Abstract
Polycomb group (PcG) proteins function as high molecular weight complexes that maintain transcriptional repression patterns during embryogenesis. The vertebrate DNA binding protein and transcriptional repressor, YY1, shows sequence homology with the Drosophila PcG protein, pleiohomeotic (PHO). YY1 might therefore be a vertebrate PcG protein. We used Drosophila embryo and larval/imaginal disc transcriptional repression systems to determine whether YY1 repressed transcription in a manner consistent with PcG function in vivo. YY1 repressed transcription in Drosophila, and this repression was stable on a PcG-responsive promoter, but not on a PcG-non-responsive promoter. PcG mutants ablated YY1 repression, and YY1 could substitute for PHO in repressing transcription in wing imaginal discs. YY1 functionally compensated for loss of PHO in pho mutant flies and partially corrected mutant phenotypes. Taken together, these results indicate that YY1 functions as a PcG protein. Finally, we found that YY1, as well as Polycomb, required the co-repressor protein CtBP for repression in vivo. These results provide a mechanism for recruitment of vertebrate PcG complexes to DNA and demonstrate new functions for YY1.
Keywords: development/Polycomb group/repression/transcription/YY1
Introduction
Polycomb group (PcG) proteins were first identified in Drosophila as proteins required to maintain repression of homeotic genes necessary for anterior–posterior development (McKeon and Brock, 1991; Simon et al., 1992). Homeotic gene expression patterns are initiated early in development by the maternal and segmentation genes such as the gap and pair-rule genes (Beinz and Muller, 1995). The gap and pair-rule genes are expressed transiently, but homeotic gene expression must be continuous for proper development. Two additional families of regulatory proteins are necessary for the maintenance of homeotic gene expression. These are the trithorax group proteins, which maintain active homeotic gene expression where the genes were originally expressed (Kennison, 1993), and the PcG proteins, which maintain the repressed state where homeotic gene expression was originally inactive (Paro, 1993; Pirrotta, 1997a,b; Schumacher and Magnuson, 1997). In PcG loss-of-function mutants, homeotic gene expression is correctly initiated, but as expression of maternal and segmentation genes decays, the anterior boundaries of homeotic gene expression are not properly maintained but are shifted toward the anterior (Duncan and Lewis, 1982). This results in posterior homeotic transformation where ectopic expression of homeotic genes takes place.
A number of vertebrate proteins homologous to Drosophila PcG proteins have been identified. These mammalian PcG proteins can regulate hox gene expression and are important for skeletal development and hematopoiesis (van der Lugt et al., 1994; Akasaka et al., 1997, 2001; Bel et al., 1998). Like their Drosophila counterparts, mammalian PcG gene mutants result in segmentation defects characterized by posterior transformations of various skeletal structures (van der Lugt et al., 1994; Alkema et al., 1995; Akasaka et al., 1996; Bel et al., 1998). A subset of mammalian and Drosophila PcG mutants result in lethality very early in embryogenesis, indicating important functions in both early and late developmental stages (van der Lugt et al., 1994; O’Carroll et al., 2001). Although mammalian and Drosophila PcG proteins are generally believed to mediate similar functions, only a single mammalian PcG protein has been shown to function in Drosophila to correct a PcG mutant phenotype (Muller et al., 1995).
PcG proteins function as high molecular weight complexes that, in Drosophila, bind to regulatory elements termed Polycomb (Pc) response elements (PRE) (Pirrotta, 1997a,b, 1999; Satijn and Otte, 1999; Brock and van Lohuizen, 2001; Francis and Kingston, 2001). No mammalian PREs have been identified, partly because hardly any PcG proteins individually bind to DNA specifically. A single Drosophila PcG protein, pleiohomeotic (PHO), has been shown to bind to DNA specifically (Brown et al., 1998), and therefore may function to nucleate PcG complexes on DNA. PHO can bind to specific sites in many PRE sequences, and mutation of either PHO DNA binding site or the PHO protein itself can reduce PcG silencing (Girton and Jeon, 1994; Brown et al., 1998; Fritsch et al., 1999; Busturia et al., 2001; Mishra et al., 2001). PHO can physically interact with some PcG proteins and can generate ternary complexes on DNA with the Pc protein (Mohd-Sarip et al., 2002). Therefore, PHO appears to be an important component of at least some PcG repression systems. Interestingly, PHO has sequence homology to the well-characterized vertebrate transcription repressor, YY1.
YY1 is a 414 amino acid, multifunctional transcription factor that can either activate or repress transcription, depending upon promoter contextual differences, or specific protein interactions (reviewed in Shrivastava and Calame, 1994; Shi et al., 1997; Thomas and Seto, 1999). Some of the domains responsible for YY1 function have been mapped, with most studies showing that sequences near the C-terminus (which overlap the YY1 zinc fingers) can repress transcription (Bushmeyer and Atchison, 1998; Thomas and Seto, 1999). However, some studies indicate that other sequences are also involved in transcriptional repression (Yang et al., 1996). YY1 sequences important for transcriptional activation reside near the N-terminus (Lee et al., 1994, 1995; Bushmeyer et al., 1995; Austen et al., 1997). The mechanism by which YY1 activates or represses transcription is presently unclear. A number of repression mechanisms have been proposed, but nearly all of the transcriptional properties of YY1 have been defined by transient expression assays. Although mouse knock-out studies show that YY1 homozygous mutants die peri-implantation (Donohoe et al., 1999), little is know about the function of YY1 in vivo.
The homology between YY1 and Drosophila PHO resides in two YY1 domains: sequences 298–414 constituting the four zinc fingers (95% identical) and a short segment between residues 205–226 (82% identity) with no defined function. Although the remainder of YY1 shows no similarity to PHO, the above homologies suggest that YY1 might be a vertebrate counterpart of PHO and thus function as a PcG protein. If so, the finding would provide a mechanism for nucleating mammalian PcG complexes to DNA and assist in the identification of mammalian PREs, since the YY1 DNA binding site is well characterized (Hyde-DeRuyscher et al., 1995). Although YY1 has not been observed as a component of the known PcG complexes, it can physically interact with the vertebrate PcG protein, EED (Satijn et al., 2001). As described above, YY1 knock-out mutants are embryonic lethal (Donohoe et al., 1999), similar to some PcG genes (Schumacher et al., 1996; O’Carroll et al., 2001). However, YY1 has never been tested in an in vivo system that would reveal PcG function. We therefore set out to address the mechanism of YY1 transcriptional repression in vivo, using a system that would enable us to test its potential PcG function.
We show here that YY1 can repress transcription in developing Drosophila embryos and in larval imaginal discs. Similar to known PcG proteins, stable repression by YY1 was observed with a promoter responsive to PcG function, but not with a PcG-non-responsive promoter. Using various PcG mutant backgrounds, we found that YY1 transcriptional repression was dependent on PcG function. We also found that human YY1 could functionally compensate for PHO to correct phenotypic defects in pho mutant flies. Taken together, our results demonstrate that YY1 functions like a PcG protein. Finally, we have identified the co-repressor protein, CtBP, as a possible link between YY1 and the PcG complex.
Results
YY1 can repress transcription in developing Drosophila embryos
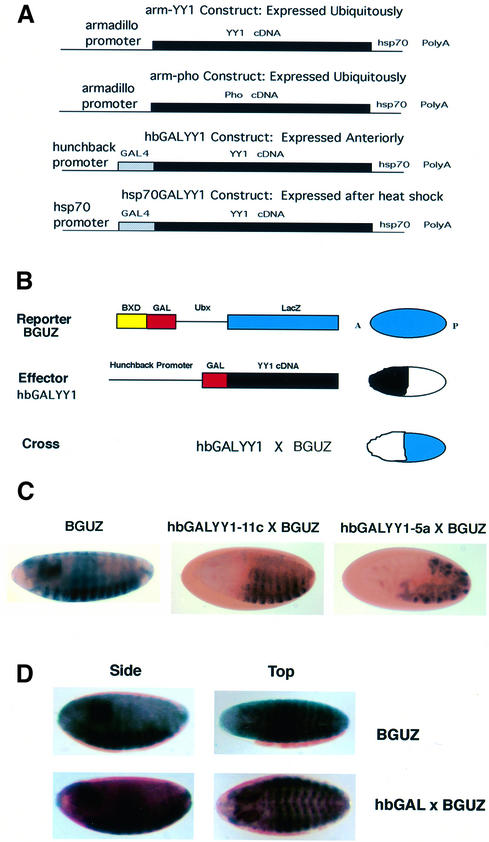
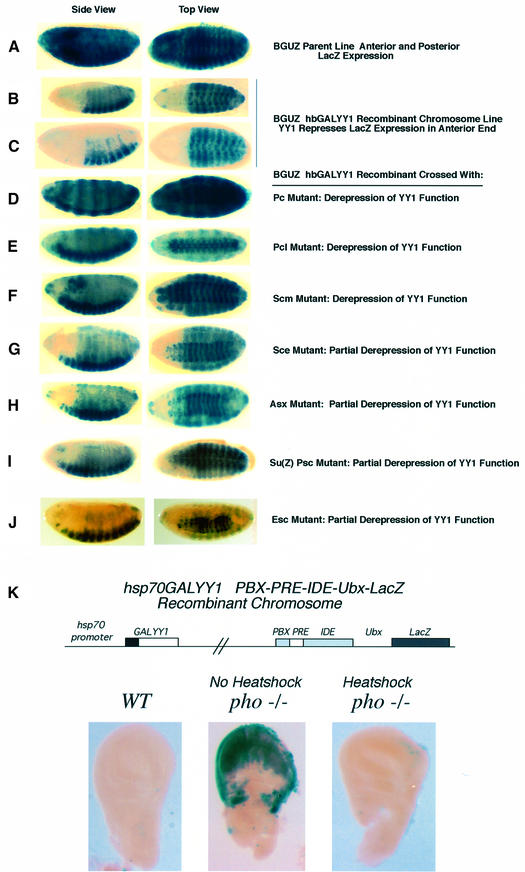
We wished to study YY1 function in a developing organism where its role in transcription and potential PcG function could be assessed in vivo. PcG function is well studied in Drosophila, and transgenic reporters are available that are repressed in a PcG-dependent fashion. Previously, it was shown that chimeric GAL-PcG proteins can nucleate PcG complexes to DNA and repress endogenous or ectopic reporter genes by a PcG-dependent mechanism (Muller, 1995). One known PcG-responsive construct consists of a LacZ gene under the control of the Ultrabithorax (Ubx) BXD enhancer and the Ubx promoter adjacent to GAL4 binding sites (BXDGALUbxLacZ; abbreviated to BGUZ; see Figure 1B) (Muller, 1995). This reporter gene is expressed ubiquitously during embryogenesis but is selectively repressed in a PcG-dependent fashion by the Pc protein linked to the GAL4 DNA binding domain (Muller, 1995). Therefore, we prepared transgenic Drosophila lines expressing a GALYY1 fusion construct driven by the hunchback promoter (hbGALYY1; Figure 1A). This construct delivers a pulse of GALYY1 in the anterior ends of developing Drosophila embryos. We crossed hbGALYY1 transgenic lines with the BGUZ reporter line and assayed the resulting embryos for LacZ expression. If GALYY1 can repress the BGUZ reporter transgene, one would expect LacZ expression only in the posterior ends where hbGALYY1 is not expressed (see Figure 1B for strategy). Indeed, two independent hbGALYY1 transgenic lines repressed LacZ expression in the embryonic anterior ends (Figure 1C), while no repression was observed with the GAL DNA binding domain alone (Figure 1D). The GALYY1 repression is similar to the repression previously observed with the hb driven GAL-Pc gene (Muller, 1995).
Fig. 1. (A) Transgenic constructs. The top construct shows the YY1 cDNA (Park and Atchison, 1991) (black rectangle) under control of the armadillo promoter at the 5′ side, with the hsp70 poly(A) site on the 3′ side (Muller et al., 1995). The same expression plasmid was also used for making the arm-pho transgene (Brown et al., 1998) (second construct). The third and fourth constructs show the YY1 cDNA (black rectangle) fused to the GAL4 DNA binding domain (sequences 1–147, cross hatched rectangle) (Bushmeyer et al., 1995) under control of either the hunchback promoter or the hsp70 promoter on the 5′ side and the hsp70 poly(A) site on the 3′ side (Muller, 1995). (B) Strategy for crosses to determine the repression activity of GALYY1 in transgenic flies. The reporter construct (Muller, 1995) in transgenic fly strain BGUZ is shown at top. The expression pattern of this gene in embryos (i.e. throughout the embryo) is shown at the right, with the ‘A’ denoting anterior, and the ‘P’ posterior ends of the embryo. The GALYY1 effector plasmid expression pattern in the anterior half of embryos is shown at the right by black shading. Anticipated pattern of expression of embros from a cross between the reporter and effector transgenic lines (should repression occur), is shown at the bottom. LacZ expression will only be observed in the posterior ends of the embryos. (C) YY1 represses transcription in Drosophila embryos. LacZ expression in embryos (blue color) is shown in the parental reporter line (BGUZ; left panel) and embryos derived from crosses with two independent hbGALYY1 transgenic lines (middle and right panels). In each cross, LacZ expression is observed to be repressed in the anterior half of the embryo in either 10 h (middle panel) or 6 h (right panel) embryos. (D) The GAL DNA binding domain alone does not repress the BGUZ reporter. LacZ expression is shown in embryos of a cross between transgene lines hbGAL and BGUZ.
YY1 repression is stable in vivo
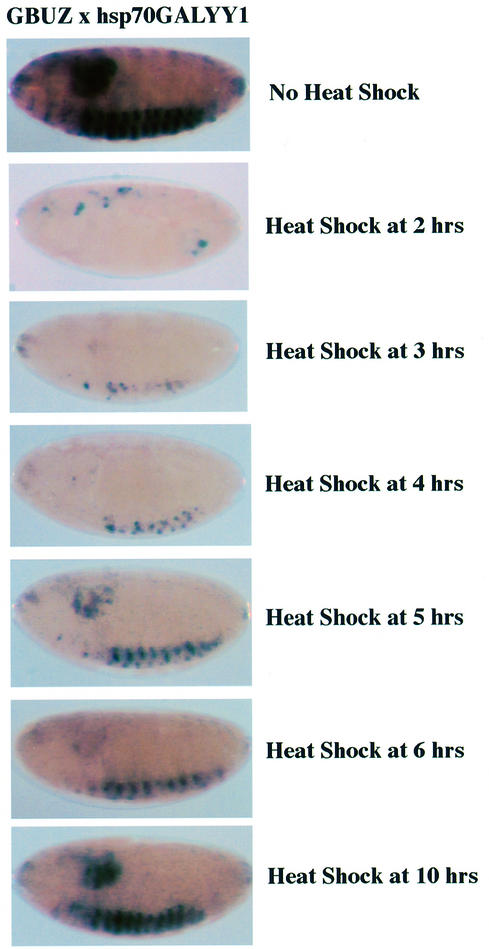
One hallmark of PcG proteins is their ability to generate stable transcriptional repression. Our results shown in Figure 1C suggest that stable repression was observed because repression persisted at the 16 h time point, long after YY1 expression had ceased. To further define the stability of YY1 repression, we placed the GALYY1 sequence under control of the heat shock protein 70 (hsp70) promoter (Figure 1A). The hsp70GALYY1 transgene was crossed with the BGUZ reporter line and embryos were heat shocked at various times after laying. All embryos were then harvested at 16 h and processed for LacZ expression. Interestingly, if embryos were heat shocked at 2 h, transcriptional repression persisted for 16 h (Figure 2). Likewise, a single heat shock treatment at either 3 or 4 h resulted in stable repression out to 16 h. Low levels of LacZ expression were observed when embryos were heat shocked at either 5 or 6 h, while somewhat higher expression was observed with the 10 h sample (Figure 2). Since the BGUZ reporter first becomes active ∼4 h post-laying, these levels of expression may be indicative of the LacZ expressed prior to GALYY1 expression. Even the 10 h heat shock sample showed less LacZ expression than the untreated control, suggesting that subsequent LacZ expression was repressed after the appearance of GALYY1. In summary, the above results indicate that YY1 can stably repress transcription similar to a PcG protein, and that YY1 appears to repress previously active genes. This feature of YY1 will be elaborated on in the Discussion.
Fig. 2. Transcriptional repression by YY1 is stable. The BGUZ reporter line was crossed with the hsp70GALYY1 line. Embryos were either untreated, or heat shocked at 37°C for 45 min at various times, and embryos were processed for staining 16 h after laying.
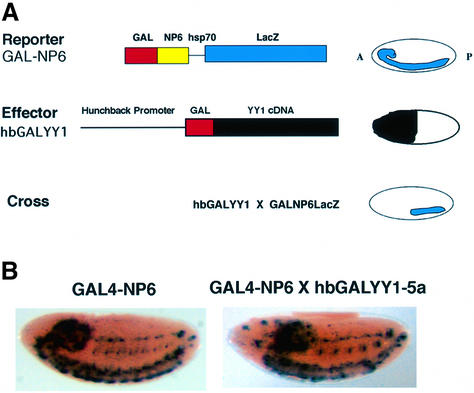
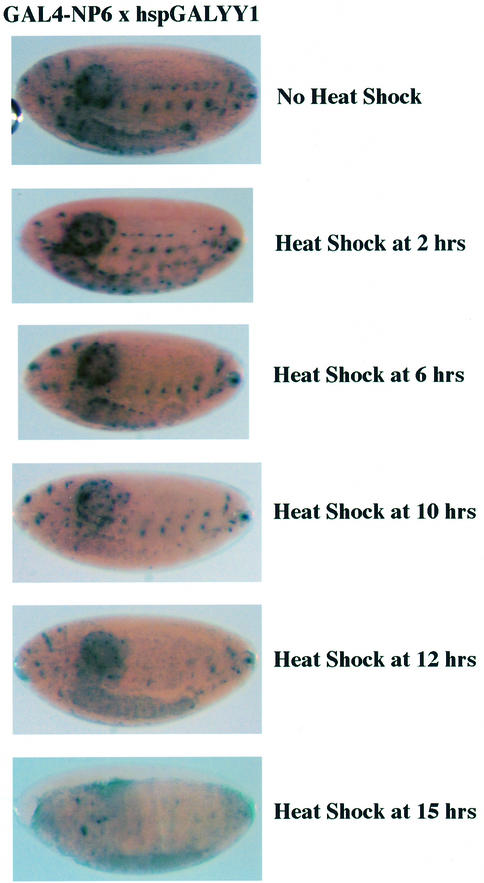
We next tested YY1 repression with a distinct LacZ reporter gene system that does not exhibit stable repression dependent upon PcG function. This LacZ transgene contains GAL4 binding sites adjacent to a synthetic NP6 enhancer and yields expression in late embryonic development (Muller, 1995). The GAL4-NP6 reporter does not respond to stable PcG-dependent repression, but can be repressed transiently by the GAL-Pc protein if expressed shortly before GAL4-NP6 expression (Muller, 1995). We first crossed the hbGALYY1 line with the GAL4-NP6 reporter transgene line. Transient GALYY1 expression from the hunchback promoter did not lead to stable repression (Figure 3A and B), similar to results with the GAL-Pc protein (Muller, 1995). To determine whether GALYY1 could repress GAL4-NP6 expression if GALYY1 was expressed later, we used the hsp70GALYY1 transgene. Embryos from GAL4-NP6 × hsp70GALYY1 crosses were heat shocked at various times after laying and embryos were harvested at 18 h. In this case, GALYY1 only repressed expression if heat shock occurred just prior to GAL4-NP6 expression (heat shock at 15 h post-laying; Figure 4). From these experiments we conclude that GALYY1 expressed prior to 15 h is not able to establish a stable repression mechanism on the GAL4-NP6 construct. Apparently, as GALYY1 levels decay after the heat shock, insufficient protein is available to mediate repression. However, if GALYY1 is expressed just prior to GAL4-NP6 expression, sufficient GALYY1 is present to repress the promoter. Therefore, YY1 repression showed specificity between the two reporter constructs (BGUZ and GAL4-NP6). This specificity is identical to that previously observed with GAL-Pc when it was assayed with the same reporter lines (Muller, 1995). Therefore, YY1 behaves in the same fashion as a known PcG protein with these reporter constructs.
Fig. 3. Early embryonic YY1 expression cannot repress the GAL4-NP6 gene. (A) Strategy for potential repression using the GAL4-NP6 reporter line. A synthetic NP6 enhancer and minimal heat shock promoter yields the expression pattern shown at the right. Anticipated potential repression with hbGALYY1 is shown at the bottom. (B) Transient YY1 expression from the hb promoter does not lead to repression of the GAL4-NP6 reporter. Similar staining patterns were observed either in the absence (left) or presence (right) of hbGALYY1.
Fig. 4. YY1 expressed late in embryonic development can repress GAL4-NP6 expression. Embryos from hsp70GALYY1 X GAL4-NP6 crosses were heat shocked at various times, harvested at 18 h after laying, then processed for LacZ staining. Only YY1 induced at 15 h was able to strongly repress GAL4-NP6 activity.
YY1 repression requires PcG function
The YY1 repression patterns observed above were the same as those obtained previously with a known PcG protein. Therefore, we asked whether YY1 repression required PcG function. To determine this, we prepared an hbGALYY1 BGUZ recombinant chromosome line and crossed this chromosome into various homozygous PcG mutant backgrounds. Since PcG proteins function as complexes, mutation of a single PcG gene often abrogates PcG-dependent repression (Jurgens, 1985; Muller, 1995). Strikingly, homozygous mutant Polycomb (Pc), Polycomb-like (Pcl) or Sex combs on midleg (Scm) backgrounds led to complete derepression of YY1 function (Figure 5D–F) compared with controls (Figure 5A–C). Even heterozygous Pc and Pcl mutants abolished YY1 repression (data not shown). Homozygous mutant Sex combs extra (Sce), Additional sex combs (Asx) or Suppressor of zeste [Su(Z)2] plus Posterior sex combs (Psc) backgrounds yielded partial derepression of YY1 activity, perhaps due to maternal effects (Figure 5G–I). Therefore, YY1 repression in vivo required PcG function.
Fig. 5. YY1 transcriptional repression requires PcG function and can occur at both embryonic and larval stages. Embryos were collected from the BGUZ parent (A), the BGUZ hbGALYY1 recombinant chromosome line (B and C), and the recombinant chromosome line in various PcG homozygous mutant backgrounds (D–J). Embryos were collected at 16 h [or 6 h; (C)] and processed for LacZ staining. Blue staining indicates LacZ expression and light colored areas indicate repression of LacZ expression by GALYY1. The source of each embryo is shown at the right. (K) GALYY1 can compensate for PHO in wing imaginal discs. A diagram of the recombinant chromosome containing the hsp70GALYY1 and PBX-PRE-IDE-LacZ transgenes is shown at the top. This recombinant chromosome was crossed into either a wild type or a pho–/– (pho1/pho1) mutant background and developing larvae were either untreated or heat shocked twice daily. In the pho–/– crosses 18% of larvae are expected to contain the recombinant chromosome and the pho1/pho1 alleles to yield derepression of the reporter gene and resultant LacZ expression. As expected, 13 out of 75 larvae (17%) yielded wing imaginal discs that expressed LacZ (a representative positive wing disc is shown in the middle panel). The half of the larvae from the same cross that were heat shocked to induce GALYY1 expression showed dramatically different results. Only a single larvae out of 83 (1%) yielded wing discs staining positive for LacZ (a representative of the 82 negative imaginal discs is shown). The single positive larvae likely represents an organism in which the two transgenes became unlinked during the second cross due to absence of the balancer chromosome.
Two distinct PcG complexes have been identified. The first complex, termed the PRC1 complex, contains Pc, Scm, Polyhomeotic (Ph) and Psc proteins (Shao et al., 1999). This complex is clearly necessary for YY1 repression since Pc and Scm mutants abolished YY1 function (Figure 5D and F). The second complex contains Esc and E(z) (Jones et al., 1998; Tie et al., 1998, 2001; Ng et al., 2000). As mentioned above, YY1 physically interacts with EED, the vertebrate homolog of Drosophila Esc (Satijn et al., 2001). Therefore, we tested the necessity of Esc for YY1 repression in vivo. Homozygous mutation of the esc gene caused partial loss of YY1 repression (Figure 5J). Thus, both complexes are needed for maximal YY1 repression, although mutations of proteins in the PRC1 complex cause more dramatic loss of YY1 repression.
YY1 can repress transcription in larval imaginal discs
The above results demonstrated that GALYY1 can repress transcription in a PcG-dependent fashion in embryos through synthetic GAL4 DNA binding sites. We wanted to test YY1 function at a later developmental stage using a reporter with native regulatory elements. Therefore, we obtained the PBX-PRE-IDE-LacZ reporter developed by Fritsch et al. (1999), which contains the PBX and IDE enhancers flanking the Ubx PRED element (see Figure 5K). In the absence of the PRED sequence, the IDE enhancer drives LacZ expression in larval wing imaginal discs, while in the presence of the PRED sequence expression is repressed. However, in a pho1/pho1 mutant background, PHO no longer binds to the six PHO binding sites in the PRE and repression is lost, resulting in LacZ expression (Fritsch et al., 1999). Since YY1 can bind to PHO binding sites (Brown et al., 1998), we sought to determine whether YY1 could repress transcription in third instar larvae through a native PRE sequence.
We prepared a fly line with a recombinant chromosome containing the PBX-PRE-IDE-LacZ reporter and our hsp70GALYY1 transgene. This recombinant chromosome was crossed into a pho1/pho1 mutant background and developing larvae were either untreated, or heat shocked twice daily. Third instar larval wing discs were then isolated and processed for LacZ expression. As expected, wing discs for wild-type larvae showed no LacZ expression (Figure 5K, bottom left panel). In a pho1/pho1 background PRE activity was lost, resulting in activation of the LacZ gene by the IDE enhancer (Figure 5K, middle panel). However, LacZ expression was repressed in wing discs isolated from larvae that were heat shocked to express GALYY1 (Figure 5K, bottom right panel). Therefore, GALYY1 can repress transcription through a native PRE sequence at late developmental stages.
YY1 can correct phenotypic defects in pho mutant flies
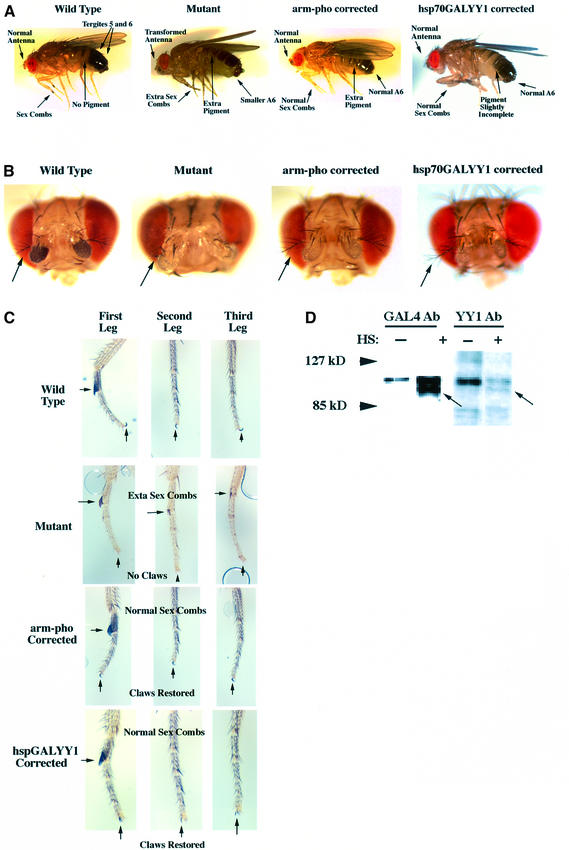
Our results suggest that YY1 is a vertebrate PcG protein. If true, YY1 might function in fly development to compensate for loss of PHO function in a pho mutant background. Drosophila mutants bearing the pho1/phocv alleles show a number of homeotic defects including partial transformation of antennal structures into legs, partial transformation of mesothoracic and methathoracic legs into prothoracic legs, and partial transformation of abdominal segments into more posterior abdominal segments (Girton and Jeon, 1994). We found that the Drosophila pho cDNA driven by the ubiquitously expressed armadillo promoter (arm-pho; Figure 1A) nearly completely corrected segmentation, and antennal and leg defects (Figure 6A–C; Table I). Only ectopic pigmentation on tergite 4 and occasional extra sex combs or antennal defects distinguished these flies from wild type.
Fig. 6. GALYY1 and PHO can partially correct phenotypic defects in pho1/phocv mutant flies. (A) PHO and YY1 partially correct segmentation defects. Wild-type, pho1/phocv mutant, arm-pho-bearing and hsp70GALYY1-bearing pho1/phocv flies are shown. Wild-type male flies are darkly pigmented on the tergites of the last two posterior segments (segments 5 and 6, left panel). Mutant flies show posterior transformation of the segmentation pattern that results in pigmentation on tergite 4 (and sometimes tergites 3 and 2; see arrows pointing to extra pigment). In addition, males lack segment A7 (the posterior-most segment) and transformation of A6 towards A7 can be detected as a smaller A6 which causes the male genitalia to protrude more than in wild-type flies (see mutant panel, smaller A6). In pho1/phocv flies bearing the arm-pho transgene, the posterior-most segment is wild type, although pigmentation is still abnormal on tergite number 4 (right panel). In pho1/phocv flies bearing the hsp70GALYY1 transgene, segmentation pattern is almost completely normal, although pigmentation is not complete. (B) The arm-pho and hsp70GALYY1 transgenes can rescue antenna development. Head mount photographs of wild-type, pho1/phocv mutant, arm-pho-corrected and hsp70GALYY1-corrected flies are shown. The arrows point to arista structures. The arista (an appendage of the antenna) is normally bushy and branch like. In pho1/phocv mutants the aristae are either absent, or are poorly developed and clumped (middle panel). In 85–90% of pho1/phocv mutant flies bearing the arm-pho transgene and 95% of flies bearing the hsp70GALYY1 transgene, the aristae were of normal appearance, indicating substantial correction of the mutant phenotype (right panel). (C) The arm-pho and hsp70GALYY1 transgenes can completely correct the sex comb and claw defects found in pho1/phocv mutant flies. Leg mounts are shown of wild-type, pho1/phocv mutant, arm-pho-corrected pho1/phocv mutant and hsp70GALYY1-corrected pho1/phocv mutant flies. Arrows point to sex comb and claw structures. (D) GALYY1 protein is expressed after heat shock of hspGALYY1 transgenic embryos. Embryos from hsp70GALYY1 flies were either untreated or heat shocked for 45 min at 37°C. Western blots of lysates were assayed with either GAL4 or YY1 specific antibodies. The arrow points to the induced GALYY1 band.
Table I. Correction of pho mutant flies by the arm-pho and hsp70GALYY1 transgenes.
| Body structure | Wild type | pho1/phocv mutants | arm-pho corrected | hsp70GALYY1 corrected |
|---|---|---|---|---|
| Arista and antenna (% bushy) | 100 | 0a | 85–90 | 95 |
| Average no. sex combs per flyb | 2 ± 0 | 5.1 ± 1.3 | 2.7 ± 1.4 (72% = 2.0) | 2 ± 0 |
| Leg claws per flyb | 6 ± 0 | 0.07 ± 0.4 | 6 ± 0 | 4.4 ± 1.6 |
Data were obtained using arm-pho transgene line 27a (18 flies), hsp70GALYY1 line 44H (19 flies) and pho mutant alleles pho1/phocv (28 flies).
aOne hundred percent missing or clumped.
bNumbers represent averages ± SD of the mean.
We tested YY1 rescue of the pho mutant phenotype using the same arm promoter system (arm-YY1; Figure 1A), but initially found no correction. However, these transgenes failed to express detectable YY1 protein (data not shown). Therefore, we used the inducible hsp70 promoter to drive expression of a GALYY1 chimera (hsp70GALYY1; Figure 1). Transgenic embryos bearing the hsp70GALYY1 transgene produced GALYY1 protein after heat shock (Figure 6D). This transgenic construct was crossed into a pho1/phocv mutant background and developing flies were heat shocked twice daily. The results on phenotypic correction were dramatic. GALYY1 expression nearly completely corrected the antennal and sex comb defects in pho1/phocv mutant flies (Figures 6A and 3B; Table I). Segmentation was largely normal, although pigmentation on tergite 5 was not always complete. Correction of claw structures was variable ranging from complete to partial correction (Table I). Flies with partial correction (three to four claws rather than six) appeared normal, but were clumsy due to inability to grasp vial walls efficiently.
We also tested the ability of GALYY1 to rescue the more severe phenotype of pho1/pho1 mutants. Homo zygous pho1/pho1 mutants are pupal lethal and fail to eclose, demonstrating a much earlier lethal phenotype that the pho1/phocv mutant combination. GALYY1 expression partially corrected the pho1/pho1 phenotype yielding flies that survived to adulthood, but which died shortly thereafter.
The above results indicate that mammalian YY1 can replace a mutant Drosophila PcG protein to phenotypically rescue pho mutant flies. Coupled with the above PcG-dependent transcriptional repression data in embryos and larvae in vivo, we conclude that YY1 is very likely a vertebrate PcG protein.
YY1 repression in vivo requires co-repressor protein CtBP
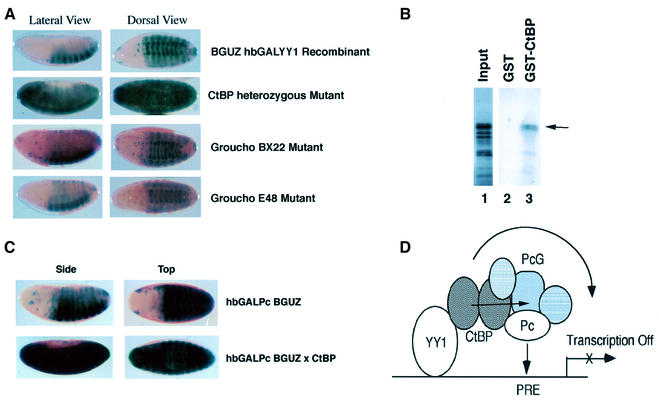
We next used the embryo repression assay system to identify other proteins needed for YY1 repression in vivo. YY1 function can be altered in transient expression assays by interaction with either histone deacetylase (HDAC), or histone acetyltransferase (HAT) proteins, and its function has been proposed to involve chromatin remodeling (Lee et al., 1995; Yang et al., 1996; Thomas and Seto, 1999; Yao et al., 2001). However, we found that mutation of HDAC (rpd3), HAT (dCBP) or chromatin remodeling (kismet) genes had no effect on YY1 repression in vivo (data not shown). We then addressed whether either of two well-characterized co-repressor proteins in Drosophila, CtBP and Groucho, showed an interaction. Whereas Groucho mutants had little effect, CtBP was absolutely essential for YY1 repression (Figure 7A). Even heterozygous loss of CtBP activity completely abolished YY1 repression (Figure 7A).
Fig. 7. (A) Heterozygous CtBP mutants completely abolished YY1 repression. The GBUZ hbGALYY1 recombinant line was crossed into the either ctbp or groucho mutant backgrounds. Heterozgous CtBP mutants completely abolished YY1 repression, whereas Groucho mutants showed little effect. (B) YY1 physically interacts with CtBP. GST pull-down experiments were performed with recombinant YY1 prepared by in vitro translation, and GST–CtBP prepared in bacteria. The arrow indicates full-length YY1. (C) Heterozygous CtBP mutation abolishes Pc repression. The hbGAL-Pc BGUZ recombinant chromosome was crossed into a heterozygous ctbp mutant background, resulting in loss of Pc repression. (D) Potential mechanism linking YY1, CtBP and the PcG complex. In this model, YY1 binds a homodimer of CtBP that interacts with the PcG complex to recruit the complex to PRE sequences, thereby repressing transcription. Arrows show potential dual functions of CtBP in recruiting PcG proteins and repressing transcription.
The striking genetic interaction between CtBP function and YY1 activity led us to investigate whether there was a physical interaction between CtBP and YY1. Indeed, GST pull-down experiments revealed binding between CtBP and YY1 (Figure 7B). CtBP can also be co-immunoprecipitated with YY1 from transfected cells (Y.Shi, personal communication). CtBP has previously been shown to interact with Pc in vivo (Sewalt et al., 1999). Therefore, the in vivo and in vitro interaction described here of CtBP with YY1 suggests a potential mechanism by which these proteins might complex with the PcG proteins in vivo to mediate repression. For instance, CtBP might tether the PcG complex to YY1 that is bound to DNA (Figure 7D). Alternatively, CtBP might play a direct role in the PcG repression mechanism. These two functions are not necessarily mutually exclusive. If CtBP plays a tethering role only, it would not be expected to influence repression by GAL-Pc, because GAL-Pc is already able to directly bind DNA through the GAL4 DNA binding domain. Therefore, we tested the effect of ctbp mutation on GAL-Pc repression of the BGUZ reporter. Interestingly, ctbp mutation abolished GAL-Pc repression of BGUZ activity (Figure 7C), indicating a function for CtBP distinct from merely tethering the PcG complex to YY1 bound to DNA.
Discussion
Our results indicate that YY1 is a vertebrate PcG protein. YY1 can generate stable transcriptional repression via a PcG-dependent mechanism in vivo, and can functionally compensate for the PcG protein, PHO, in pho mutant flies. Most biochemical studies have not revealed a physical association of YY1 with the known PcG complexes (reviewed in Satijn and Otte, 1999; Brock and van Lohuizen, 2001; Francis and Kingston, 2001), although substoichiometric levels are observed in human Pc complexes (Levine et al., 2002), and some associations have been documented for Drosophila PHO (Poux et al., 2001b; Mohd-Sarip et al., 2002). The transient nature of the Drosophila associations (Poux et al., 2001b) suggests that an intermediary protein exists. Here we demonstrate genetic and physical associations between YY1 and CtBP, which link YY1 to PcG function and provide a mechanism for the recruitment of vertebrate PcG complexes to DNA. Since CtBP is able to homodimerize (Sewalt et al., 1999), it may interact with Pc by one dimer partner and with YY1 by the other dimer partner (Figure 7D). These interactions could define the mechanism by which YY1 functions to repress transcription in both a PcG- and CtBP-dependent fashion. On the other hand, our CtBP and Pc experiments (Figure 7C) indicate that CtBP plays a more direct role in PcG repression. Thus, CtBP may perform more than one function in the repression mechanism.
The PcG function of YY1 that we identify here extends a list of YY1 functions including transcriptional activation and repression via apparently non-PcG pathways. YY1 binds to numerous promoters and can mediate repression by a variety of mechanisms including binding site competition, DNA bending and interference with activator interactions with the basal transcription machinery (Gualberto et al., 1992; Nateson and Gilman, 1993; Lu et al., 1994; Zhou et al., 1995; Ye et al., 1996; Galvin and Shi, 1997; Shi et al., 1997). YY1 repression can be influenced by interactions with proteins such as adenoviral E1A and the co-activator p300 (Shi et al., 1991; Lee et al., 1995). YY1 can also interact with histone deacetylase proteins (Yao et al., 2001) and is speculated to play a role in chromatin remodeling (Thomas and Seto, 1999). Thus, the PcG function of YY1 identified here may be one of numerous functions mediated by this complex transcription factor. It may not be surprising that YY1 carries out multiple functions, because diverse functions of other PcG proteins are now being elucidated. For example, the PcG proteins Bmi-1 and Mel-18 play roles in controlling the cell cycle and their mutation leads to proliferative defects that impact the hematopoietic system (van der Lugt et al., 1994; Akasaka et al., 1997; Jacobs et al., 1999; Lessard et al., 1999). Therefore, PcG proteins play roles in multiple processes in addition to body axis formation.
We observed stable transcriptional repression by YY1, but also found that YY1 appeared to repress expression of a previously active gene (see Figure 2, 10 h heat shock). Generally, PcG proteins are believed to be maintenance repressors that do not initiate de novo repression. However, YY1 has the feature that it can repress de novo and may be able to repress transcription by multiple mechanisms that include PcG-dependent and -independent mechanisms. This is in agreement with the multiple YY1 repression mechanisms that have already been identified (Shi et al., 1997; Thomas and Seto, 1999).
The peri-implantation lethal phenotype of YY1 knock-out mice (Donohoe et al., 1999) is similar to the phenotype of eed–/– mice. In contrast, pho mutant Drosophila show a phenotype much later in development, potentially indicating some differences between YY1 and PHO. Our phenotypic rescue experiments demonstrate considerable functional similarity between these proteins, but 75% of vertebrate YY1 and Drosophila PHO protein sequences contain no discernable homology, suggesting some distinct functions. PHO appears insufficient for repression at early embryonic stages in Drosophila, since a LexA-Pho chimeric protein is incapable of repressing transcription of a LexA-Ubx-LacZ reporter (Poux et al., 2001a), and a GAL-Pho chimeric protein is incapable of repressing the identical BGUZ construct we used here in embryos (Fritsch, 2002). Thus, unlike YY1, PHO does not repress transcription in early embryos. However, PHO is necessary for repression at later stages of development, since mutating PHO binding sites in the Ubx PRE results in loss of silencing in wing imaginal discs (Fritsch et al., 1999). Here we show that YY1 can clearly repress transcription at both early embryonic stages, as well as at later larval stages in wing imaginal discs. The early function of YY1 is consistent with its early lethal phenotype in YY1 mutant mice (Donohoe et al., 1999). This repression indicates that YY1 can mediate embryonic functions lacking in the PHO protein. Specifically, the association of YY1 with CtBP may provide a bridging function not mediated by PHO. Most proteins that bind to CtBP contain a canonical PXDLS motif (reviewed in Chinnadurai, 2002). While YY1 contains a similar sequence, this motif is absent from PHO.
The precise role of CtBP in PcG repression is unclear. CtBP mutants in flies show segmentation defects (Poortinga et al., 1998), but homeotic derepression has not been observed. Similarly, mouse ctbp1 and ctbp2 null mutants show a variety of defects including skeletal abnormalities (Hildebrand and Soriano, 2002), but these defects do not precisely match the skeletal posterior transformations seen with mammalian PcG mutants (Akasaka et al., 1996; Schumacher et al., 1996; Bel et al., 1998). It is quite possible that YY1 and CtBP are necessary for a subset of PcG functions. Similarly, it has been proposed that multiple distinct PcG complexes exist to regulate distinct genes (Satijn and Otte, 1999). An additional potential link between YY1 and the PcG complex is the protein RYBP. Similar to CtBP, RYBP can physically interact with both YY1 and PcG proteins (Garcia et al., 1999). The absence of a corresponding mutant in Drosophila precluded our testing the necessity of RYBP for YY1 repression.
Our demonstration that YY1 functions as a PcG protein predicts that vertebrate PREs should contain YY1 binding sites. YY1/PHO binding sites (CGCCATNTT) are indeed present within many Drosophila PRE sequences (Mihaly et al., 1998), and are required for function (Fritsch et al., 1999). Since the YY1 binding motif is well characterized (Hyde-DeRuyscher et al., 1995), our results should facilitate the identification of vertebrate PRE regions, which thus far have proved elusive. Our experiments linking YY1 to PcG function reveal mechanistic features of YY1-mediated transcriptional repression, with implications for PcG activity in mammals. It will be very interesting in the future to determine whether YY1 heterozygous mice augment mutant phenotypes in PcG mutant heterozyotes.
Materials and methods
Transgene construction
DNA constructs (also bearing the ry+ gene as a selectable marker) were co-injected with a transposase expressing plasmid (phsπ) into the posterior ends of dechorionated 30 min-old embryos from the ry506 strain to generate transgenics (Rubin and Spradling, 1982). Transgenes were mapped with respect to chromosome insertion and were stabilized by crosses with appropriate balancer lines.
Drosophila lines and crosses
The BGUZ, GAL4-NP6, hbGAL-Pc BGUZ and hbGAL transgene lines were provided by J.Mueller (Muller, 1995). PcG mutant lines included flies with the following mutant alleles: PcXT, PclD5, Sce1, AsxXF23, Su(Z)21.68, Esc6 and ScmD1. Co-repressor mutants included CtBP03464, GrouchoBX22 and GrouchoE48. To generate a line containing a recombinant hbGALYY1 BGUZ chromosome, homozygous hbGALYY1 females were crossed with BGUZ males. The resulting females were crossed with ry506 males to determine recombination frequency. Individual male progeny were then crossed with an FM7c balancer strain and the males were subsequently individually genotyped by PCR for the two transgenes. Females from the recombinant hbGALYY1 BGUZ line were crossed with males of each PcG mutant, and resulting males were crossed with virgin females from the balanced PcG mutant stocks. Embryos were collected from grape plates after either 6, 10 or 16 h. For correction of the pho1/phocv or pho1/pho1 phenotypes with hspGALYY1, organisms bearing the appropriate genes were heat shocked for 45 min at 37°C every 12 h throughout development. To generate a line containing a recombinant hsp70GALYY1 PBX-PRE-IDE-LacZ chromosome, hsp70GALYY1/CyO males were crossed with homozygous PBX-PRE-IDE-LacZ females. Resulting females were crossed with ry506 males to determine recombination frequency. Individual male progeny were crossed with a BcE1p/CyO balancer line and males were subsequently individually genotyped by PCR for the two transgenes. Balanced females from the recombinant chromosome line were crossed with pho1/CID males and resulting Cyo– CID– males and virgin females were crossed to generate larvae with the recombinant chromosome in a pho1/pho1 background.
LacZ staining of embryos and imaginal discs
Embryos were dechorionated with chlorox, fixed for 15 min in phosphate-buffered saline (PBS) containing 4% formaldehyde, then incubated at 37°C in 0.01 M NaPO4 pH 7.2, 0.15 M NaCl, 0.1 mM MgCl2, 11 mM K4Fe(CN)6, 11 mM K4Fe(CN)6, 0.03% Triton X-100 and 0.1% X-gal (O’Kane and Gehring, 1987). Imaginal discs from larvae were dissected and fixed for 30 min in PBS containing 1% glutaraldehyde, washed four times in 100 mM Tris pH 7.5, 130 mM NaCl, 3 mM KCl, 5 mM sodium azide and 1 mM EGTA, and incubated in X-gal solution as described above.
GST pull-down assays
Reactions consisted of GST fusion protein, or an equivalent amount of GST protein alone, incubated with 5–15 µl of 35S-labeled YY1 prepared by in vitro transcription and translation in a 100 µl reaction containing 20 mM Tris–HCl pH 8, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40 (NETN). Samples were rocked for 2 h at 4°C and washed at least five times with 450 µl NETN. Samples were electrophoresed on 10% SDS–polyacrylamide gels for 1 h at 160 V, dried and subjected to autoradiography.
Western blots
Embryos were collected for 2 h on grape plates, cured for 1 h at room temperature, and then either left untreated or heated for 45 min at 37°C. Embryos were harvested, dechorionated with chlorox and then lysed in SDS sample buffer. After boiling for 5 min, samples were fractionated by SDS–PAGE and then subjected to the western blot procedure with either anti-GAL4 (Santa Cruz Biotechnologies) or anti-YY1 antibodies (Geneka).
Acknowledgments
Acknowledgements
We thank Juerg Muller for helpful advice and for transgene vector constructs, transgenic lines BGUZ, GAL-NP6, hbGAL-Pc BGUZ and hbGAL, and for PcG mutant flies. We are grateful to Judith Kassis for the pho cDNA and pho mutant flies, and to Albert Courey for groucho mutant flies. We thank Susan Parkhurst for the GST–CtBP plasmid, Michael O’Connor for rpd3 mutant flies and John Tamkun for kismet mutant flies. We also thank John Warrick for help with leg mounts. This work was supported by NIH grant GM42415 and March of Dimes grant FY02-173 to M.L.A., grant R29-Ey11259 to N.B., support from training grant T32 DK07708 to F.W. and sabbatical support from Chestnut Hill College to L.A.
References
- Akasaka T., Kanno,M., Balling,R., Mieza,M.A., Taniguchi,M. and Koseki,H. (1996) A role for mel-18, a polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development, 122, 1513–1522. [DOI] [PubMed] [Google Scholar]
- Akasaka T. et al. (1997) The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity, 7, 135–146. [DOI] [PubMed] [Google Scholar]
- Akasaka T. et al. (2001) Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development, 128, 1587–1597. [DOI] [PubMed] [Google Scholar]
- Alkema M.J., van der Lugt,N.M.T., Bobeldijk,R.C., Berns,A. and van Lohuizen,M. (1995) Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature, 374, 724–727. [DOI] [PubMed] [Google Scholar]
- Austen M., Luscher,B. and Luscher-Firzlaff,J.M. (1997) Characterization of the transcriptional regulator YY1. J. Biol. Chem., 272, 1709–1717. [DOI] [PubMed] [Google Scholar]
- Beinz M. and Muller,J. (1995) Transcriptional silencing of homeotic genes in Drosophila. BioEssays, 17, 775–784. [DOI] [PubMed] [Google Scholar]
- Bel S., Core,N., Djaball,M., Kieboo,K., van der Lugt,N., Alkema,M.J. and Van Lohuizen,M. (1998) Genetic interactions and dosage effects of Polycomb group genes in mice. Development, 125, 3543–3551. [DOI] [PubMed] [Google Scholar]
- Brock H.W. and van Lohuizen,M. (2001) The Polycomb group—no longer an exclusive club? Curr. Opin. Genet. Dev., 11, 175–181. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Mucci,D., Whiteley,M., Dirksen,M.-L. and Kassis,J.A. (1998) The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell, 1, 1057–1064. [DOI] [PubMed] [Google Scholar]
- Bushmeyer S.M. and Atchison,M.L. (1998) Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell. Biochem., 68, 484–499. [PubMed] [Google Scholar]
- Bushmeyer S., Park,K. and Atchison,M.L. (1995) Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem., 270, 30213–30220. [DOI] [PubMed] [Google Scholar]
- Busturia S., Lloyd,A., Bejarano,F., Zavortink,M. and Sakonju,S. (2001) The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development, 128, 2163–2173. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell, 9, 213–224. [DOI] [PubMed] [Google Scholar]
- Donohoe M.E., Zhang,X., McGinnis,L., Biggers,J., Li,E. and Shi,Y. (1999) Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol., 19, 7237–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan I. and Lewis,E.B. (1982) Genetic Control of Body Segment Differentiation in Drosophila. Alan R.Liss, Inc., New York, NY.
- Francis N.J. and Kingston,R.E. (2001) Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol., 2, 409–421. [DOI] [PubMed] [Google Scholar]
- Fritsch C. (2002) Funktionelle analyse des Polycomb gruppen proteins pleiohomeotic in Drosophila. Dissertation Thesis, der Fakultät für Biologie der Eberhard Karls Universität Tübingen, Tübingen, Germany.
- Fritsch C., Brown,J.L., Kassis,J.A. and Muller,J. (1999) The DNA-binding Polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development, 126, 3905–3913. [DOI] [PubMed] [Google Scholar]
- Galvin K.M. and Shi,Y. (1997) Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol., 17, 3723–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E., Marcos-Gutierrez,C., del Mar Lorente,M., Moreno,J.C. and Vidal,M. (1999) RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex and with the transcription factor YY1. EMBO J., 18, 3404–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton J.R. and Jeon,S.H. (1994) Novel embryonic and adult homeotic phenotypes are produced by pleiohomeotic mutations in Drosophila. Dev. Biol., 161, 393–407. [DOI] [PubMed] [Google Scholar]
- Gualberto A., LePage,D., Pons,G., Mader,S.L., Park,K., Atchison,M.L. and Walsh,K. (1992) Functional antagonism between YY1 and the serum response factor. Mol. Cell. Biol., 12, 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D. and Soriano,P. (2002) Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol., 22, 5296–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher R.P., Jennings,E. and Shenk,T. (1995) DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res., 23, 4457–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.J.L., Kieboom,K., Marino,S., DePinho,R.A. and van Lohuizen,M. (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature, 397, 164–168. [DOI] [PubMed] [Google Scholar]
- Jones C.A., Ng,J., Peterson,A.J., Morgan,K., Simon,J. and Jones,R. (1998) The Drosophila esc and E(z) proteins are direct partners in Polycomb Group-mediated repression. Mol. Cell. Biol., 18, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. (1985) A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature, 316, 153–155. [Google Scholar]
- Kennison J.A. (1993) Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet., 9, 75–78. [DOI] [PubMed] [Google Scholar]
- Lee J.-S., Galvin,K.M., See,R.H., Eckner,R., Livingston,D., Moran,E. and Shi,Y. (1995) Relief of YY1 transcriptional repression by adenovirus E1A is mediated by EIA-associated protein p300. Genes Dev., 9, 1188–1198. [DOI] [PubMed] [Google Scholar]
- Lee T.-C., Zhang,Y. and Schwartz,R.J. (1994) Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene, 9, 1047–1052. [PubMed] [Google Scholar]
- Lessard J., Schumacher,A., Thorsteinsdottir,U., van Lohuizen,M., Magnuson,T. and Sauvageau,G. (1999) Functional antagonism of the Polycomb Group genes eed and Bmi1 in hematopoietic cell proliferation. Genes Dev., 13, 2691–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S.S., Weiss,A., Erdjument-Bromage,H., Shao,Z., Tempst,P. and Kingston,R.E. (2002) The core of the Polycomb Repressive Complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol., 22, 6070–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.-Y., Rodriguez,M. and Liao,W.S.L. (1994) YY1 represses rat serum amyloid A1 gene transcription and is antagonized by NF-κB during acute-phase response. Mol. Cell. Biol., 14, 6253–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon J. and Brock,H.W. (1991) Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Rouxs Arch. Dev. Biol., 199, 387–396. [DOI] [PubMed] [Google Scholar]
- Mihaly J., Mishra,R.K. and Karch,F. (1998) A conserved sequence motif in Polycomb response elements. Mol. Cell, 1, 1065–1066. [DOI] [PubMed] [Google Scholar]
- Mishra R.K., Mihaly,J., Barges,S., Spierer,A., Karch,F., Hagstrom,K., Schweinsberg,S.E. and Schedl,P. (2001) The iab-7 Polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activitiy. Mol. Cell. Biol., 21, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Sarip A., Venturini,F., Chalkley,G.E. and Verrijzer,C.P. (2002) Pleiohomeotic can link Polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol., 22, 7473–7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J. (1995) Transcriptional silencing by the polycomb protein in Drosophila embryos. EMBO J., 6, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Guant,S. and Lawrence,P.A. (1995) Function of the polycomb protein is conserved in mice and flies. Development, 121, 2847–2852. [DOI] [PubMed] [Google Scholar]
- Nateson S. and Gilman,M.Z. (1993) DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev., 7, 2497–2509. [DOI] [PubMed] [Google Scholar]
- Ng J., Hart,C.M., Morgan,K. and Simon,J.A. (2000) A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol., 20, 3069–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Carroll D., Erhardt,S., Pagani,M., Barton,S.C., Surani,M.A. and Jenuwein,T. (2001) The Polycomb-Group gene Ezh2 is required for early mouse development. Mol. Cell. Biol., 21, 4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane C.J. and Gehring,W.J. (1987) Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl Acad. Sci. USA, 84, 9123–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. and Atchison,M.L. (1991) Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain µE1 site. Proc. Natl Acad. Sci. USA, 88, 9804–9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. (1993) Mechanisms of heritable gene repression during development of Drosophila. Curr. Opin. Cell Biol., 5, 999–1005. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1997a) Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet., 13, 314–318. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1997b) PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev., 7, 249–258. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. (1999) Polycomb silencing and the maintenance of stable chromatin states. Results Probl. Cell Differ., 25, 205–228. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S., McCabe,D. and Pirrotta,V. (2001a) Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development, 128, 75–85. [DOI] [PubMed] [Google Scholar]
- Poux S., Melfi,R. and Pirrotta,V. (2001b) Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev., 15, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Satijn D.P. and Otte,A.P. (1999) Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim. Biophys. Acta, 1447, 1–16. [DOI] [PubMed] [Google Scholar]
- Satijn D.P., Hamer,K.M., den Blaauwen,J. and Otte,A.P. (2001) The polycomb group protein EED interacts with YY1 and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol., 21, 1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A. and Magnuson,T. (1997) Murine polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet., 13, 167–170. [PubMed] [Google Scholar]
- Schumacher A., Faust,C. and Magnuson,T. (1996) Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature, 383, 250–253. [DOI] [PubMed] [Google Scholar]
- Sewalt R.G.A.B., Gunster,M.J., van der Vlag,J., Satijn,D.P.E. and Otte,A.P. (1999) C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol., 19, 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Raible,F., Mollaaghababa,R., Guyon,J.R., Wu,C., Bender,W. and Kingston,R.E. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell, 98, 37–46. [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto,E., Chang,L.-S. and Shenk,T. (1991) Transcriptional repression by YY1, a human GLI-Kruppel-related protein and relief of repression by adenovirus E1A protein. Cell, 67, 377–388. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lee,J. and Galvin,K.M. (1997) Everything you have ever wanted to know about Yin Yang1. Biochim. Biophys. Acta, 1332, F49–F66. [DOI] [PubMed] [Google Scholar]
- Shrivastava A. and Calame,K. (1994) An analysis of genes regulated by the multi-functional transcriptional regulatory Yin Yang-1. Nucleic Acids Res., 22, 5151–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J., Chiang,A. and Bender,W. (1992) Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development, 114, 493–505. [DOI] [PubMed] [Google Scholar]
- Thomas M.J. and Seto,E. (1999) Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene, 236, 197–208. [DOI] [PubMed] [Google Scholar]
- Tie F., Furuyama,T. and Harte,P.J. (1998) The Drosophila Polycomb-Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development, 125, 3438–3496. [DOI] [PubMed] [Google Scholar]
- Tie F., Furuyama,T., Prasad-Sinha,J., Jane,E. and Harte,P.J. (2001) The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development, 128, 275–286. [DOI] [PubMed] [Google Scholar]
- van der Lugt N.M.T. et al. (1994) Posterior transformation, neurological abnormalities and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev., 8, 757–769. [DOI] [PubMed] [Google Scholar]
- Yang W.-M., Inouye,C., Zeng,Y., Bearss,D. and Seto,E. (1996) Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl Acad. Sci. USA, 93, 12845–12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.-L., Yang,W.-M. and Seto,E. (2001) Regulationof transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol., 21, 5979–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Zhang,X. and Dong,Z. (1996) Characterization of the human granulocyte–macrophage colony-stimulating factor gene promoter: an AP1 complex and an Sp1-related complex transactivate the promoter activity that is suppressed by a YY1 complex. Mol. Cell. Biol., 16, 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Gedrich,R.W. and Engel,D.A. (1995) Transcriptional repression of the c-fos gene by YY1 is mediated by direct interaction with ATF/CREB. J. Virol., 69, 4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]