thermolabile Rio2-1p protein (Supplementary figure 2A) can be purified as part of 40S pre-ribosomes from cells grown at permissive, but not at restrictive temperature (Supplementary figure 2B). Interestingly, we find that the composition of the purified Tsr1p-TAP complex (for comparison see Figure 4) is not affected in rio2-1 ts mutants at 37°C, except that the thermolabile Rio2-1p protein is absent from the particle (Supplementary figure 2B). Thus, the stability of the 40S pre-ribosomes does not depend on the presence of Rio2p. Notably, Rio2-1p–GFP is inhibited in its shuttling behavior, and has the tendency to accumulate in the nucleus of an LMB-sensitive xpo1 mutant, even when no drug is added (Supplementary figure 2C).
When the rio2-1 ts mutant was analyzed for ribosomal export defects, we found that nuclear export of 40S subunits, but not of 60S subunits, was impaired after shift to the restrictive temperature (Figure 7A), and a significant decrease in free 40S subunits was observed on polysomal gradients (Figure 7B). To find out whether the Rps2p–GFP reporter remains associated with pre-ribosomal particles under restrictive conditions, we performed sucrose gradient centrifugation of WCLs derived from rio2-1 and enp1-1 ts mutants, respectively, grown at either 23°C or shifted to 37°C for 4 h (Supplementary figure 3). Western blot analysis of the resulting gradient fractions revealed no free Rps2p–eGFP reporter in the upper part of the gradient. Instead, Rps2p–eGFP was found exclusively in the lower part, which contains ribosomes and pre-ribosomal particles. These data show that the small subunit reporter Rps2p–GFP stays associated with 40S pre-ribosomes even if the rio2 ts mutant is shifted to the non-permissive temperature. We conclude that nuclear export of 40S but not 60S pre-ribosomes requires Rio2p. However, we do not know whether rio2-1 has an exclusive export defect, or affects the maturation of the pre-40S particles, which then impairs nuclear export.
As Rio2p interacts with Tsr1p in pre 40S-particles (Figure 4, see also Table II), we wanted to know if this protein is also involved in 40S ribosomal export. Therefore, a GAL1::TRS1 depletion strain (Gelperin et al., 2001) was transformed with either the 40S or the 60S subunit reporter. When shifted from galactose to glucose medium to deplete for Tsr1p, Rps2p–GFP accumulated inside the nucleus whereas export of Rpl25p–GFP was not affected (Figure 7A). We conclude that Enp1p, Rio2p and Tsr1p are each required for normal export of the pre-40S particles from the nucleus to the cytoplasm.
Discussion
We have combined biochemical assays with intracellular localization studies to investigate 40S subunit biogenesis from assembly in the nucleolus until export to the cytoplasm. All of the factors analyzed here are likely to be associated with the pre-40S particles in the cytoplasm, but differ in their time of interaction. Several proteins (Enp1p, Dim1p, Hrr25p and probably Rrp12p) initially associate with 90S pre-ribosomes, while Nob1p, Rio2p and Tsr1p appear to associate with pre-40S only after the rRNA cleavages at sites A0–A2 that generate 20S pre-rRNA and separate pre-40S and pre-60S particles.
Interpretation of the data was aided greatly by the separation of 90S and 40S pre-ribososmal particles by sedimentation centrifugation, prior to a TAP purification of Enp1p. This strategy of performing a pre-fractionation step prior to a consecutive TAP purification could also be applicable to isolate other assembly intermediates from cell lysates.
The highly complex 90S pre-ribosomes, composed of 35S pre-rRNA, small subunit r-proteins, U3 snoRNP and numerous 40S subunit processing and assembly factors (Grandi et al., 2002), appear to be directly converted into much simpler 40S pre-ribosomes. The U3 snoRNP is base-paired to the 5′-ETS, and the resulting structure (also known as the terminal knob or SSU processome) remains associated with this region even under the fairly stringent conditions of chromatin spreads (Saffer and Miller, 1986; Beltrame and Tollervey, 1995; Dragon et al., 2002). Analysis of TAP-tagged components of the 90S pre-ribosome shows that many of these remain associated with the excised 5′-ETS region that is released by cleavage at site A0 (Grandi et al., 2002; E.Petfalski and D.Tollervey, unpublished observations). This suggests that many or all of the 90S-specific factors are released as a complex still bound to the 5′-ETS. This gross removal of ∼30 factors leaves a relatively simple pre-40S particle with only a few major non-ribosomal factors.
Amongst the proteins analyzed in this study, only Hrr25p–GFP is significantly present in the nucleoplasm, but this protein kinase is reported to have additional functions in transcriptional activation in response to DNA damage (Hoekstra et al., 1991; Ho et al., 1997). All other pre-40S particles studied here show little nucleoplasmic accumulation, suggesting that they quickly transit through the nucleoplasm (Figure 8). Ultrastructural studies in yeast showed that 40S and 60S subunits travel through the nucleolar and nucleoplasmic compartments, respectively, with different kinetics (Gleizes et al., 2001), and cell fractionation studies also indicated that pre-40S particles are exported more rapidly to the cytoplasm than pre-60S particles (Udem and Warner, 1973).
Fig. 8. Model of 40S and 60S ribosomal biogenesis and export. For description, see text.
This analysis has identified late pre-40S particles that have already reached the cytoplasm, for which Rio2p is a useful marker. Although its exact function is not known, Rio2p may perform a role similar to a homologous protein termed Rio1p/Rrp10p, which is required for processing of the 20S pre-rRNA to 18S rRNA and is associated with 40S pre-ribosomes in the cytoplasm (Vanrobays et al., 2001). Rio1p was not, however, identified in our proteomic analyses or in the analyses of other putative pre-40S particles carried out as part of a high-throughput screen (Fatica and Tollervey, 2002; Gavin et al., 2002), suggesting that it is weakly or very transiently associated with the pre-40S particles. Rio1p shows clear homology to Ser/Thr protein kinases and may be a member of a novel family of kinases involved in cell cycle progression (Angermayr et al., 2002). Thus, it is possible that Rio2p is also a protein kinase with a role in cytoplasmic cleavage of 20S pre-rRNA. Several other ribosome synthesis factors are linked to cell cycle progression; whether this also applies to these putative kinases remains to be determined.
Tsr1p also appears to associate with the pre-40S particle only after formation of 20S pre-rRNA. However, whereas Rio2p–eGFP is predominantly cytoplasmic, Tsr1p is both nucleolar and cytoplasmic at steady state. The difference in their apparent subcellular locations presumably reflects the time during which they remain associated with the pre-40S particles in the nucleus and cytoplasm. Tsr1p is related to the putative GTPase Bms1p, but may lack GTPase activity (Gelperin et al., 2001). In general, the pre-40S subunits notably lack GTPases and AAA-type ATPases, in marked contrast to the pre-60S particles (see Nissan et al., 2002; and references therein). This suggests that structural rearrangements during 40S formation may be less prevalent than for 60S biogenesis.
Dim1p is a methyltransferase that dimethylates the 3′ end of the 18S rRNA within the 20S pre-rRNA (Lafontaine et al., 1995, 1998). This modification is thought to take place in the cytoplasm (Udem and Warner, 1973). Hence the co-precipitation of the methylated form of the 20S pre-rRNA with each of the TAP-tagged proteins studied here provides evidence that all remain associated with the cytoplasmic pre-40S particle. However, Dim1p is also required for earlier, nucleolar pre-rRNA cleavage at sites A1 and A2 (Lafontaine et al., 1995). Consistent with this, Dim1p–GFP localized predominantly to the nucleolus, and purified Dim1p-TAP revealed a set of associated proteins typical for 90S pre-ribosomes. Recovery of the modified 20S pre-rRNA with Dim1p-TAP was relatively weak when compared with co-purified 35S and 18S rRNA, which might be indicative of alterations in affinity and binding constraints as the methylation reaction occurs. The role of Nob1 in the pre-40S particles is not clear. Remarkably, Nob1 was reported to be required for biogenesis of the 26S proteasome and degraded upon its maturation in yeast (Tone and Toh, 2002).
It was shown recently that Nmd3p, which is exported via the general nuclear export receptor Crm1/Xpo1p, and another export factor Mtr2p join the Arx1p-containing 60S pre-ribosome before nuclear export to the cytoplasm (Nissan et al., 2002). Nuclear export of 40S subunits was inhibited in both mutants of the Xpo1p export receptor and Ran-cycle mutants (Moy and Silver, 1999). It thus appears that pre-40S export is also mediated by a NES-containing adaptor protein, but a prime candidate has not been identified yet. A weak NES activity was detected in a sequence of Rio2p (residue 318–425), which can partly replace the endogenous NES of the Nmd3p adaptor protein (T.Schäfer and E.Hurt, unpublished data). The pre-40S subunit that actually undergoes nuclear exit may be closely related to the major forms of the Tsr1p- and Hrr25p-associated particles. Further characterization of these particles may lead to the identification of factors, possibly present only in substoichiometric amounts, which act as nuclear export factors for 40S pre-ribosomes.
Together, our data allow us to draw a model of the maturation and export of 40S ribosomal subunits (Figure 8). The initial ribosomal precursor particle is a 90S structure composed of the huge U3 processome and other 40S synthesis factors, but few 60S processing and assembly factors. After cleavage at site A2, 90S breaks up into 40S and 60S pre-subunits. The pre-60S ribosome acquires a very large number of factors and undergoes a complicated set of processing, assembly and maturation steps in both the nucleolus and nucleoplasm (Fatica and Tollervey, 2002; Nissan et al., 2002). In contrast, the newly synthesized pre-40S subunit has a relatively simple protein composition and acquires few additional factors before nuclear exit. In the cytoplasm, the late 40S pre-ribosome is associated with only a few factors; functional characterization has been reported only for Tsr1p, which is required for 20S to 18S rRNA processing. Analyses of the other factors are underway.
Materials and methods
Yeast strains and plasmids
C-terminal genomic integration of GFP (HIS3MX6-Marker) into yeast strains DS1-2b (MATa, ura3, trp1, his3, leu2) and the LMB-sensitive xpo1 mutant (Neville and Rosbash, 1999) resulting in fusion proteins of Enp1p (YBR247c), Dim1p (YPL266w), Nob1p (YOR056c), Hrr25p (YPL204w), Rrp12p (YPL012w), Rio2p (YNL207w), Tsr1p (YDL060w) and YOR145c was performed as described (Longtine et al., 1998). Strains were also transformed with pRS314-DsRed-NOP1 as a marker for the nucleolus. Genomic TAP integration resulting in C-terminal fusion proteins of Enp1p, Rio2p, Tsr1p, Nob1p, Dim1p, Hrr25p, Rrp12p and Yor145p was performed as described (Grandi et al., 2002). Thus, all constructs are under the control of the authentic promoters.The strain used for these integrations is MATa, ade2, arg4, leu2-3,112, trp1-289, ura3-52. A haploid RIO2 shuffle strain was constructed by tetrad dissection of a heterozygous rio2::kanMX4/RIO2 diploid strain containing a pRS416-RIO2 plasmid (Euroscarf, accession Nos Y22005 and P20053). RIO2 was isolated by PCR from yeast genomic DNA using primers 5′-TTTCTCGAGTGGCGGTCAAGGTTTCGGAAGAC-3′ and 5′-TTTGCGGCCGAAGCTGAGAGCGACCTTT-3′, and cloned into plasmid pRS315. The rio2 ts mutant was generated by random PCR-mediated mutagenesis as described (Muhlrad et al., 1992; Santos-Rosa et al., 1998), and the DNA sequence of the rio2-1 ts allele was determined (see text). Plasmid-borne pRS315-rio2-1 was C-terminally TAP tagged and expressed in the rio2::KAN null strain. Moreover, Tsr1p-TAP was expressed in the rio2-1 ts mutant. For subcellular localization, pRS316-RIO2-GFP and pRS316-rio2-1-GFP (both alleles under the authentic promoter) were constructed and transformed into strain DS1-2b (wild-type) and the LMB-sensitive xpo1 mutant. The enp1-1 ts strain (Chen et al., 2003) and GAL1::TRS1 depletion strain (Gelperin et al., 2001) were described previously. Finally, strains rio2-1 and enp1-1 were transformed with a pRS316-RPS2-GFP reporter construct.
Sucrose density gradient centrifugation
Low salt isolation of ribosomes by sucrose gradient centrifugation was performed essentially as described (Baßler et al., 2001). Briefly, no or 100 µg/ml cycloheximide was added to yeast cultures grown to an OD600 of 0.7. After 30 min incubation at room temperature, cells were harvested, washed with cold buffer A [20 mM HEPES pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 1 mM EGTA and 1 mM dithiothreitol (DTT)] and lysed. Cell lysates were centrifuged for 5 min at 14 000 r.p.m. and 4°C, before the supernatant was loaded on a 10–50% sucrose gradient prepared in buffer A. After ultracentrifugation for 16 h at 27 000 r.p.m., a gradient collector was used to record the UV profile and to collect fractions for further analysis.
TAP-purification of Enp1p from 90S and 40S pools
For the fractionation of pre-ribosomal particles, 2 l cultures were grown to an OD600 of 3.5, before 100 µg/ml cycloheximide was added. Cells were incubated for an additional 30 min at room temperature and harvested by centrifugation. Pellets were washed in 50 ml of ice-cold buffer A, resuspended in buffer A and broken by glass bead lysis (Fritsch pulverisette 6). The lysate was centrifuged for 10 min at 4000 r.p.m. and further for 30 min at 14 000 r.p.m.. Then 12 × 3 ml of the supernatant were loaded on 20–40% sucrose gradients in buffer A (3.5 inch Beckmann centrifuge tubes) and subjected to ultracentrifugation for 16 h at 27 000 r.p.m. and 4°C in a SW28 rotor. A gradient collector was used to record the UV profile and to collect the gradient fractions. Fractions corresponding to 40S and 90S peaks were pooled and subjected to TAP purification. Eluates were precipitated by trichloroacetic acid and analyzed by SDS–PAGE.
RNA analysis
The rRNA content of purified bait proteins, isolated according to Grandi et al. (2002) with minor modifications to minimize RNA degradation, was analyzed by northern hybridization and primer extension. Briefly, TAP purifications were performed in 0.1% diethylpyrocarbonate (DEPC) buffers using 1 mM ribonucleoside vanadyl complex (Sigma) as RNase inhibitor with time reduction in centrifugation and TEV cleavage steps. TEV protease was pre-incubated with RNase inhibitor from human placenta. Northern hybridization and primer extension were performed as described (Beltrame and Tollervey, 1992). Oligonucleotides used were 004 (CGGTTTTAATTGTCCTA), 008 (CATGGCTTAATCTTTGA GAC), 020 (TGAGAAGGAAATGACGCT) and 200 (UUAUGG GACUUGUU).
Miscellaneous
Microbiological techniques and DNA recombinant work were performed according to Maniatis et al. (1982). Affinity purification of TAP-tagged bait proteins was carried out as described (Nissan et al., 2002 and references therein). SDS–PAGE and immunoblotting were performed according to Siniossoglou et al. (1996). Mass spectrometry using tryptic digests of bands excised from the Coomassie Blue-stained SDS–polyacrylamide gels was performed according to Baßler et al. (2001). The fluorescence-based visual assay to analyze nuclear export of large and small ribosomal subunits using the Rlp25p–eGFP and Rps2–GFP reporters, respectively, was performed according to Gadal et al. (2001b) and Milkereit et al. (2003). Fluorescence microscopy was performed as described (Gadal et al., 2001a), and LMB treatment of the LMB-sensitive xpo1 mutant was according to Neville and Rosbash (1999) and Gadal et al. (2001b).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to Dr M.Rosbash (Brandeis University, Waltham) for providing the LMB-sensitive xpo1 strain, and Dr M.Yoshida (University of Tokyo, Japan) for generously providing the LMB used in this study. The excellent technical assistance of Sabine Merker and Petra Ihrig under the supervison of Dr J.Lechner (Mass Spectrometry Unit, BZH, Heidelberg) is acknowledged. We would also like to acknowledge Dr R.Sternglanz (Stony Brook University) for providing the enp1 ts strain, and Dr S.Lemmon (Case Western Reserve University) for a TSR1 depletion strain. E.H. is a recipient of grants from the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm ‘Funktionelle Archi tektur des Zellkerns’ and Gottfried Wilhelm Leibniz Program) and Fonds der Chemischen Industrie.
References
- Angermayr M., Roidl,A. and Bandlow,W. (2002) Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol. Microbiol., 44, 309–324. [DOI] [PubMed] [Google Scholar]
- Baßler J., Grandi,P., Gadal,O., Leßmann,T., Tollervey,D., Lechner,J. and Hurt,E.C. (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol. Cell, 8, 517–529. [DOI] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey,D. (1995) Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Bucaria,J., Band,D.A., Sutton A. and Sternglanz,R. (2003) Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res., 31, 690–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F. et al. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature, 417, 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A. and Tollervey,D. (2002) Making ribosomes. Curr. Opin. Cell Biol., 14, 313–318. [DOI] [PubMed] [Google Scholar]
- Fatica A., Cronshaw,A.D., Dlakic M. and Tollervey,D. (2002) Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell, 9, 341–351. [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauß,D., Braspenning,J., Hoepfner,D., Petfalski,E., Philippsen,P., Tollervey,D. and Hurt,E.C. (2001a) A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J., 20, 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauß,D., Kessl,J., Trumpower,B., Tollervey,D. and Hurt,E. (2001b) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a NES-containing factor Nmd3p that associates with the large subunit protein Rpl10p. Mol. Cell. Biol., 21, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.-C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Gelperin D., Horton,L., Beckman,J., Hensold,J. and Lemmon,S.K. (2001) Bms1p, a novel GTP-binding protein and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA, 7, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizes P.E., Noaillac-Depeyre,J., Leger-Silvestre,I., Teulieres,F., Dauxois,J.Y., Pommet,D., Azum-Gelade,M.C. and Gas,N. (2001) Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J. Cell Biol., 155, 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P. et al. (2002) 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell, 10, 105–115. [DOI] [PubMed] [Google Scholar]
- Harnpicharnchai P. et al. (2001) Composition and functional characterization of yeast 66s ribosome assembly intermediates. Mol. Cell, 8, 505–515. [DOI] [PubMed] [Google Scholar]
- Ho J.H.N., Kallstrom,G. and Johnson,A.W. (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol., 151, 1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Mason,S., Kobayashi,R., Hoekstra,M. and Andrews,B. (1997) Role of the casein kinase I isoform, Hrr25 and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra M.F., Liskay,R.M., Ou,A.C., DeMaggio,A.J., Burbee,D.G. and Heffron,F. (1991) HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science, 253, 1031–1034. [DOI] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and De La Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D., Vandenhaute,J. and Tollervey,D. (1995) The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev., 9, 2470–2481. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L.J., Preiss,T. and Tollervey,D. (1998) Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol. Cell. Biol., 18, 2360–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Fritsch,E.T. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Milkereit P., Strauß,D., Bassler,J., Gadal,O., Kühn,H., Schütz,S., Lechner,J., Hurt,E. and Tschochner,H. (2003) A Noc-complex specifically involved in maturation and nuclear export of ribosomal 40S subunits. J. Biol. Chem., 278, 4072–4081. [DOI] [PubMed] [Google Scholar]
- Miller O.L. and Beatty,B.R. (1969) Visualization of nucleolar genes. Science, 164, 955–957. [DOI] [PubMed] [Google Scholar]
- Mougey E.B., O’Reilly,M., Osheim,Y., Miller,O.L.,Jr, Beyer,A. and Sollner-Webb,B. (1993) The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev., 7, 1609–1619. [DOI] [PubMed] [Google Scholar]
- Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy T.I. and Silver,P.A. (2002) Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci., 115, 2985–2995. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Neville M. and Rosbash,M. (1999) The NES–Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J., 18, 3746–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T.A., Baßler,J., Petfalski,E., Tollervey,D. and Hurt,E.C. (2002) 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J., 21, 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffer L.D. and Miller,O.L.,Jr (1986) Electron microscopic study of Saccharomyces cerevisiae rDNA chromatin replication. Mol. Cell. Biol., 6, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Moreno,H., Simos,G., Segref,A., Fahrenkrog,B., Panté,N. and Hurt,E. (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol., 18, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C., Bienvenu,D., Namane,A., Gleizes,P.E., Gas,N., Jacquier,A. and Fromont-Racine,M. (2001) Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J., 20, 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer,C., Rieger,M., Doye,V., Tekotte,H., Weise,C., Emig,S., Segref,A. and Hurt,E.C. (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell, 84, 265–275. [DOI] [PubMed] [Google Scholar]
- Stevens A., Hsu,C.L., Isham,K.R. and Larimer,F.W. (1991) Frag ments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J. Bacteriol., 173, 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y. and Toh,E.A. (2002) Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev., 16, 3142–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapman J., Retèl,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- Udem S.A. and Warner,J.R. (1972) Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol., 65, 227–242. [DOI] [PubMed] [Google Scholar]
- Udem S.A. and Warner,J.R. (1973) The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem., 248, 1412–1416. [PubMed] [Google Scholar]
- Vanrobays E., Gleizes,P.E., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Caizergues-Ferrer,M. and Gelugne,J.P. (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J., 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- Warner J.R. (2001) Nascent ribosomes. Cell, 107, 133–136. [DOI] [PubMed] [Google Scholar]