Abstract
DNA-binding proteins are generally thought to locate their target sites by first associating with the DNA at random and then translocating to the specific site by one-dimensional (1D) diffusion along the DNA. We report here that non-specific DNA conveys proteins to their target sites just as well when held near the target by catenation as when co-linear with the target. Hence, contrary to the prevalent view, proteins move from random to specific sites primarily by three-dimensional (3D) rather than 1D pathways, by multiple dissociation/re-association events within a single DNA molecule. We also uncover a role for DNA supercoiling in target-site location. Proteins find their sites more readily in supercoiled than in relaxed DNA, again indicating 3D rather than 1D routes.
Keywords: diffusion/DNA–protein interaction/EcoRV/restriction enzyme/sliding
Introduction
Proteins that interact with specific DNA sequences must locate their target sites among very large numbers of alternative sequences. Yet they can bind to specific sites in long DNA molecules at extremely rapid rates (Riggs et al., 1970). The rates can exceed the theoretical limit for random diffusion in solution (Berg and von Hippel, 1985). For this to occur, the protein must find its target by facilitated rather than random diffusion, by binding initially to the DNA anywhere along the chain and then translocating to a specific site by an intramolecular process (von Hippel and Berg, 1989). Non-specific sequences in the DNA can thus capture the protein and convey it to the specific sequence (Shimamoto, 1999).
Various mechanisms have been proposed for facilitated diffusion (von Hippel and Berg, 1989; Halford and Szczelkun, 2002). In one scheme, known as ‘sliding’, the protein moves along the DNA by linear (one-dimensional; 1D) diffusion between adjacent non-specific sites without losing contact with the DNA (Richter and Eigen, 1974; Berg et al., 1981). In another scheme, the protein moves through three-dimensional (3D) space, dissociating from the DNA before re-associating elsewhere on the same molecule (Berg et al., 1981; Stanford et al., 2000). The majority of re-associations will be at or near the site from which the protein dissociated: following the dissociation, the protein will remain close to its initial site on DNA for some time, due to its relatively slow diffusion rate (Berg and von Hippel, 1985). However, a DNA molecule in dilute solution occupies a discrete volume that contains a high concentration of DNA segments, but these are separated by large volumes that are ‘empty’ of DNA (Winter et al., 1981). Hence, the protein may occasionally diffuse away from its initial site yet remain inside the volume occupied by that DNA. The re-association can then be to a site distant from the point of dissociation on the 1D contour. The distances between the molecules are much larger than those between different segments of the same chain, so a protein that dissociates from one site is more likely to re-associate with the same molecule than with a different one. Indeed, it can be difficult for a protein to escape from a long DNA. For example, after cutting its recognition site in a 3.6 kb plasmid, the EcoRV endonuclease dissociates from its recognition site with a half-time of ∼0.25 s, yet the half-time for its liberation from the cleaved plasmid into bulk solution is ∼50 s (Baldwin et al., 1999). EcoRI behaves similarly (Wright et al., 1999).
In a third scheme, ‘intersegmental transfer’ (Berg et al., 1981), the protein moves from one site to another by binding transiently to both sites, as in a DNA looping interaction, before dissociating from the initial site. This scheme applies only to proteins with two DNA-binding sites, such as the Lac repressor or the SfiI nuclease (Halford et al., 2000). It is inapplicable to the main test system used here, the EcoRV restriction enzyme. EcoRV is a dimeric protein with one DNA-binding site at the subunit interface (Winkler et al., 1993). In the presence of Mg2+, it cleaves double-stranded DNA at one particular sequence, normally cutting both strands before dissociating from the DNA (Taylor and Halford, 1989). Yet EcoRV binds to DNA initially in a non-specific manner (Taylor et al., 1991; Reid et al., 2001). It then translocates to its recognition site by an intramolecular process (Taylor et al., 1991; Stanford et al., 2000).
At present, it is widely believed that proteins locate their target sites by 1D diffusion (Shimamoto, 1999). This view comes mainly from analysing how the binding of proteins to specific sites varies with the length of the DNA (Winter et al., 1981; Jack et al., 1982; Jeltsch and Pingoud, 1998). However, a longer DNA always provides a larger target for the initial collision with the protein. Consequently, while increased binding with increasing lengths of DNA indicates that the non-specific DNA is on the route to the specific site, the different pathways for facilitated diffusion cannot be distinguished from each other solely by length dependencies. Such data are often correlated to 1D sliding without excluding 3D routes. Nonetheless, 3D routes may be more general than considered previously (Misteli, 2001). In mammalian cell nuclei, proteins with fluorescent tags were observed moving from one location to another in the DNA via free solution (Lever et al., 2000; Phair and Misteli, 2000). In vivo, the large number of proteins bound to DNA may prevent 1D motion over any significant distance, but in vitro studies on EcoRV also support 3D routes (Stanford et al., 2000). On a series of DNA molecules with two EcoRV sites separated by different lengths of DNA, the processivity of EcoRV (the extent to which it cleaved both sites during one encounter with the DNA molecule) declined with increasing inter-site spacing in a manner that was inconsistent with 1D sliding, but which matched that for 3D motion. Even so, it has yet to be shown that proteins move from one site to another in the same DNA through 3D space.
DNA communications that follow the 1D contour of the DNA can be distinguished unequivocally from those that operate through 3D space by using catenanes containing two interlinked rings of DNA (Adzuma and Mizuuchi, 1989). If a protein bound to a site in one ring of a catenane operates by a 1D pathway, it will never contact the DNA in the other ring: the type I restriction enzymes act in this way (Szczelkun et al., 1996). However, sites in the separate rings of a catenane are held almost as close to each other in 3D space as two sites in the same chain of DNA (Levene et al., 1995). Consequently, if a protein bound to one ring of the catenane acts through 3D space, it will be able to contact sequences in the other ring almost as readily as sequences in the same ring: DNA looping proteins show this behaviour (Szczelkun and Halford, 1996). We report here that a 3 kb segment of non-specific DNA can recruit restriction enzymes to recognition sites just as well when held close to the target site in 3D space, in a catenane, as when co-linear with the site. The enzymes therefore cannot travel from non-specific to specific sites solely by 1D steps, but must instead move through 3D space. We also uncover a role for DNA supercoiling in aiding the transfer of proteins from non-specific to specific sites.
Results
Experimental strategy
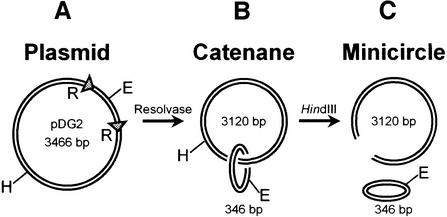
EcoRV and several other restriction enzymes were tested against three substrates. The first was the plasmid pDG2 (Figure 1A), a 3466 bp circle of covalently closed DNA with one recognition site for each restriction enzyme. This was used initially in its native supercoiled state, as isolated from Escherichia coli. The second was a catenane derived from pDG2 (Figure 1B). Site-specific recombination between the res sites on pDG2, by Tn21 resolvase in vitro (Oram et al, 1997), converts this circular DNA to two circles, interlinked once to form a catenane: a 346 bp minicircle that carries the recognition sites for all of the enzymes tested here and a larger ring of 3120 bp that lacks sites for these enzymes. The catenane thus has the same array of DNA sequences as the plasmid, but while the non-specific sequences in the plasmid are contiguous with the recognition sites, the majority of the non-specific sequences in the catenane are tethered to the recognition sites only by topological means. The third substrate was the free minicircle, released from the catenane by cutting the larger ring with another restriction enzyme, HindIII (Figure 1C). To maintain a constant ratio of specific to non-specific sequences, the reactions on the free minicircle used the mixture of circular and linear DNA directly from the HindIII digest.
Fig. 1. DNA substrates. (A) The plasmid pDG2 contains recognition sites for EcoRV and HindIII, at the positions marked E and H, respectively, and two res sites from the transposon Tn21 (triangles, marked R). The 346 bp segment of DNA between the res sites also contains single sites for BsaAI, ClaI, Eco47III, PstI and XhoI. (B) Recombin ation at the res sites in pDG2, by Tn21 resolvase, generates a catenane with two interlinked rings of 3120 and 346 bp; the sites for EcoRV (marked E), BsaAI, ClaI, Eco47III, PstI and XhoI are in the 346 bp circle and that for HindIII (H) in the 3120 bp circle. (C) The 346 bp minicircle is liberated from the 3120 bp ring by cleaving the latter with HindIII.
To see whether the catenane is as effective as either the plasmid or the minicircle at capturing EcoRV for its recognition site, the enzyme was added to mixtures containing equal concentrations of two of these substrates. Both DNA were at much higher concentrations than the enzyme, so as to prohibit two or more molecules of EcoRV binding to one DNA. Both DNA were also at higher concentrations (25 nM) than the Km of EcoRV for plasmid substrates (0.5 nM; Taylor and Halford, 1989). The reaction velocities thus correspond to the maximal rates at saturation of the enzyme. Under these conditions, each molecule of EcoRV carries out multiple turnovers: as soon as it is liberated after cleaving one DNA molecule, it will commence its next reaction on another DNA molecule. The question thus asked repeatedly in this assay is which of the two DNA substrates does the enzyme go for in its next reaction? Even if one substrate gives a faster Vmax, the extent of cleavage of each DNA still provides a direct measure of the partitioning of the enzyme between the two substrates. The partitioning is governed by the specificity constants for the two substrates, their values for kcat/Km (Fersht, 1998).
Supercoiled substrates
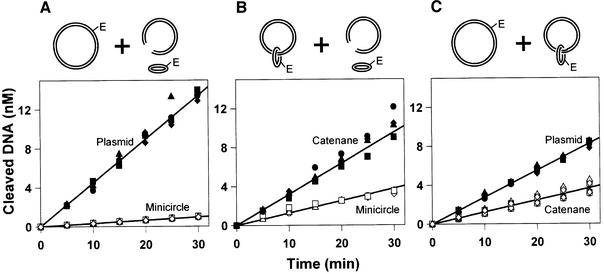
When EcoRV was added to the three possible pair-wise combinations of substrates, the total amounts of DNA cleaved during the reactions were similar in each case (Figure 2). In all three mixtures, the sum of the two initial rates, one for cutting each DNA, came to 1.8 ± 0.2 mol DNA cleaved/mol enzyme/min. This sum equals the Vmax of EcoRV under these conditions (Taylor and Halford, 1989), indicating that the enzyme is indeed operating at its maximal rate at complete saturation with substrate. However, in all three mixtures, more of one substrate was cleaved than the other. Moreover, even though the total amount of DNA cleavage was similar in each case, the extent of cutting any particular DNA varied with the nature of the second DNA in the mixture. For example, the plasmid was cleaved to a greater extent when mixed with the minicircle (Figure 2A) than when mixed with the catenane (Figure 2C). This observation thus validates the experimental design, namely that the extent of cleavage of each DNA is a direct consequence of the partitioning of the enzyme between the two substrates. The DNA that is cleaved preferentially must be better at capturing EcoRV for its recognition site than the other DNA is for its own site.
Fig. 2. Supercoiled DNA. The reactions contained two DNA substrates, both at 25 nM, and 0.2 nM EcoRV endonuclease in reaction buffer at 37°C. Samples were removed at various times after adding the EcoRV and analysed as in Materials and methods for the extent of cleavage of each DNA. Data from four repeats are shown: circles, squares, triangles and diamonds mark the individual experiments. The two substrates in each reaction are as indicated above the respective panel: (A) supercoiled plasmid (filled data points) and minicircle (unfilled data points); (B) supercoiled catenane (filled points) and minicircle (unfilled points); and (C) plasmid (filled points) and catenane (unfilled points), both supercoiled.
When faced with the mixture of plasmid and free minicircle, EcoRV showed a strong preference for the plasmid over the minicircle (Figure 2A). This is as expected: restriction enzymes are known to prefer long substrates to short substrates (Jack et al., 1982; Ehbrecht et al., 1985; Jeltsch and Pingoud, 1998). One might also expect the ratio of the rates on the long and the short DNA to be equal to or smaller than the ratio of their lengths. If both DNA molecules are shorter than the mean path length traversed by the protein during a single DNA-binding event, then the cleavage ratio should equal the length ratio. For example, if the mean path length is 10 000 bp, then a restriction enzyme bound anywhere on a 2000 bp DNA can reach the recognition site. Hence, given one DNA of 2000 bp and another of 500 bp, the enzyme should show a 4-fold preference for the longer DNA, matching the length ratio. On the other hand, if the length of the DNA exceeds the mean path length, the ratio of the cleavage rates will be smaller than the length ratio. For instance, if the mean path length is 100 bp, the enzyme can reach the recognition site only from initial binding events within 100 bp of the site: the ratio of the rates on the 2000 bp DNA and the 500 bp DNA will then be 1:1, smaller than the length ratio of 4:1. In this scheme, the cleavage ratio cannot be larger than the length ratio. Yet the rates at which EcoRV cleaved the 3466 bp plasmid and the 346 bp minicircle differed by more than the length ratio; 14-fold against 10-fold (Table I).
Table I. Substrate selection.
| Partition ratios |
|||
|---|---|---|---|
| Plasmid/minicircle | Catenane/minicircle | Plasmid/catenane | |
| (A) Supercoiled DNA | |||
| EcoRV | 14.0 | 2.5 | 2.2 |
| ClaI | 11.5 | 2.9 | 2.2 |
| BsaAI | 13.0 | 3.0 | 2.4 |
| Eco47III | 15.9 | 3.2 | 2.0 |
| PstI | 10.9 | 3.4 | 2.0 |
| XhoI | 14.4 | 3.8 | 1.9 |
| (B) Relaxed DNA | |||
| EcoRV | 3.5 | 2.4 | 1.1 |
| ClaI | 3.4 | 2.7 | 1.1 |
The mixtures contained two DNA substrates (both 25 nM) and a restriction enzyme in reaction buffer at 37°C. The partitioning of the enzyme between the substrates was assessed as in Materials and methods: the ratios are given as means from ≥4 independent experiments. In (A), the plasmid and the catenane were in their native supercoiled states. In (B), the plasmid and the catenane had been relaxed with topoisomerase I.
EcoRV cleaves both the catenane and the minicircle in the same 346 bp ring of DNA. In the catenane, this ring is tethered to a circular DNA of 3120 bp that has only non-specific sequences for EcoRV. The preparations of free minicircle also contain both the 346 bp circle and the 3120 bp DNA, but as separate moieties (Figure 1C). If the sole pathway for target-site location by EcoRV is 1D sliding, then it must react equally with the catenane and the minicircle since it can reach the site in the catenane only after binding first to the 346 bp circle. Yet, from the mixture of catenane and minicircle, EcoRV cleaved more of the catenane than the minicircle (Figure 2B). Non-specific DNA can therefore channel a protein to a target site even when it is just held near the target in 3D space. To achieve this, the protein must translocate from the large to the small ring of the catenane. This cannot occur by 1D steps along the DNA. Instead, the transfer from non-specific to specific DNA must involve steps through 3D space.
The above experiment used a buffer that lacked NaCl. The experiments were repeated under identical conditions, but with various concentrations of NaCl in the reaction buffer. Salt affected the partitioning of the EcoRV enzyme between the catenane and the minicircle: EcoRV had shown a 2.5-fold preference for the catenane over the minicircle in the absence of NaCl (Table I), but this preference fell to 1.6-fold at 50 mM NaCl and no preference was observed at NaCl concentrations ≥100 mM (data not shown). Hence, as in many previous studies with EcoRV and with many other DNA-binding proteins (Winter et al., 1981; Terry et al., 1985; Stanford et al., 2000), the ability of non-specific DNA to recruit the protein to its specific site declines with increasing salt concentrations. All subsequent experiments in this study were therefore carried out in the absence of NaCl. However, MgCl2 was always present at a concentration (10 mM) sufficient to neutralize essentially all of the DNA charge (Vologodskii and Cozzarelli, 1994).
With the mixture of plasmid and catenane, EcoRV cleaved more of the plasmid than the catenane (Figure 2C). This concurs with the observation that the degree of preference for the plasmid over the minicircle (14-fold) was higher than that for the catenane over the minicircle (2.5-fold). Since the plasmid and the catenane contain the same amounts of non-specific DNA, the catenane ought to be as effective as the plasmid for the initial capture of the enzyme. In addition, once a protein dissociates from one site in a DNA, the probability of it colliding with another specified site in the same DNA is inversely proportional to the distance between the sites in 3D space (Stanford et al., 2000). The spatial separation of two DNA sites in the separate rings of a catenane is similar to that between the equivalent sites in a single ring of DNA (Levene et al., 1995). Hence, if the enzyme travels from non-specific to specific sites by dissociation/re-association events through 3D space, then not only should the initial collision of the enzyme with the catenane be as frequent as that with the plasmid, the subsequent transfer to the target site should also occur almost as readily in the catenane as the plasmid. Yet this is not the case. We return below to examine why EcoRV prefers the plasmid to the catenane.
In addition to its EcoRV site, pDG2 has single sites for several other restriction enzymes in the 346 bp section between the res sites (Figure 1). The catenane generated from pDG2 carries these sites in the minicircle. Equivalent experiments to those described above with EcoRV (Figure 2) were conducted with each of these enzymes: BsaI, ClaI, Eco47III, PstI and XhoI. In all cases, the results were similar to those with EcoRV (Table I). When added to the mixture of plasmid and minicircle, the enzymes all preferred the plasmid to the minicircle, by factors in the range from 11- to 16-fold (c.f. 14-fold with EcoRV). They also prefer the catenane to the minicircle by similar factors to EcoRV, from 3- to 4-fold (c.f. 2.5-fold). Again like EcoRV, they all show a ∼2-fold preference for the plasmid over the catenane. Hence, the mechanisms that cause EcoRV to favour certain substrates over others are not unique to EcoRV. They apply to all of the enzymes tested here. In particular, these enzymes all cleave the 346 bp circle of DNA more readily when it is catenated to another DNA than when it is free from the other DNA. En route to their recognition sites in the catenane, these enzymes must all translocate between the rings, a process that cannot occur by 1D steps.
Relaxed substrates
Why do all of the enzymes tested here cleave the plasmid in preference to the catenane, rather than cleaving these equally? Moreover, why is the partition ratio between the plasmid and the minicircle larger than the ratio of the lengths of these DNA molecules? The latter observation deviates from previous studies with EcoRI and EcoRV, where ∼10-fold increases in DNA length, from ∼300 to ∼3000 bp, had enhanced the specific interaction by factors of ∼4 (Jack et al., 1982; Ehbrecht et al., 1985). In this study, a similar increase in the length of the DNA confers a 14-fold advantage to the longer DNA (Table I). However, the earlier studies, and indeed virtually all other in vestigations on the mechanism of target-site location (Shimamoto, 1999), employed linear DNA substrates, while the substrates used here were supercoiled. If a protein moves from an initial non-specific site to a specific site solely by 1D steps along the DNA, it will find the specific site just as readily in supercoiled DNA as in relaxed DNA, since the contour separation of the sites is not altered by supercoiling. But if the protein moves by 3D steps, supercoiling could facilitate target-site location.
In a relaxed plasmid, the mean distance between any two sites in 3D space is related to the distance between the sites along the contour, in the characteristic manner for a worm-like coil polymer (Doi and Edwards, 1984). Sites that are close together along the 1D contour are also close in 3D space: for example, in Figure 3, the initial landing site LS1 and the EcoRV site E. Sites that are far apart from each other along the DNA, for example LS2 and E (Figure 3), will usually be distant from each other in 3D space. However, in a supercoiled plasmid, the mean distance between two sites in 3D space is largely independent of their contour separation. Well-separated sites along the DNA, for example LS2 and E, will sometimes be juxtaposed across the superhelix and close in 3D space (Figure 3). Moreover, supercoiling compresses the volume occupied by the DNA, so two sites will generally be closer together in supercoiled DNA than in relaxed DNA (Vologodskii and Cozzarelli, 1994). As a result of the reductions in inter-site distances, the transfer of a protein from a random to a specific site by dissociation/re-association steps should be more efficient in a supercoiled plasmid than in a relaxed plasmid.
Fig. 3. Potential effects of supercoiling on target-site location. The upper pictures show the relaxed and the supercoiled forms of a plasmid with a single recognition site for EcoRV (marked E). Two landing sites for the initial binding of the protein are marked LS1 and LS2: the 1D distance along the DNA between E and LS1 is shorter than that between E and LS2. In the relaxed DNA, the mean 3D distance between E and LS1 is also shorter than that between E and LS2. In supercoiled DNA, the spatial separation of two sites is largely independent of their contour separation so E will sometimes be closer to LS2 than LS1. The lower pictures show catenanes containing one large and one small ring of DNA, with the larger ring in either its relaxed or supercoiled configurations. In the relaxed form, the small ring is free to move around the large ring in both rotational and lateral senses. In the supercoiled form, the small ring is located mainly at an apex of the superhelix and is largely blocked from lateral motion around the large ring.
On the other hand, supercoiling may curtail transfers from the large ring to the small ring of the catenane. In a singly interlinked catenane of supercoiled DNA, as formed by Tn21 resolvase (Castell et al., 1986), the interlink is usually located at the tip(s) of the superhelix: it is rarely found in the interwound segments of the superhelix (Levene et al., 1995). One ring in a catenane may therefore be able to move around and along the other ring more readily when the DNA is relaxed than when it is supercoiled (Figure 3). If so, a protein bound to the large ring of a catenane has more access to the small ring in the relaxed form of the DNA than in its supercoiled form. Supercoiling thus confines the position of the EcoRV site within the overall (3D) structure of the DNA molecule more acutely in the catenane than in the intact plasmid. Hence, transfer from random to specific sites by dissociation/re-association events may be less efficient in the supercoiled catenane than in a relaxed catenane, though this effect might be nullified by the general effect of supercoiling noted above.
To examine these possibilities, topoisomerase I was used to remove all of the supercoils from the plasmid and the catenane. The relaxed samples were then mixed with each other or with the free minicircle and the partitioning of the EcoRV enzyme was measured as before (Figure 4). The free minicircle used here came from samples of the supercoiled catenane that had not been treated with topoisomerase I. The minicircle thus constitutes a fixed frame of reference with which to compare the supercoiled and relaxed forms of the plasmid (Figures 2A and 4A) and likewise the two forms of the catenane (Figures 2B and 4B). Moreover, the electrophoretic mobility of the minicircle was unaltered by topoisomerase I (data not shown), so it lacks supercoils (the topoisomerase reaction on the catenane affects only the large ring). A circular DNA of 346 bp ought to have ∼2 supercoils, given the standard superhelical density of E.coli DNA (Bates and Maxwell, 1993). However, recombination by resolvase causes a topological change equivalent to the loss of 4 supercoils (Stark and Boocock, 1995). If resolvase takes 2 supercoils from the 346 bp DNA, it might generate directly the relaxed form of the minicircle. Alternatively, the lack of supercoils in the minicircle could be due to the topoisomerase activity of resolvase (Castell et al., 1986; Falvey et al., 1988).
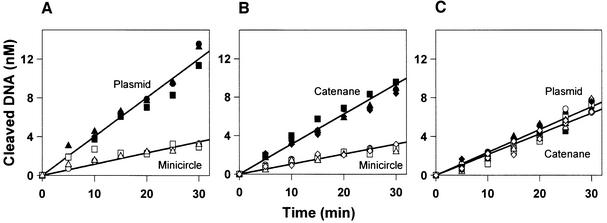
Fig. 4. Relaxed DNA. The reactions contained two DNA substrates, both at 25 nM, and 0.2 nM EcoRV in reaction buffer at 37°C. Samples were removed at various times after the addition of EcoRV and analysed as in Materials and methods for the extents of cleavage of both DNA. Data from four repeats are shown: circles, squares, triangles and diamonds denote the individual experiments. The two substrates are: in (A), relaxed plasmid (filled data points) and minicircle (unfilled data points); in (B), relaxed catenane (filled points) and minicircle (unfilled points); and in (C), relaxed plasmid (filled points) and relaxed catenane (unfilled points). The plasmid and the catenane used here had been treated with topoisomerase I.
In the three mixtures containing relaxed DNA (Figure 4), the sums of the two initial rates were similar to each other: all three fell between 1.7 and 2.2 mol DNA/mol enzyme/min. They were also similar to the sums of rates from the mixtures containing supercoiled substrates (1.8 mol DNA/mol enzyme/min; Figure 2). Nevertheless, with some mixtures, the partition ratios between the relaxed substrates differed from those with the supercoiled substrates (Table I). While EcoRV prefers the supercoiled plasmid to the minicircle by a factor of 14, it prefers the relaxed plasmid to the minicircle by a factor of just 3.5. The supercoiled plasmid is thus four times better than the relaxed plasmid at bringing EcoRV to its recognition site. Moreover, in contrast to the supercoiled DNA, the degree of preference for the relaxed plasmid over the minicircle no longer exceeds the ratio of the lengths of these two DNA molecules. Instead, the 3.5-fold enhancement for a 10-fold increase in length matches previous observations with linear DNA fragments varying from ∼300 to ∼3000 bp (Jack et al., 1982; Ehbrecht et al., 1985). However, while supercoiling aided target-site location in the plasmid, it had no impact on the catenane: the degree of preference for the relaxed catenane over the minicircle was the same as that for the supercoiled catenane over the minicircle (Table I). The general effect of supercoiling on target-site location seems to be counter-balanced by the constraint on the relative motion of the catenane rings (Figure 3).
The above comparison, the supercoiled plasmid versus the minicircle relative to the relaxed plasmid versus the minicircle, is an indirect measure of the effect of supercoiling on target-site location. To examine this effect directly, EcoRV was added to mixtures containing equal concentrations of the supercoiled and relaxed forms of pDG2, and the extent of cleavage of each DNA measured as above. For these experiments, the relaxed form was generated by using a nicking enzyme, N.BstNBI, to convert the supercoiled form of pDG2 to its open-circle form. In this direct assay, the EcoRV enzyme showed a 3.3-fold preference for the supercoiled plasmid over the relaxed plasmid (data not shown), in close agreement with the 4-fold preference in the indirect comparison.
Faced with the supercoiled forms of the plasmid and the catenane, EcoRV had shown a 2-fold preference for the plasmid (Figure 2C). But when faced with the relaxed forms of these two DNA molecules, it no longer had any significant preference for the plasmid over the catenane (Figure 4C). The same selectivity on supercoiled DNA, but lack of selectivity on relaxed DNA, was noted with ClaI (Figure 5; Table I). (The other restriction enzymes examined here were not tested against relaxed substrates.) However, while the removal of the supercoils from the plasmid and the catenane leaves these two DNA equally susceptible to these enzymes, the relaxed plasmid and the relaxed catenane are both cleaved preferentially to the minicircle: in mixtures containing the minicircle and the relaxed forms of either the plasmid or the catenane, both EcoRV and ClaI went for the larger DNA (Table I). Hence, these enzymes must still find their recognition sites in the relaxed substrates via the 3120 bp of non-specific DNA that is present in the plasmid and the catenane, but which is absent from the minicircle. Once the factors due to super coiling are eradicated, this 3120 bp segment of non-specific DNA is every bit as good a conduit when held near the recognition site in 3D space as when co-linear with the site.
Fig. 5. ClaI on supercoiled and relaxed DNA. The reactions contained the plasmid pDG2 and the catenane (both at 25 nM) and ClaI (5 U/ml) in reaction buffer at 37°C. Samples were removed at various times and analysed as in Materials and methods for the extents of cleavage of both substrates. Data from two reactions are shown: circles and squares denote the individual experiments. The filled data points mark the cleavage of the plasmid and the unfilled points the catenane. In (A), both DNA contained their natural levels of supercoiling. In (B), both DNA had been relaxed with topoisomerase I.
Discussion
One or three dimensions
For many years (Riggs et al., 1970; Richter and Eigen, 1974), DNA-binding proteins were thought to locate their target sites by ‘sliding’: first binding to the DNA at random, wherever they happen to first collide with the chain; then translocating to their target sites by 1D diffusion along the DNA (von Hippel and Berg, 1989; Shimamoto, 1999). Many proteins are capable of 1D motion on DNA in an energy-dependent manner, hydrolysing nucleoside triphosphates in order to move along the chain in a particular direction: for example, DNA and RNA polymerases, helicases, mismatch repair enzymes and type I restriction enzymes (Halford and Szczelkun, 2002). However, energy-dependent motion in a specified direction is distinct from motion by free (thermally driven) diffusion. If a protein moves between adjacent sites by free diffusion, as it must do so in the absence of an energy input, each step has the same probability for ‘backward’ or ‘forward’ motion. The mean position of the protein remains its initial locus, but the successive 1D steps result in progressively larger excursions from the initial locus. Half of the excursions will be in the ‘right’ direction and one of these may eventually take the protein to the target site. A search by free diffusion confined to the 1D contour of the DNA is thus a highly redundant—and hence inefficient—process, since it results in the protein repetitively inspecting just one segment of the DNA chain (Gerland et al., 2002). Even so, the constraint to one dimension can, under certain conditions, enhance the rate of target-site location (Berg, 1993).
In an alternative scheme, a protein moves from its initial random site to its final specific site through 3D space, by repeatedly dissociating from the DNA before binding back to another site elsewhere in the same molecule of DNA. The majority of re-associations will be at or very near the point of departure (Berg and von Hippel, 1985), so, as with 1D routes, 3D routes lead the protein to every potential binding site in the DNA, including the adjacent sites 1 bp apart from each other. After binding to DNA at a non-specific site, proteins usually interact with the nearest copy of the target site rather than a more distant copy (Terry et al., 1985; Jeltsch et al., 1994; Stanford et al., 2000), but both 1D and 3D transfers can account for this. However, 3D pathways also include long-range steps between distant sites in the same molecule of DNA. The fundamental distinction between 1D and 3D pathways is thus whether the protein remains in continual contact with the DNA as it moves from one site to another. Some DNA-binding proteins have toroidal structures and encircle the DNA: for example, the hexameric helicases (Soultanas and Wigley, 2001) and various processivity factors for DNA replication (Jeruzalmi et al., 2002). Another example is a modified form of the EcoRV restriction enzyme where the two sides of the DNA-binding cleft were cross-linked over the DNA (Schulze et al., 1998). These proteins are constrained to 1D motion on DNA, but this constraint does not apply to proteins that are not topologically linked to the DNA.
When the native form of EcoRV was given the choice between a solitary minicircle of DNA and the same circle catenated to a ring of non-specific DNA, it went for the catenated circle rather than the solitary circle (Figures 2B and 4B). The same preference was observed with several other restriction enzymes (Table I). If these enzymes had been able to reach their recognition sites on the minicircle only by sliding after initially binding to that circle, they would have shown no preference for the catenane over the minicircle. Hence, the preference for the catenane demonstrates unequivocally that these enzymes must travel to their recognition sites in the 346 bp ring of the catenane via the ring of non-specific DNA. Translocation between the rings must involve steps through 3D space.
The preference for the catenane over the minicircle does not, however, reveal the proportions of 1D and 3D steps. For instance, it is possible that the vast majority of translocation steps are 1D events, but that these are intermingled with an occasional step through 3D space. Such a theory could account for why these enzymes prefer the supercoiled plasmid to the supercoiled catenane (Table I). However, the relaxed plasmid has no advantages at all over the relaxed catenane (Figures 4C and 5B). In principle, the EcoRV enzyme could reach its recognition site in the plasmid by linear diffusion from sites up to 1733 bp away from the recognition site (i.e. half the length of the 3466 bp circle of DNA), but the maximal distance for linear diffusion to the EcoRV site in the catenane is only 173 bp (i.e. half the length of the 346 bp minicircle). Hence, if target-site location occurs by a 1D pathway covering more than 173 bp, the relaxed plasmid would have been favoured over the relaxed catenane. The fact that no such preference was observed excludes 1D transfers over distances of ≥173 bp. Yet the plasmid was still preferred to the free minicircle, which indicates translocation events over >173 bp. Furthermore, under these reaction conditions, it acts processively on DNA molecules with two EcoRV sites separated by >750 bp (Stanford et al, 2000). EcoRV is thus capable of intramolecular transfers between sites in the same DNA molecule that are separated by kilobase distances. Hence, while sliding remains a possibility for short distances of ∼100 bp, perhaps in the final docking of the protein against its recognition sequence, the large-scale motion of all of the proteins examined here, over distances >100 bp, occurs solely by dissociation/re-association steps through 3D space.
Supercoiling
Supercoiling has long been known to govern many aspects of the structure and function of cellular DNA, and to affect the activities of many enzymes acting on DNA (Bates and Maxwell, 1993; Vologodskii and Cozzarelli, 1994). For instance, all processes that perturb the helical twist of DNA, such as replication and transcription, are affected by supercoiling (Bates and Maxwell, 1993). In addition, since the probability of juxtaposition of two sites is much higher in supercoiled DNA than in relaxed DNA (Vologodskii and Cozzarelli, 1996), reactions that involve the juxtaposition of distant sites are stimulated by supercoiling (Huang et al., 2001): for example, DNA re-arrangements by site-specific recombination and DNA cleavage by the type II restriction enzymes that interact with two sites (Halford et al., 2000). However, it had not been noted before that supercoiling also affects the primary process of getting the protein to its target site. Indeed, when it was thought that proteins found their target sites by 1D sliding, there would have been no reason to envisage a role for supercoiling in this process. The 1D distance between initial and final sites must be the same in supercoiled and relaxed DNA, yet we have found that proteins find their target sites more readily in supercoiled DNA than in relaxed DNA.
One explanation for the effect of supercoiling on target-site location is that some proteins bind more tightly to supercoiled DNA than to relaxed DNA and thus might increase the length of DNA they scan per binding event. However, the proteins that bind more tightly to supercoiled DNA than to relaxed DNA alter the twist of the DNA as they bind to it (Bates and Maxwell, 1993), such as RNA polymerase (Amouyal and Buc, 1987). EcoRV, on the other hand, binds to non-specific DNA without altering the twist (Winkler et al., 1993). Moreover, if the effect of supercoiling had been due to different affinities for supercoiled or relaxed DNA, it would have affected target-site location equally on the catenane and on the plasmid. Hence, the observation that supercoiling enhances target-site location on the plasmid, but not on the catenane, shows that this effect must be due to the global structures of the DNA.
The reason why supercoiling facilitates target-site location is unlikely to be due to an effect on the bimolecular rate for the initial binding of the enzyme to the DNA: the enzyme should collide with the supercoiled substrate at much the same frequency as the relaxed substrate. Instead, it is more likely to be caused by increasing the efficiency of the 3D transfer from the initial non-specific site to the recognition site. Following the dissociation of the protein from a non-specific site, the probability of re-association with another site in the DNA is, as noted above, inversely proportional to the distance between the sites in 3D space (Stanford et al., 2000). The distance between two sites will almost always be smaller in supercoiled DNA than in relaxed DNA (Vologodskii and Cozzarelli, 1996; Huang et al., 2001). Hence, unlike 1D pathways, 3D pathways can readily explain why target-site location should be more efficient in supercoiled DNA than in relaxed DNA.
Materials and methods
Proteins
EcoRV endonuclease and Tn21 resolvase were purified and used as described previously (Oram et al., 1997; Stanford et al., 2000). Their concentrations were determined by absorbance at 280 nm and are cited as molarities for the dimeric proteins. Eco47III and wheat germ topoisomerase I were from Promega. All other enzymes were from New England Biolabs. Concentrations of commercial enzymes are in terms of units of activity, as specified by the supplier.
DNA
The plasmid pDG2 (Figure 1A) was constructed from pMLE1 (Embleton et al., 2001) by standard procedures (Sambrook et al., 1989). Transformants of E.coli HB101 with pDG2 were grown in M9 minimal media (Sambrook et al., 1989) with 37 MBq/l [methyl-3H]thymidine. Cells were harvested, lysed and the plasmid purified by density gradient centrifugations (Embleton et al., 2001). Typically, 90–95% of the DNA in the preparations was the monomeric form of the supercoiled plasmid, and 5–10% either the nicked or the dimeric forms of the plasmid. DNA concentrations were determined by absorbance at 260 nm.
To make the catenane, 2 nmol Tn21 resolvase was added to 0.2 nmol pDG2 in 1 ml of 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2 and 100 µg/ml BSA. After 5 h at 37°C, 25 µg proteinase K was added. Thirty minutes later, the DNA was extracted with phenol/chloroform, precipitated with ethanol, washed and resuspended in TE (Sambrook et al., 1989). Assays for the extent of recombination (Oram et al., 1997) showed that ∼95% of the DNA in these samples was catenane: the ∼5% unrecombined plasmid is due to the open-circle DNA in the plasmid preparations that is not recombined by resolvase.
To generate the free minicircle, 0.1 nmol catenane was incubated with 100 U HindIII in 500 µl of 10 mM Tris–HCl (pH 7.5) and 10 mM MgCl2 for 5 h at 37°C, prior to heating at 67°C for 30 min. Agarose gel electrophoresis of this DNA showed that all of the catenane had been cleaved in the large ring. These samples were used directly as the free minicircle substrate, without further manipulation.
Relaxation reactions typically contained 50 pmol DNA (either pDG2 or the catenane) and 250 U topoisomerase I in 500 µl of 12.5 mM Tris–HCl (pH 7.5), 25 mM NaCl, 10 mM MgCl2, 5 mM β-mercaptoethanol and 50 µg/ml BSA. After 5 h at 37°C, 25 µg proteinase K was added. Thirty minutes later, the DNA was extracted with phenol/chloroform, precipitated with ethanol, washed and resuspended in TE. When analysed by electrophoresis through agarose gels lacking ethidium bromide, all of the DNA possessed the mobility characteristic of the fully relaxed form.
Reactions
Reactions at 37°C contained two DNA substrates, both at 25 nM, in 100 µl reaction buffer [10 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 5 mM β-mercaptoethanol and 50 µg/ml BSA]. Reactions were started by adding 1 µl of the restriction enzyme diluted to the requisite concentration. For EcoRV, a 20 nM stock was used to give a final concentration of 0.2 nM, so that ∼1% of the total DNA was cleaved per minute (Figures 2 and 4). The amounts of the other restriction enzymes (0.5 U for ClaI and PstI, 1 U for BsaI and XhoI and 4 U for Eco47III) were selected so as to also give ∼1% total DNA cleavage per minute. (The differing amounts of the enzymes are a reflection of their activities in reaction buffer, which lacks NaCl.) Aliquots (12 µl) were taken from the reactions at various times after adding the enzyme and mixed immediately with 10 µl stop-mix (Embleton et al., 2001). A zero time point was taken before adding the enzyme. The aliquots were heated at 67°C for 15 min and cooled to room temperature prior to electrophoresis through agarose: one part (10 µl) of each aliquot was applied to a 1% gel to separate the supercoiled and linear forms of the plasmid and the catenane, and another part (10 µl) to a 2% gel to separate the circular and linear forms of the minicircle. In both cases, electrophoresis was at 7 V/cm for 2–3 h. Segments of the gels encompassing each form of the DNA were excised and analysed by scintillation counting (Embleton et al., 2001). The d.p.m. values were corrected for the different lengths of the DNA fragments and are expressed relative to the substrate at zero time.
The reactions on the plasmid and the minicircle contain the following forms of DNA: circular and linear DNA of 3466 bp; circular and linear DNA of 346 bp; or linear DNA of 3120 bp (from the preparation of the minicircle). When these were analysed by electrophoresis, the two circular substrates and the two linear products were all resolved from the other DNA species in the mixture. The concentrations of both the 3466 bp and the 346 bp substrates and their respective products were measured at each time point. The extents of cleavage of the plasmid were calculated from the mean changes in the concentrations of the circular and the linear forms of the 3466 bp DNA, and the extents of cleavage of the minicircle from the two forms of the 346 bp DNA. To determine the initial rates, the increases in the extents of cleavage with time, from ≥4 repeat reactions, were fitted to linear slopes by using GRAFIT 5 (Erithacus Software). In reactions on the catenane and the minicircle, both substrates give rise to a linear DNA of 346 bp: the extent of cleavage of the minicircle was therefore assessed solely from the decrease in the concentration of its circular form, while the extent of cleavage of the catenane was taken from the decrease in the concentration of the intact catenane together with the increase in the concentration of the 3120 bp circular product (the latter has a different mobility from the linear 3120 bp DNA in the minicircle preparations). In reactions on the plasmid and the catenane, the two 3466 bp substrates have the same mobility: the extent of cleavage of the plasmid was determined from the increase in the concentration of the linear DNA of 3446 bp, while the cleavage of the catenane was measured from the increases in the concentrations of both the circular product of 3120 bp and the linear product of 346 bp.
Acknowledgments
Acknowledgements
We thank Neil Stanford, Mark Oram, Mark Szczelkun and David Scott for proteins and/or advice. This work was funded by grant 063111 from the Wellcome Trust.
References
- Adzuma K. and Mizuuchi,K. (1989) Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell, 57, 41–47. [DOI] [PubMed] [Google Scholar]
- Amouyal M. and Buc,H. (1987) Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and UV5 sites of Escherichia coli. J. Mol. Biol., 195, 795–808. [DOI] [PubMed] [Google Scholar]
- Baldwin G.S., Sessions,R.B., Erskine,S.G. and Halford,S.E. (1999) DNA cleavage by the EcoRV restriction endonuclease: roles of divalent metal ions in specificity and catalysis. J. Mol. Biol., 288, 87–103. [DOI] [PubMed] [Google Scholar]
- Bates A.D. and Maxwell,A. (1993) DNA Topology. IRL Press, Oxford, UK.
- Berg H.C. (1993) Random Walks in Biology. Princeton University Press, Princeton, NJ.
- Berg O.G. and von Hippel,P.H. (1985) Diffusion-controlled macro molecular interactions. Annu. Rev. Biophys. Biophys. Chem., 14, 131–160. [DOI] [PubMed] [Google Scholar]
- Berg O.G., Winter,R.B. and von Hippel,P.H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry, 20, 6929–6948. [DOI] [PubMed] [Google Scholar]
- Castell S.E., Jordan,S.L. and Halford,S.E. (1986) Site-specific recombination and topoisomerization by Tn21 resolvase: role of metal ions. Nucleic Acids Res., 14, 7213–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M. and Edwards,S.F. (1984) The Theory of Polymer Dynamics. Oxford University Press, Oxford, UK.
- Ehbrecht H.-J., Pingoud,A., Urbanke,C., Maass,G. and Gualerzi,C. (1985) Linear diffusion of restriction endonucleases on DNA. J. Biol. Chem., 260, 6160–6166. [PubMed] [Google Scholar]
- Embleton M.L., Siksnys,V. and Halford,S.E. (2001) DNA cleavage reactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol., 311, 503–514. [DOI] [PubMed] [Google Scholar]
- Falvey E., Hatfull,G.F. and Grindley,N.D.F. (1988) Uncoupling of the recombination and topoisomerase activities of the γδ resolvase by a mutation at the crossover point. Nature, 332, 861–863. [DOI] [PubMed] [Google Scholar]
- Fersht A.R. (1998) Structure and Mechanism in Protein Science. W.H. Freeman, New York, NY, pp. 117.
- Gerland U., Moroz,J.D. and Hwa,T. (2002) Physical constraints and functional characteristics of transcription factor-DNA interaction. Proc. Natl Acad. Sci. USA, 99, 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford S.E. and Szczelkun,M.D. (2002) How to get from A to B: strategies for analysing protein motion on DNA. Eur. Biophys. J., 31, 257–267. [DOI] [PubMed] [Google Scholar]
- Halford S.E., Gowers,D.M. and Sessions,R.B. (2000) Two are better than one. Nat. Struct. Biol., 7, 705–707. [DOI] [PubMed] [Google Scholar]
- Huang J., Schlick,T. and Vologodskii,A. (2001) Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl Acad. Sci. USA, 98, 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W.E., Terry,B.J. and Modrich,P. (1982) Involvement of outside sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc. Natl Acad. Sci. USA, 79, 4010–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. and Pingoud,A. (1998) Kinetic characterisation of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochemistry, 37, 2160–2169. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Alves,J., Wolfes,H., Maass,G. and Pingoud,A. (1994) Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry, 33, 10215–10219. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2002) Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol., 12, 217–224. [DOI] [PubMed] [Google Scholar]
- Levene S.D., Donahue,C., Boles,T.C. and Cozzarelli,N.R. (1995) Analysis of the structure of dimeric DNA catenanes by electron microscopy. Biophys. J., 69, 1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M.A., Th’ng,J.P.H., Sun,X. and Hendzel,M.J. (2000) Rapid exchange of histone H1.1 on chromatin in living human cells. Nature, 408, 873–876. [DOI] [PubMed] [Google Scholar]
- Misteli T. (2001) Protein dynamics: implications for nuclear architecture and gene expression. Science, 291, 843–847. [DOI] [PubMed] [Google Scholar]
- Oram M., Marko,J.F. and Halford,S.E. (1997) Communications between distant sites on supercoiled DNA from non-exponential kinetics for DNA synapsis by resolvase. J. Mol. Biol., 270, 396–412. [DOI] [PubMed] [Google Scholar]
- Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Reid S.L., Parry,D., Liu,H.-H. and Connolly,B.A. (2001) Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarization. Biochemistry, 40, 2484–2494. [DOI] [PubMed] [Google Scholar]
- Richter P.H. and Eigen,M. (1974) Diffusion controlled reaction rates in spheroidal geometry. Application to repressor-operator association and membrane bound enzymes. Biophys. Chem., 2, 255–263. [DOI] [PubMed] [Google Scholar]
- Riggs A.D., Bourgeois,S. and Cohn,M. (1970) The lac repressor-operator interaction. III. Kinetic studies. J. Mol. Biol., 53, 401–417. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schulze C., Jeltsch,A., Franke,I., Urbanke,C. and Pingoud,A. (1998) Crosslinking the EcoRV restriction endonuclease across the DNA-binding site reveals transient intermediates and conformational changes of the enzyme during DNA binding and catalytic turnover. EMBO J., 17, 6757–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto N. (1999) One-dimensional diffusion of proteins along DNA. J. Biol. Chem., 274, 15293–15296. [DOI] [PubMed] [Google Scholar]
- Soultanas P. and Wigley,D.B. (2001) Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci., 26, 47–54. [DOI] [PubMed] [Google Scholar]
- Stanford N.P., Szczelkun,M.D., Marko,J.F. and Halford,S.E. (2000) One- and three-dimensional pathways for proteins to reach specific sites. EMBO J., 19, 6546–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark W.M. and Boocock,M.R. (1995) Topological selectivity in site-specific recombination. In Sherratt,D.J. (ed.), Mobile Genetic Elements. Oxford University Press, Oxford, UK, pp. 101–129.
- Szczelkun M.D. and Halford,S.E. (1996) Recombination by resolvase to analyse DNA communications by the SfiI restriction endonuclease. EMBO J., 15, 1460–1469. [PMC free article] [PubMed] [Google Scholar]
- Szczelkun M.D., Dillingham,M.S., Janscak,P., Firman,K. and Halford,S.E. (1996) Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J., 15, 6335–6347. [PMC free article] [PubMed] [Google Scholar]
- Taylor J.D. and Halford,S.E. (1989) Discrimination between DNA sequences by the EcoRV restriction endonuclease. Biochemistry, 28, 6198–6207. [DOI] [PubMed] [Google Scholar]
- Taylor J.D., Badcoe,I.G., Clarke,A.R. and Halford,S.E. (1991) EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry, 30, 8743–8753. [DOI] [PubMed] [Google Scholar]
- Terry B.J., Jack,W.E. and Modrich,P. (1985) Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J. Biol. Chem., 260, 13130–13137. [PubMed] [Google Scholar]
- Vologodskii A.V. and Cozzarelli,N.R. (1994) Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct., 23, 609–643. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. and Cozzarelli,N.R. (1996) Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J., 70, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P.H. and Berg,O.G. (1989) Facilitated target location in biological systems. J. Biol. Chem., 264, 675–678. [PubMed] [Google Scholar]
- Winkler F.K. et al. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter R.B., Berg,O.G. and von Hippel,P.H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor-operator interaction: kinetic measurements and conclusions. Biochemistry, 20, 6961–6977. [DOI] [PubMed] [Google Scholar]
- Wright D.J., Jack,W.E. and Modrich,P. (1999) The kinetic mechanism of EcoRI endonuclease. J. Biol. Chem., 274, 31896–31902. [DOI] [PubMed] [Google Scholar]