Abstract
In tailed bacteriophages and herpes viruses, the viral DNA is packaged through the portal protein channel. Channel closure is essential to prevent DNA release after packaging. Here we present the connector structure from bacteriophage SPP1 using cryo-electron microscopy and single particle analysis. The multiprotein complex comprises the portal protein gp6 and the head completion proteins gp15 and gp16. Although we show that gp6 in the connector has a fold similar to that of the isolated portal protein, we observe conformational changes in the region of gp6 exposed to the DNA-packaging ATPase and to gp15. This reorganization does not cause closure of the channel. The connector channel traverses the full height of gp6 and gp15, but it is closed by gp16 at the bottom of the complex. Gp16 acts as a valve whose closure prevents DNA leakage, while its opening is required for DNA release upon interaction of the virus with its host.
Keywords: bacteriophage SPP1/connector/cyclical oligomers/electron cryo-microscopy/portal protein
Introduction
Viral assembly is a multistep process determined by a specific sequence of protein–protein and protein–DNA interactions. Tailed bacteriophages and herpes viruses package their DNA into a pre-formed procapsid struc ture (Hendrix, 1999; Figure 1). Double-stranded DNA (dsDNA) translocation occurs through the channel of the portal protein, a central component of the DNA encapsidation apparatus that is localized at a single vertex of the viral procapsid (Valpuesta and Carrascosa, 1994; Newcomb et al., 2001). The DNA-packaging ATPase (terminase) bound to the viral chromosome docks in the portal vertex, forming the DNA translocation machine (Figure 1). The portal protein was proposed to be a rotary motor that pumps DNA into the capsid interior against a steep concentration gradient in a reaction fuelled by the terminase ATPase activity (Hendrix, 1978; Dube et al., 1993; Simpson et al., 2000). Upon termination of DNA packaging, one end of the DNA remains bound to the portal vertex prior to leaving the virion at the onset of infection (Tavares et al., 1996). DNA is packed in a dense arrangement, reaching concentrations of ∼500 mg/ml that could exert a pressure of 6 MPa within the capsid shell (Earnshaw and Casjens, 1980; Smith et al., 2001). This strong internal pressure can cause ejection of DNA from the phage particles triggered in vitro (Earnshaw and Casjens, 1980; Tavares et al., 1996; and references therein). To avoid chromosome leakage, the portal channel has to be closed shortly after encapsidation of the DNA. This can be achieved by a conformational change in the portal protein (φ29; Hagen et al., 1976; Donate et al., 1988) or by binding of head completion proteins that plug the portal pore to form the connector structure, i.e. T4 (Coombs and Eiserling, 1977), λ (Perucchetti et al., 1988), P22 (Strauss and King, 1984) and SPP1 (Lurz et al., 2001; this work). We define the connector as the complete knob structure assembled at the capsid portal vertex prior to tail attachment, and distinct from the portal protein cyclical oligomer (gp6 in SPP1). The additional feature in bacteriophages T3 and T7 is an internal core that extends from the portal structure to the procapsid interior (Steven and Trus, 1986). Interestingly, the T3 portal protein pore appears partially closed after DNA packaging (Valpuesta et al., 1992). Closure of the portal channel involves a valve mechanism that is reversed for ejection (Bazinet and King, 1985; Tavares et al., 1996). Viral DNA delivery to the host cytoplasm is a complex and regulated process that probably engages a variety of phage and host factors (Molineux, 2001; and references therein).
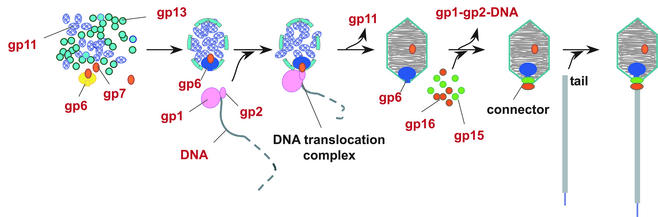
Fig. 1. SPP1 morphogenesis. Current knowledge of the sequence of assembly reactions during SPP1 capsid assembly (Dröge et al., 2000; Gual et al., 2000; Lurz et al., 2001; and references therein). Sizes are not scaled, to emphasize locations of minor capsid components. Gp1-gp2, terminase complex; gp6, portal protein; gp7, minor capsid protein; gp11, scaffolding protein; gp13, major capsid protein; gp15 and gp16, head completion proteins. The connector components are shown in the same colour code as in Figure 3. The assembly-naïve gp6 is shown in yellow, while the gp6 oligomer embedded in the capsid lattice is in blue to illustrate the different radial symmetry of the two gp6 forms (Lurz et al. 2001).
The connector of Bacillus subtilis bacteriophage SPP1 is composed of the portal protein gp6 (subunit molecular mass of 57.3 kDa) and the two head completion proteins gp15 (11.6 kDa) and gp16 (12.5 kDa; Lurz et al., 2001). It was found that the connector complex has 12-fold cyclical symmetry (Lurz et al., 2001), though isolated gp6 is a closed cyclical 13mer in equilibrium with a small population of open curvilinear oligomers (…9, 10, 11, 12, 13mers; van Heel et al., 1996b). Reassociation and refolding–reassociation experiments showed that formation of closed rings of 13 subunits is an intrinsic property of gp6 (Jekow et al., 1999; our unpublished results). The portal protein participates in the early reactions of procapsid assembly (Dröge et al., 2000). Co-production of gp6 with the two other essential procapsid proteins of SPP1 in the same Escherichia coli strain that is used to produce gp6 13mers led to formation of biologically active procapsids in vivo (Dröge and Tavares, 2000; Dröge et al., 2000). These procapsids contain a functional portal protein that is a 12mer at late stages of morphogenesis (Figure 1; Lurz et al., 2001). To reconcile the finding of the two different symmetries of the SPP1 portal protein, it was suggested that the gp6 oligomers competent for the procapsid assembly reaction are open curvilinear forms found in equilibrium with isolated 13mers. These open oligomers would form closed 12mers when they are surrounded by the major capsid protein and interact with the scaffolding protein (Lurz et al., 2001). After procapsid assembly, gp6 participates in the reactions required for viral DNA packaging. Packaging is terminated by cleavage of the DNA concatemer, generating unit-length virus chromosome molecules (Tavares et al., 1995). Encapsidation of the DNA is followed by binding of gp15 and gp16 to the portal vertex, leading to formation of the connector (Figure 1). The whole complex consists of gp6, gp15 and gp16 annular oligomers (Lurz et al., 2001; this work). The phage tail attaches to the gp16 ring, whereas the DNA extremity, which is packaged last, remains attached to the connector structure (Tavares et al., 1996). Initiation of phage infection requires the opening of the connector to enable the release of the viral chromosome through the tail channel into the host cytoplasm. Here we present a structural analysis by cryo-electron microscopy and angular reconstitution of the ∼900 kDa connector complex and its comparison with the isolated portal protein. The new structure provides a framework for understanding how the connector controls the final stages of DNA encapsidation and DNA release at the onset of viral infection.
Results and discussion
Gp15 and gp16 are required to prevent release of packaged DNA
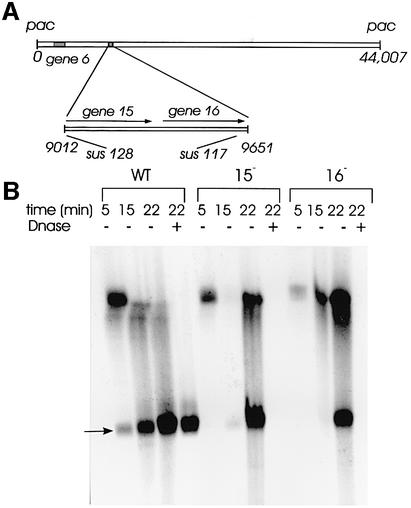
The portal protein gp6 is necessary for SPP1 DNA packaging, but the additional components of the connector, which prevent the release of the DNA that is held at high pressure inside the capsid, were not identified. Electron microscopy studies suggested that gp15 and gp16 present in the portal vertex of SPP1 capsids might serve to lock the connector base (Lurz et al., 2001). To check this idea, we have infected non-permissive B.subtilis YB886 cells with SPP1sus128 and SPP1sus117 that do not produce gp15 and gp16, respectively (Figure 2; Becker et al., 1997). Total DNA was extracted from infected cells at different times after infection and was resolved by pulse-field gel electrophoresis (PFGE; Figure 2). Unit-length viral chromosomes (∼45.9 kbp) produced after each DNA encapsidation cycle were detected after 15 min of infection, and the quantity increased significantly at 22 min post-infection. DNA packaging thus occurs efficiently when gp15 and gp16 are absent. However, when the cell extracts are treated with DNase, the lack of gp15 or gp16 correlates with complete degradation of viral chromosomes, showing that they are not protected by the capsid structure (Figure 2). Protection of mature SPP1 DNA is observed in infections leading to formation of phage particles (Figure 2) or DNA-filled capsids (e.g. in SPP1sus9 infections) that can both be purified as stable nucleoprotein complexes in caesium chloride gradients after DNase treatment with comparable yields (data not shown). Electron microscopy of structures partially purified from extracts of cells infected with the gp15– or gp16– phages showed that a significant number of capsids have undergone expansion, a conformational change that occurs during SPP1 DNA packaging (Figure 1; Isidro, 2002; data not shown), but that they contained no DNA inside (data not shown). Therefore, gp15 and gp16 are not required for the DNA packaging reactions, but both are essential for retention of DNA inside the capsid.
Fig. 2. Production of mature viral chromosomes during infection with SPP1 wild type and with SPP1 mutants defective in production of the connector proteins gp15 and gp16. (A) Position of the genes coding for the connector proteins in the SPP1 genome (black boxes) (nucleotide coordinates are according to Alonso et al., 1997). The enlarged region shows the position of mutations sus128 (codon Q4→stop; Becker et al., 1997) and sus117 (Q94→stop; this work) in genes 15 and 16, respectively. DNA packaging is from left to right, leading to encapsidation of a DNA molecule with ∼45.9 kbp (Tavares et al., 1996), which is 104% the size of the SPP1 genome (44 007 bp; Alonso et al., 1997). (B) Total DNA extracted from B.subtilis YB886 cells infected with SPP1 wild type (wt), SPP1sus128 (gp15–) and SPP1sus117 (gp16–) at the time points after infection indicated and resolved by PFGE. Samples treated (+) and untreated (–) with DNase were analysed. The position to where SPP1 unit-length chromosomes migrate is indicated by an arrow (45.9 kbp). The band observed at this position at 5 min post-infection is attributed to input phages used for infection that were not eliminated since SPP1 DNA packaging is only detected after 12 min of infection (our unpublished results). The gel was stained with ethidium bromide. A Southern blot hybridized with a probe specific for the SPP1 genome showed a pattern of SPP1 DNA distribution in the gel similar to that observed with total DNA staining (data not shown).
Connector structure
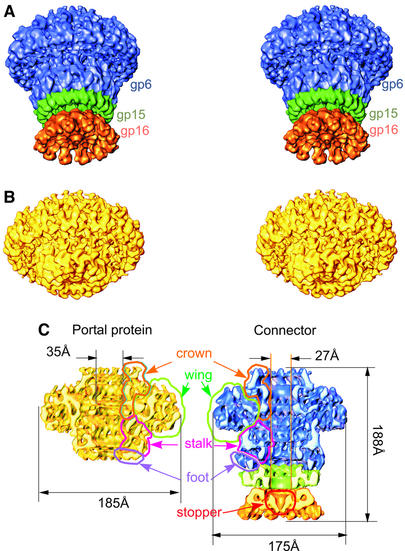
Connector complexes were isolated from DNA-filled capsids (Figure 1) produced by infection with an SPP1 mutant deficient in tail assembly (SPP1sus9) (Lurz et al., 2001). Disruption of the capsids yielded connectors composed of gp6, gp15 and gp16. These were purified by sedimentation through glycerol gradients (Lurz et al., 2001). Statistical analysis of views along the rotational axis revealed that gp6 has 12-fold symmetry within the connector (Lurz et al., 2001), in contrast to the 13mer symmetry found in isolated gp6 (Dube et al., 1993). The structure of the connector was determined at 10 Å resolution, using images from ice-embedded samples and the angular reconstitution technique (van Heel, 1987) (Figure 3A and C).
Fig. 3. Structure of the SPP1 connector and portal protein. The stereo view of the SPP1 connector structure at 10 Å resolution is shown in (A). The portal protein gp6 in the connector is shown in blue, gp15 in green and gp16 in orange. The structure of the SPP1 portal protein at 9 Å resolution is shown in (B) (stereo view). (C) Cut-away views of the connector and portal protein. The outlines show the location of the gp6 main domains: the crown of tentacles is in orange, the wing is outlined in green, the steam stalk is in magenta, the stem foot is in purple and the gp16 stopper is in red.
The connector has a mushroom-like shape and a channel along the central axis (Figure 3A and C). Its upper part has a structural organization similar to the isolated portal protein oligomer (Orlova et al., 1999; Figure 3B and C) in spite of the different rotational symmetry (Dube et al., 1993; Lurz et al., 2001). The bottom part of the connector has additional rings, which are identified as the gp15 and gp16 oligomers (Lurz et al., 2001). The three different proteins in the connector are depicted in different colours in Figures 3 and 4 (gp6 is shown in blue, gp15 in green and gp16 in orange). Areas of low densities between the rings, where the links were relatively thin, defined the boundaries of each oligomer. The volumes of each of the three proteins account for molecular masses of 640, 120 and 140 kDa, and these correspond well to the masses expected for 12mers of gp6, gp15 and gp16, respectively.
Fig. 4. Comparison between the structure of bacteriophage SPP1 portal protein, the SPP1 portal protein in the connector and bacteriophage φ29 portal protein. (A) Comparison of the gp6 12mer from the connector (in blue) with the model of the portal protein 12mer (in pink). Thin lines show alignment of structures. (B) Single subunits from gp6 in the SPP1 connector and isolated gp6. The upper row shows side views of subunits with fitted cylinders, corresponding to putative helices, superimposition of helices and the side view of the φ29 portal protein. In the bottom row are shown front views for each subunit from the two forms. The four cylinders are highlighted by colours matching the helices in the φ29 portal protein crystal structure (Simpson et al., 2000). The cylinders are labelled according to the nomenclature of Simpson et al. (2000). The most significant changes are observed in the area corresponding to helix a3.
Both the isolated portal protein and the gp6 oligomer in the connector have the same three main domains in common: the wing, the crown and the stem (Figure 3; Orlova et al., 1999). Wings radiate outwards from the stem, giving the structures a turbine-like appearance (top views in figure 5 of Lurz et al., 2001; data not shown). The crown on the connector extends 20 Å above the upper rim of the wing and towards the interior of the viral capsid (Figure 3A and C). The most extensive inter-subunit contacts are observed in the stem and crown regions. The gp6 stem can be divided into a ‘stalk’ and a ‘foot’. The stalk is composed of rods of density that connect the wing to the foot. A set of three rods with diameters ∼10 Å can be assigned to each individual subunit, a feature that can be appraised best when a single subunit is extracted from the 12mer (Figure 4B). Using the program Helixhunter, we analysed the density map to see whether the rods could be interpreted as α-helical elements (Jiang et al., 2001). Because the resolution of the reconstruction is limited to 10 Å, we could only search for rather long cylinders of density (length ≥12 Å and diameter 10 Å). A preliminary search throughout the complete structure revealed some helical elements in the gp6 stem. The search was then refined further to concentrate on the region forming the central channel below the crown of gp6 and the area close to it (see Materials and methods). The positions of putative helices are shown in Figure 4B. The gp6 foot has a complex distribution of densities with areas where the inter-subunit separation remains uncertain.
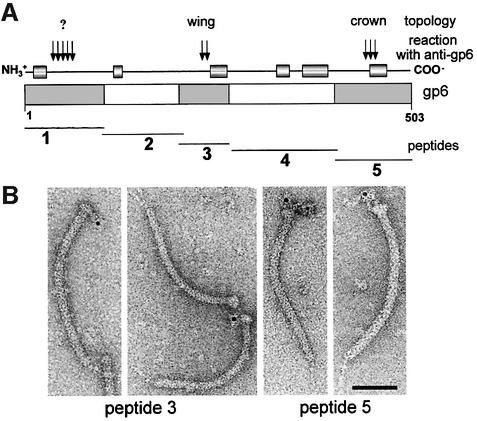
Fig. 5. Correlation between gp6 amino acid sequence and immunoreactive regions of the gp6 oligomer. The thick bar in (A) represents the gp6 amino acid sequence. The position of putative α-helices (>8 amino acids and with a reliability index of prediction ≥7) based on the secondary structure prediction PHD program (Rost, 1996) are shown by cylinders connected by a thin line above the thick bar. The thin lines below represent the peptides of gp6 used to define the regions of the protein that react with anti-gp6 polyclonal serum. Immunoreactive peptides in a western blot developed with anti-gp6 serum (peptides 1, 3 and 5) are shadowed in the thick bar. The relative strength of the western blot signal is indicated by vertical arrows on top of the bar. Affinity-purified antibodies specific for peptides 3 and 5 were used to identify the position of those peptides in gp6 present in tails with a connector by immunoelectron microscopy (B). More than 90% of the tail–connector complexes observed in the EM were immunolabelled, and the antibody–connector complexes revealed reproducibly the topology presented in (A). The topology found is indicated at the top of (A). Antibodies against the first 100 N-terminal amino acids, the most antigenic peptide of gp6 (A), could not be obtained due to lack of dissociation of the antibody–peptide 1 complexes. The bar on the micrograph represents 50 nm.
The foot of the gp6 assembly forms a sleeve over the top of the gp15 ring (Figure 3A and C). This region of gp15 facing gp6 has a wider opening (∼40 Å) than its lower part (∼27 Å), which interacts with gp16 (Figure 3C). Twelve individual petals of gp15 spread out to form the outer part of the ring. Each petal makes a contact with gp6. The inner part of gp15 makes the most extensive contacts with gp16 (Figure 3C). Twelve subunits of gp16 stretch outwards from the symmetry axis. The central part of the gp16 oligomer forms an inverted conical ‘stopper’ that closes the connector channel (Figure 3A and C). The bottom of the gp16 ring provides the interface for interaction with the phage tail (Lurz et al., 2001).
Structure of the assembly-naïve portal protein
The structure of the SPP1 portal protein produced in the absence of other viral proteins (assembly-naïve) was refined to 9 Å resolution, using images of ice-embedded gp6 (see Materials and methods). The present reconstruction provides considerably more detail than our previous reconstructions using negative staining (Tavares et al., 1995) or the cryogenic high-contrast embedding technique (Orlova et al., 1999). The wing, crown and stem domains that previously appeared as continuous density have now been resolved into a network of structural elements. The stem is divided into stalk and foot regions, as described for gp6 in the connector (Figure 3C). In addition, the organization of the crown is now clearly visible because gp6 was imaged in the presence of magnesium (Figure 3C; Tavares et al., 1995). In the absence of divalent cations, this region appears as a short fringe whose upper part was only seen at low threshold values, due to structural flexibility (figure 2 in Orlova et al., 1999). Millimolar amounts of magnesium stabilize and compact the oligomer, causing a reduction of its hydrodynamic radius in solution (Jekow et al., 1999). The most significant effect observed in the portal structure is the stabilization of the crown structural elements and a reduction in the gp6 maximum diameter from 205 to 185 Å (Figure 3C; Tavares et al., 1995).
Comparison of gp6 structures
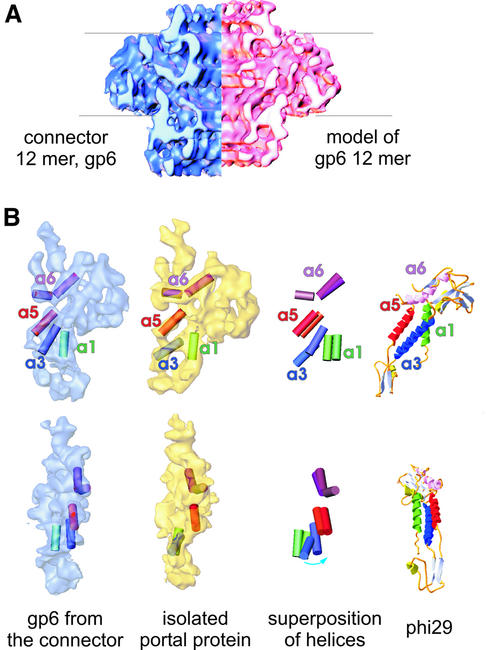
The structures of assembly-naïve gp6 (13mer; Figure 3B and C) and of gp6 assembled in the connector (12mer; Figure 3A and C) were compared in order to assess the structural changes that the protein undergoes during viral assembly. Based on the structure of a subunit isolated from the 13mer, we generated an artificial 12mer to compare it with the gp6 12mer within the connector. To make the model of the 12mer, one subunit of the 13mer was aligned as a rigid body with one subunit of gp6 from the connector using three-dimensional cross-correlation (see Materials and methods). Alignment showed that the subunit from the portal protein has to be rotated ∼5° to make it more vertical, and that it was also shifted inward by ∼4 Å. Afterwards, the subunit was replicated with 12-fold symmetry. The two dodecameric structures are shown in Figure 4A. The crowns are similar in both structures, and the wings, though slightly more upright in the connector, keep the same appearance in both structures. Significant differences are found in the gp6 stem. The cut-away views show that the compact region of the foot that contacts gp15 is wider in the connector when compared with isolated gp6, implying that parts of the polypeptide chain move outwards to accommodate the inner ring of gp15 inside the bottom of the gp6 stem (Figure 3C). This structural change increases the height of gp6 in the connector.
The set of three rods that were assigned to the stalk of individual gp6 subunits in the connector structure can be identified readily in the subunit of assembly-naïve gp6. These were also interpreted as α-helices using the program Helixhunter for density analysis (Jiang et al., 2001; see Materials and methods). The rods of density labelled as a1 (green, a length of ∼15 Å) and a5 (orange, ∼18 Å) adopt in general very similar though slightly more vertical orientations in the connector than in the portal protein. The density corresponding to a3 (blue, ∼22 Å) in the connector is rotated by a rather large angle (∼35°) to a significantly more vertical position relative to the a3 density in the portal protein (Figure 4B). The general arrangement of putative α-helical elements in the gp6 stalk shows an intriguing similarity to the long α-helices that line the central channel in the corresponding region of the X-ray crystal structure of the φ29 portal protein (Simpson et al., 2000; Figure 4B). The wing domain of the gp6 subunit is larger than that of the φ29 subunit (∼40 kDa), which is consistent with the larger molecular mass of the gp6 subunit (57.3 kDa).
Three segments of the gp6 amino acid sequence are exposed at the protein surface (Figure 5A). Antibodies purified by affinity for two of those peptides were used to localize their position by immuno-electron microscopy of connector–tail complexes (Figure 5B). Antibodies against peptide 3 (residues 200–269 of gp6) bind to the tail– connector complexes on the side of gp6 and cross-link the complexes mainly side by side. This binding suggests that the peptide is localized in the gp6 wing. Antibodies against the gp6 C-terminal peptide 5 (residues 404–503) reproducibly label the top of the connector where the crown is located (Figure 5B). Correlation of this topology information with the arrangement of predicted α-helices derived from the gp6 amino acid sequence in the C-terminus (Figure 5A) is consistent with the arrangement of putative helices within gp6 and helices 3, 5 and 6 in the φ29 portal protein (Simpson et al., 2000), further supporting the structural similarity shown in Figure 4B.
Biological implications
The structure of the connector highlights the function of this multiprotein complex as a gatekeeper of the packaged viral DNA. Although the portal protein is the only connector component responsible for DNA packaging into the capsid, attachment of gp15 and gp16 to the portal vertex after termination of DNA encapsidation is required to retain the packed DNA inside the capsid (Figure 2). The connector represents an intermediate in assembly of the phage head–tail interface, whose function is to prevent DNA leakage immediately after the packaging reaction (Figure 1).
Comparison of the structure of gp6 from the connector complex, which is assembled at a late stage of viral assembly, and of assembly-naïve gp6 shows that the general appearance of the molecule is similar in both structures. The portal protein wings and crown are stable, although the stem undergoes noticeable conformational changes. Changes in densities that correspond to putative α-helical elements of the stem show alterations in the tilt angle, and the base of the stem endures a large motion outwards, creating an interface to accommodate gp15 below gp6. At present, we cannot specify the point at which these changes occur during the viral assembly pathway (Figure 1). An interesting possibility is that the change in orientation of the stem helices might be related to movements involved in DNA translocation (see also Simpson et al., 2000). Comparison of portal protein structures from different phages (φ29 in Simpson et al., 2000; Guasch et al., 2002; SPP1 here; and T3 in Valpuesta et al., 2000) exhibits the common structural organization. Conservation of the structural elements lining the central channel and their relatively similar dimensions might suggest association with a common function in DNA packaging. Other portal structure domains show more variable structure and size (e.g. large wing and crown domains in gp6).
The conformational change at the bottom of the stem that leads to exposure of the gp6 interface for interaction with gp15 is most probably concomitant with termination of DNA packaging. If the gp6 interface for interaction with gp15 were exposed in the portal structure at an early morphogenetic step, gp15 could bind strongly to the portal at any of the morphogenetic steps that precede DNA packaging, or could compete with the terminase–DNA complex for interaction with the portal vertex (Figure 1). These situations would lead to abortive morphogenesis. Furthermore, gp15 and gp16 are found exclusively in DNA-filled capsids, showing that those proteins do not bind stably to assembly intermediates before DNA packaging is achieved (Lurz et al., 2001). These observations support the idea that the portal protein undergoes a conformational change to trigger gp15 binding at the appropriate moment, shortly after termination of DNA packaging (Figure 1).
The connector structure shows that gp15 serves as an extension of the portal protein channel where gp16 binds. The central channel is closed by gp16 physically blocking the exit from the DNA-filled capsid (Figure 3). The subsequent attachment of the phage tail to the connector leads to proteolysis of a few amino acids in gp16. Then the DNA terminus, which is packaged last, becomes tightly bound to the head–tail interface (Tavares et al., 1996; Lurz et al., 2001). A DNA fragment of length 210 ± 20 Å is protected by the head–tail interface (Tavares et al., 1996). These observations demonstrate that tail binding is accompanied by some rearrangement in the connector structure. This restructuring may represent the first step of preparation for DNA ejection. The fragment of gp16 that is cleaved off when the tail binds could be part of the ‘stopper’ allowing the end of the viral DNA to make a strong bond with the head–tail interface. This would create the structural context for DNA release through the tail channel once it is triggered by virus–host interaction.
The ubiquity of the portal vertex in tailed bacteriophages and probably in herpes viruses (Newcomb et al., 2001; and references therein) strongly suggests that a number of the molecular mechanisms discussed here, essential for SPP1 assembly, also apply to many viruses.
Materials and methods
Preparation of total DNA from infected cells and PFGE
Bacteriophages SPP1 wild type, SPP1sus117 and SPP1sus128 were from our strain collection (Behrens et al., 1979). The positions of mutations sus117 and sus128 were mapped by marker rescue in genes 16 (this work) and 15 (Becker et al., 1997). Their exact position was determined by DNA sequencing (Figure 2).
To assay DNA packaging, 20 ml cultures of the non-permissive strain B.subtilis YB886 were grown in rich medium to a density of ∼108 colony-forming units/ml. The culture was supplemented with 10 mM CaCl2 and infected with the phage strain under analysis to an input multiplicity of 10. Aliquots of 5 ml of infected culture were taken at 5, 15 and 22 min post-infection, and phage multiplication was stopped by mixing the culture with an identical volume of ice-cold TBT (100 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2) supplemented with 1% (w/v) sodium azide (Chai et al., 1992). Cells were sedimented by centrifugation and the supernatant that contained input phages used for infection was removed carefully. The pellet was resuspended in 1 ml of low melting point agarose (SeaPlaque agarose, FMC BioProducts) prepared in 6 mM Tris–HCl pH 8.0. The suspended material was distributed in a block form. When the agarose was solidified, the blocks were poked out from the form and half of the blocks were incubated in 40–50 vols of lysis buffer (6 mM Tris–HCl pH 8.0, 0.1 M EDTA pH 8.0, 1 M NaCl, 30 µg/ml lysozyme, 3 µg/ml RNase). The other half of the blocks was incubated in 40–50 vols of lysis buffer containing DNase (6 mM Tris–HCl pH 8.0, 50 mM NaCl, 10 mM MgCl2, 1% tergitol NP-40, 30 µg/ml lysozyme, 1 µg/ml RNase, 1 µg/ml DNase) for 4 h at 37°C and then EDTA was added to 0.1 M and NaCl to 1 M. Blocks were incubated overnight at 37°C. The lysis buffer was replaced by proteinase K buffer (0.5 M EDTA, 30 mM N-lauroyl-sarkosine, 1 µg/ml proteinase K), and the samples were incubated at 37°C for 4–6 h followed by four washes for 30 min with TE buffer (5 mM Tris–HCl pH 7.5, 0.1 mM EDTA). The agarose blocks were melted for 10 min at 65°C, and 10 µl were loaded in 1% agarose gels (SeaKem GTG agarose, FMC BioProducts). Electrophoresis conditions were as described by Tavares et al. (1995). SPP1 mature chromosomes were visualized as a discrete band in ethidium bromide-stained gels and their relative size was estimated by comparison with mature DNA purified from different SPP1siz mutants (Tavares et al., 1995).
Production of gp6 peptides, affinity purification of anti-peptide antibodies, and antibody labelling procedures
Segments of gene 6 coding for five peptides that span the complete gp6 amino acid sequence (Figure 5A) were amplified by PCR and cloned in vector pRSET A (Invitrogen) using a strategy similar to that described by Lurz et al. (2001) to generate proteins tagged on their N-terminus with polyhistidine. Peptide overproduction was as described in Lurz et al. (2001) except that 2.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was used for culture induction, and overproduction was carried out for 2 h in the absence of rifampicin. Fusion peptides were purified in batch by affinity chromatography to Ni-NTA resin under denaturing conditions, as described by the manufacturers (Qiagen, The QIAexpressionist 03/99). Peptides were checked by SDS–PAGE stained with Coomassie Blue, and by western blotting developed with polyclonal anti-gp6 serum (data not shown).
Semi-purified peptides were run in preparative SDS–polyacrylamide gels and electro-transferred to Immobilon-P membranes (Millipore). Each peptide was visualized in the membrane by Ponceau S staining, and a strip containing the immobilized peptide was cut out. The strips were blocked with phosphate-buffered saline (PBS) containing 5% non-fat milk and 0.1% Tween-20 for 30 min. Loosely bound peptide was eliminated by incubation with stripping solution (150 mM NaCl, 100 mM glycine pH 2.5) for 15 min and then washed with PBS with 0.1% Tween-20 (wash buffer). Membrane strips were incubated individually for 1 h with anti-gp6 rabbit polyclonal serum diluted 1:10 in PBS containing 5% non-fat milk and 0.1% Tween-20 for antibody affinity binding. The serum solution was removed carefully and each membrane was washed five times, each for 5 min, with wash buffer. Bound antibodies were eluted with a small volume of stripping solution for 15 min, followed by immediate titration with Tris buffer pH 8.0, to a final concentration of 100 mM. Each solution of affinity-purified antibody was kept at 4°C for >48 h and then used for western blot and immunoelectron microscopy labelling as described in Lurz et al. (2001).
Electron microscopy and image processing
Gp6 and connectors were purified as described previously (Jekow et al., 1999; Lurz et al., 2001). Connectors were concentrated and washed to eliminate glycerol in Centricon-30 microconcentrators (Amicon). Low-dose images (∼10 e–/Å2) of the purified portal protein and connector were taken in a Philips CM200 FEG electron cryomicroscope at 200 kV using a Gatan side-entry cryo holder. The magnification was 50 000× and defocus was in the range 1.7–3 µm. High quality micrographs selected by optical diffraction were digitized using a patchwork densitometer (van Heel et al., 2000) (step size of 2.1 Å on the specimen scale). Particles on each micrograph were selected interactively within frames of 256 × 256 pixels (see Table I) and corrected for the contrast transfer function (CTF) by flipping phases. Images were band-pass filtered (half-widths of ∼0.005 Å–1 and ∼0.5 Å–1 for the low and high frequency cut off) and the frames were then cropped to 190 × 190 pixels. Alignment and classification were performed as described previously (Orlova et al., 1999; van Heel et al., 2000). The relative orientation of class averages was determined by angular reconstitution (van Heel, 1987). Orientations of class averages were refined iteratively against the three-dimensional model obtained from the classes with the lowest errors in the reconstruction, which were equally distributed on the Euler sphere (Serysheva et al., 1995; Orlova, 2000). Classes with errors in the angle orientation search >40% were rejected in the course of refinement. For the final steps of analysis, classes with reconstruction errors >50% were also rejected. The average error for all classes used in the final reconstruction was 30%, corresponding to the average correlation coefficient (CC) of 0.7 between classes and reprojections from the reconstruction for the portal protein, and 35% (CC = 0.65) for the connector. Seventy percent of all images were included for the reconstruction so that the reconstruction can be considered as representative of the data set. Three-dimensional maps were calculated using the exact-filter back projection algorithm (Harauz and van Heel, 1986; Radermacher, 1988).
Table I. EM data processed at the analysis.
| Sample | No. of films processed | No. of particles selected | No. of classes analysed | No. of classes in the final 3D | Average error of the reconstruction | Resolution (Å) at ∼20σ |
|---|---|---|---|---|---|---|
| Gp6 | 15 | ∼8000 | 650 | 430 | 30% | 9 |
| Connector | 21 | ∼4700 | 320 | 260 | 35% | 10 |
Three-dimensional alignment of subunits between maps was based on cross-correlation of densities. First we performed an approximate translational alignment in three-dimensional space. This was followed by a search for rotational orientation with the best matching of the two structures. The procedure was iterated three times. All image analysis was performed using the IMAGIC-5 package (van Heel et al., 1996a).
Boundaries between subunits were defined by gaps between clusters of high densities, though there were some areas where the inter-subunit separation was uncertain. At the resolution obtained, we were not able to determine accurately the subunit boundary in the base of the portal protein stem and in some central areas of the connector. However, the gradient of densities between subunits helped us to extract hypothetical subunits from both structures. Calculation of molecular masses of the extracted rings was an additional test for checking the boundaries chosen. Molecular masses of rings and oligomers were calculated from the volumes at the threshold of 1σ above the average density level in reconstructions, using the specific density of 0.84 Da/Å3 for proteins.
Identification of density regions corresponding to helices was performed using the program ‘Helixhunter’ (Jiang et al., 2001). A prototype helix of length ∼12 Å and of diameter ∼10 Å was used for the search procedure. At rough increment (∼7°), the program found some helical elements in the stem region and below the crown of gp6. To make the analysis somewhat faster and more accurate, we extracted the region of the gp6 subunit that lines the internal channel for the next search. The increment was reduced to 5°. The correlation coefficients found for the potential helices varied between 0.44 and 0.71 for the portal protein and between 0.35 and 0.53 for the connector, with lengths in the range of 12–22 Å. The analysis clearly revealed the positions of possible helices in the stem area, but the positions of helical elements were ambiguous in the wing area. The accuracy of positioning of each putative helix is about ±3°, ±4 Å depending on the angular increment and on filtering of the three-dimensional map. The most convincing results were achieved when low frequencies corresponding to details larger then the width of the main wing domain (≤1/30 Å–1) were suppressed.
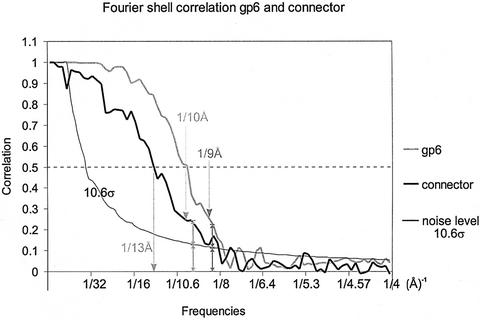
The resolution of the portal protein map was determined to be 9 Å as assessed by the Fourier shell correlation (FSC) (van Heel and Harauz, 1986), where the correlation curve was twice as high as the K∗σ background level (K depends on the symmetry and, for the portal protein, K = 3√13σ ≌10.6σ; see Orlova et al., 1997). From our experience, structural details at the resolution level defined in this way appear reliable. The resolution at 0.5 correlation level was 10 Å. The resolution for the connector reconstruction was estimated to be 10 Å by the above-mentioned criterion, and 13 Å at the 0.5 correlation level (Figure 6).
Fig. 6. Fourier shell correlation curves of gp6 and of the connector in grey and black, respectively. The threshold curve is in thin black and corresponds to 10.6σ. The arrows are pointing to the frequency level, where the correlation twice exceeds the background level. The lower resolution for the connector as compared with the portal protein structure resulted from the smaller number of molecular images available for the analysis.
Accession codes
The EM maps of the SPP1 portal protein and of the connector have been deposited in the macromolecular structure database (EBI) under accession codes EMD-1020 and EMD-1021, respectively.
Acknowledgments
Acknowledgements
We thank T.A.Trautner for continuous interest in and support of the project and for critically reading the manuscript, H.Saibil for insightful suggestions during manuscript preparation, M.Schatz and R.Schmidt for IMAGIC software support, and A.A.Antson for fruitful discussions. The research was supported in part by grants from BBSRC to M.v.H.
References
- Alonso J.C., Lüder,G., Stiege,A.C., Chai,S., Weise,F. and Trautner,T.A. (1997) Analysis of the complete nucleotide sequence and functional organisation of Bacillus subtilis bacteriophage SPP1. Gene, 204, 201–212. [DOI] [PubMed] [Google Scholar]
- Bazinet C. and King,J. (1985) The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol., 39, 109–129. [DOI] [PubMed] [Google Scholar]
- Becker B., de la Fuente,N., Gassel,M., Günther,D., Tavares,P., Lurz,R., Trautner,T.A. and Alonso,J. (1997) Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol., 268, 822–839. [DOI] [PubMed] [Google Scholar]
- Behrens B., Lüder,G., Behnke,M. and Trautner,T.A (1979) The genome of B.subtilis phage SPP1, physical arrangement of phage genes. Mol. Gen. Genet., 175, 351–357. [DOI] [PubMed] [Google Scholar]
- Chai S., Bravo,A., Luder,G., Nedlin,A., Trautner,T.A. and Alonso,J.C. (1992) Molecular analysis of the Bacillus subtilis bacteriophage SPP1 region encompassing genes 1 to 6. The products of gene 1 and gene 2 are required for pac cleavage. J. Mol. Biol., 224, 87–102. [DOI] [PubMed] [Google Scholar]
- Coombs D.H. and Eiserling,F.A. (1977) Studies on the structure, protein composition and assembly of the neck of bacteriophage T4. J. Mol. Biol., 116, 375–405. [DOI] [PubMed] [Google Scholar]
- Donate L.E., Herranz,L., Secilla,J.P., Carazo,J.M., Fujisawa,H. and Carrascosa,J.L. (1988) Bacteriophage T3 connector: three-dimensional structure and comparison with other viral head–tail connecting regions. J. Mol. Biol., 201, 91–100. [DOI] [PubMed] [Google Scholar]
- Dröge A. and Tavares,P. (2000) In vitro packaging of DNA of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol., 296, 103–115. [DOI] [PubMed] [Google Scholar]
- Dröge A., Santos,M.A., Stiege,A., Alonso,J.C., Lurz,R., Trautner,T.A. and Tavares,P. (2000) Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J. Mol. Biol., 296, 117–132. [DOI] [PubMed] [Google Scholar]
- Dube P., Tavares,P., Lurz,R. and van Heel,M. (1993) Bacteriophage SPP1 portal protein: a DNA pump with 13-fold symmetry. EMBO J., 12, 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C. and Casjens,S. (1980) DNA packaging by the double-stranded DNA bacteriophages. Cell, 21, 319–331. [DOI] [PubMed] [Google Scholar]
- Gual A., Camacho,A.G. and Alonso,J.C. (2000) Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem., 275, 35311–35319. [DOI] [PubMed] [Google Scholar]
- Guasch A., Pous,J., Ibarra,B., Gomis-Rüth,F.X., Valpuesta,J.M., Sousa,N., Carrascosa,J.L. and Coll,M. (2002) Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophages φ29 connector particle. J. Mol. Biol., 315, 663–676. [DOI] [PubMed] [Google Scholar]
- Hagen E.W., Reilly,B.E., Tosi,M.E. and Anderson,D.L. (1976) Analysis of gene function of bacteriophage φ29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J. Virol., 19, 501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harauz G. and van Heel,M. (1986) Exact filters for general geometry three dimensional reconstruction. Optik, 73, 146–156. [Google Scholar]
- Hendrix R.W. (1978) Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl Acad. Sci. USA, 75, 4779–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R.W. (1999) The long evolutionary reach of viruses. Curr. Biol., 9, R914–R917. [DOI] [PubMed] [Google Scholar]
- Isidro A. (2002) Analysis of the SPP1 portal protein function during prohead assembly and DNA packaging—a bridge between function and structure. PhD thesis, Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Portugal.
- Jekow P. Behlke,J., Tichelaar,W., Lurz,R., Regalla,M., Hinrichs,W. and Tavares,P. (1999) Role of the ionic environment in the assembly of bacteriophage SPP1 portal protein. Eur. J. Biochem., 264, 724–735. [DOI] [PubMed] [Google Scholar]
- Jiang W., Baker,M.L., Ludtke,S.J. and Chiu,W. (2001) Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol., 308, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Lurz R., Orlova,E.V., Günther,D., Dube,P., Dröge,A., Weise,F., van Heel,M. and Tavares,P. (2001) Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol., 310, 1027–1037 [DOI] [PubMed] [Google Scholar]
- Molineux I.J. (2001) No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol., 40, 1–8. [DOI] [PubMed] [Google Scholar]
- Newcomb W.W., Juhas,R.M., Thomsen,D.R., Homa,F.L., Burch,A.D., Weller,S.K. and Brown,J.C. (2001) The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol., 75, 10923–10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova E.V. (2000) Structural analysis of non-crystalline macromolecules: the ribosome. Acta Crystallogr. D, 56, 1253–1258. [DOI] [PubMed] [Google Scholar]
- Orlova E.V,. Dube,P., Harris,J.R., Beckman,E., Zemlin,F., Markl,J. and van Heel,M. (1997) Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol., 271, 417–437. [DOI] [PubMed] [Google Scholar]
- Orlova E.V., Dube,P., Beckmann,E., Zemlin,F., Lurz,R., Trautner,T.A., Tavares,P. and van Heel,M. (1999) Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol., 6, 842–846. [DOI] [PubMed] [Google Scholar]
- Perucchetti R., Parris,W., Becker,A. and Gold,M. (1988) Late stages in bacteriophage λ head morphogenesis: in vitro studies on the action of the bacteriophage λD-gene and W-gene products. Virology, 165, 103–114. [DOI] [PubMed] [Google Scholar]
- Radermacher M. (1988) Three-dimensional reconstruction of single particles from random and non-random tilt series. J. Electron Microsc. Tech., 9, 359–394. [DOI] [PubMed] [Google Scholar]
- Rost B. (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Serysheva I.I., Orlova,E.V., Chiu,W., Sherman,M.B., Hamilton,S.L. and van Heel,M. (1995) Electron microscopy and angular reconstitution used to visualize the skeletal muscle calcium release channel. Nat. Struct. Biol., 2, 18–24. [DOI] [PubMed] [Google Scholar]
- Simpson A.A. et al. (2000) Structure of the bacteriophage φ29 DNA packaging motor. Nature, 408, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.E., Tans,S.J., Smith,S.B., Grimes,S., Anderson,D.L. and Bustamante,C. (2001) The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature, 413, 748–752. [DOI] [PubMed] [Google Scholar]
- Steven A.C. and Trus,B.L. (1986) The structure of bacteriophage T7. In Harris,J.R. and Horne,R.W. (eds), Electron Microscopy of Proteins, Volume 5, Viral Structure. Academic Press, New York. pp. 1–35.
- Strauss H. and King,J. (1984) Steps in the stabilisation of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol., 172, 523–543. [DOI] [PubMed] [Google Scholar]
- Tavares P., Dröge,A., Lurz,R., Graeber,I., Orlova,E., Dube,P. and van Heel,M. (1995) The SPP1 connection. FEMS Microbiol. Rev., 17, 47–56. [Google Scholar]
- Tavares P., Lurz,R., Stiege,A., Rückert,B. and Trautner,T.A. (1996) Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol., 264, 954–967. [DOI] [PubMed] [Google Scholar]
- Valpuesta J.M. and Carrascosa,J.L. (1994) Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys., 27, 107–155. [DOI] [PubMed] [Google Scholar]
- Valpuesta J.M., Fujisawa,H., Marco,S., Carazo,J.M. and Carrascosa,J.L. (1992) Three-dimensional structure of T3 connector purified from overexpressing bacteria. J. Mol. Biol., 224, 103–112. [DOI] [PubMed] [Google Scholar]
- Valpuesta J.M., Sousa,N., Barthelemy,I., Fernandez,J.J., Fujisawa,H., Ibarra,B. and Carrascosa,J.L. (2000) Structural analysis of the bacteriophage T3 head-to-tail connector. J. Struct. Biol., 131, 146–155. [DOI] [PubMed] [Google Scholar]
- van Heel M. (1987) Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy, 21, 111–124. [DOI] [PubMed] [Google Scholar]
- van Heel M. and Harauz,G. (1986) Resolution criteria for three dimensional reconstruction. Optik, 73, 119–122. [Google Scholar]
- van Heel M., Harauz,G., Orlova,E.V., Schmidt,R. and Schatz,M. (1996a) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- van Heel M., Orlova,E.V., Dube,P. and Tavares,P. (1996b) Intrinsic versus imposed curvature in cyclical oligomers: the portal protein of bacteriophage SPP1. EMBO J., 15, 4785–4788. [PMC free article] [PubMed] [Google Scholar]
- van Heel M. et al. (2000) Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys., 33, 307–369. [DOI] [PubMed] [Google Scholar]