Abstract
Plant mitochondria are remarkable with respect to their content in foreign, alien and plasmid-like DNA, raising the question of the transfer of this information into the organelles. We demonstrate the existence of an active, transmembrane potential-dependent mechanism of DNA uptake into plant mitochondria. The process is restricted to double-strand DNA, but has no obvious sequence specificity. It is most efficient with linear fragments up to a few kilobase pairs. When containing appropriate information, imported sequences are transcribed within the organelles. The uptake likely involves the voltage-dependent anion channel and the adenine nucleotide translocator, i.e. the core components of the mitochondrial permeability transition pore complex in animal cells, but it does not rely on known mitochondrial membrane permeabilization processes. We conclude that DNA import into plant mitochondria might represent a physiological phenomenon with some functional relevance.
Keywords: adenine nucleotide translocator/genetic flux/plasmid like/potato/voltage-dependent anion channel
Introduction
Systematic sequence analyses predict a highly compact mitochondrial genome in the ancestor of green plants (Turmel et al., 2002). Nevertheless, higher plant, especially angiosperm, mitochondrial genomes have a strikingly large size [usually 300–800 kilobase pairs (kb)], as compared with the DNA of animal or fungal mitochondria (usually 16–85 kb). From that wealth of sequences, only 11–18% correspond to protein or structural RNA genes, >5% originate from plastids, nuclei or viruses and, even more puzzling, about half have no recognizable function and origin (Marienfeld et al., 1999; Kubo et al., 2000). Unlike those of mammals and most other eukaryotes, mitochondria of numerous plant species have also acquired, in addition to the main high molecular weight genome, one or several types of extrachromosomal plasmids or replicons composed of either DNA or RNA, with a size ranging from 0.7 to >20 kb (Brown and Zhang, 1995). In the plant species studied so far, mitochondrial DNA plasmids are arranged as species-specific or even line-specific sets of circular and linear DNA molecules with essentially unknown functions, and replicate independently of the main mitochondrial genomic DNA. Most of the mitochondrial plasmids have no sequence homology to the main mitochondrial genome and seem to be dispensable. Nevertheless, according to the developmental stage, they can be present at a high stoichiometry relative to the main genome: a ca 6:1 ratio was shown in the case of the S1 and S2 linear plasmids in maize mitochondria. This somehow resembles the situation occurring with chromosomal DNA and plasmids in bacteria. Linear mitochondrial DNA plasmids may carry expressed sequences encoding proteins or transfer RNAs (Leon et al., 1989, 1992). The origin of mitochondrial plasmids is unknown. It was suggested that double-strand DNA plasmids might have been introduced into higher plant cells by symbionts or pathogens (Douce and Neuburger, 1989). Supporting this hypothesis, the double-strand linear plasmids with 5′-associated proteins are reminiscent of the structure of some viral genomes. As a whole, all these data suggest that plant mitochondria have a high capacity to capture and integrate foreign sequences.
The presence of integrated retrotransposons from the nuclear compartment, of sequences derived from RNA viruses and of RNA plasmids in plant mitochondria emphasizes the possibility of intercompartmental transfers via RNA intermediates (Marienfeld et al., 1999). In line with such considerations, the existence of specific tRNA import into plant mitochondria (Maréchal-Drouard et al., 1993; Glover et al., 2001) has indeed been established, in particular in our laboratory, but possible uptake of larger RNAs has received little attention and cannot account for all genetic information capture events and for the acquisition of DNA plasmids. We therefore asked the question of the existence of a mitochondrial mechanism responsible for controlled DNA transport. Using the well defined maize mitochondrial linear DNA plasmid of 2.3 kb (Leon et al., 1989) as a model substrate, we showed that plant mitochondria are able to take up double-strand linear DNA through a sequence-independent active mechanism, establishing a novel mitochondrial transport process of macromolecules. The incorporated DNA can be transcribed in the organelles and serve as a template for repair synthesis. Inhibition studies of the uptake point to an involvement of the voltage-dependent anion channel (VDAC) and the adenine nucleotide translocator (ANT), which are considered as the core components of the mitochondrial permeability transition pore complex (PTPC) in animal cells (Zamzami and Kroemer, 2001). However, DNA import into plant mitochondria does not seem to be related to mitochondrial permeability transition (MPT).
Results
Plant mitochondria can incorporate large size DNA
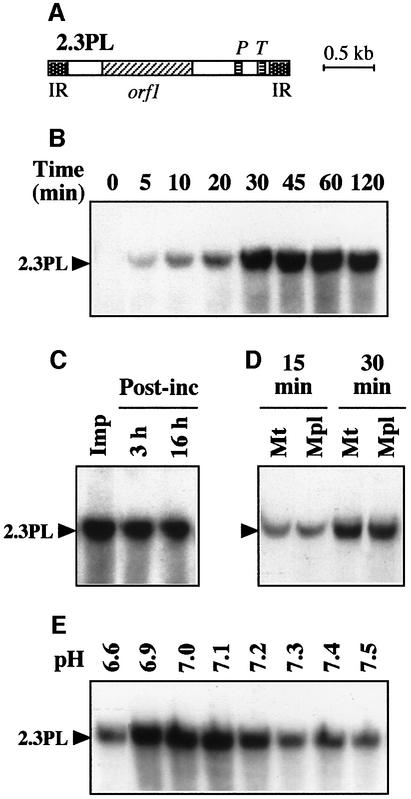
Tests for DNA import into isolated plant mitochondria were developed using radioactively labeled, double-strand maize (Zea mays) mitochondrial 2.3 kb linear plasmid (Figure 1A). Functional and intact potato (Solanum tuberosum) mitochondria were used throughout the present studies. Their respiratory control ratio (RCR; see Materials and methods) was 4.5–5 with succinate- supported respiration and their integrity was >97%. Mitochondrial incorporation was characterized by acquired resistance to extensive DNase digestion. Various experimental conditions were tested, and DNA uptake into plant mitochondria was obtained in a minimal medium containing an osmoticum (0.4 M sucrose) and a buffer (40 mM potassium phosphate pH 7.0). The detailed protocol is described in Materials and methods. According to time-course analyses, incorporation of the 2.3 kb plasmid into the mitochondrial fraction was progressive and reached a plateau after ∼30 min (Figure 1B). To test the stability of the incorporated DNA in mitochondria, the import reaction was stopped at 30 min by eliminating the excess of labeled exogenous DNA through DNase treatment and washing before further incubation of the organelles in the same conditions without import substrate. Even when mitochondria were re-isolated and extracted after 16 h of post-incubation, the amount of incorporated DNA lost by export (if any) or degradation was low (Figure 1C), indicating that the process leads to rather stable DNA integration into the organelles. In further assays, the mitochondrial outer membrane was broken by an osmotic shock after the import step, generating mitoplasts and leaving accessible to DNase digestion any labeled DNA that would not have crossed the mitochondrial inner membrane. Such a treatment had no influence on the DNase resistance of the incorporated DNA, as compared with a mock treatment (Figure 1D), implying that the DNA was protected by the mitochondrial inner membrane. Considering the large size of the DNA used in the tests, such a protection was likely to require a complete transfer of the DNA to the matrix side. It therefore appears that DNA is able to cross both mitochondrial membranes. The transport by itself is probably fast, as we never observed the appearence of fragments with intermediate sizes or of a smear upon electrophoretic analysis of the incorporated material. The efficiency was estimated at 1–5 pg DNA incorporated for an amount of mitochondria corresponding to 300 µg protein. The process was saturated at a DNA concentration of ∼20 ng/ml and was dependent on the pH (Figure 1E), with an optimal efficiency arround pH 7.0.
Fig. 1. Plant mitochondria can incorporate large size DNA. (A) Organization of the 2.3 kb maize mitochondrial plasmid (2.3PL). The plasmid contains two copies of a 170 nt inverted repeat (IR), a 885 nt open reading frame (orf1) and chloroplast-like genes for a tRNAPro (P) and a tRNATrp (T). (B) Time course of DNA uptake. Labeled linear maize 2.3 kb plasmid was incubated for different times in the presence of purified potato tuber mitochondria prior to DNase digestion. Mitochondrial nucleic acids were subsequently extracted, fractionated by agarose gel electrophoresis and transferred onto a nylon membrane, which was autoradiographed. (C) Stability of the incorporated DNA. Uptake of labeled maize 2.3 kb plasmid into potato mitochondria was run for 30 min. Following DNase treatment and washing, a sample was kept for direct analysis of the 30 min import (Imp), whereas the rest of the mitochondria was further incubated (Post-inc) for 3 or 16 h in the absence of added DNA. (D) DNase protection of incorporated plasmid is resistant to outer membrane breaking. Following incorporation of labeled maize 2.3 kb plasmid for 15 or 30 min, mitochondria were mock treated (Mt) or submitted to osmotic shock (Mpl) before DNase treatment. (E) DNA uptake is pH dependent. Incorporation of labeled maize 2.3 kb plasmid in the presence of potato mitochondria was run with a series of potassium phosphate buffers with different pH values. Incubation time for uptake was 30 min. Migration of the incorporated plasmid (2.3PL) is indicated.
DNA incorporation into plant mitochondria is not sequence specific
To characterize the specificity of DNA incorporation into plant mitochondria, different substrates were tested. When the labeled 2.3 kb maize plasmid was heated for 2 min at 95°C and chilled on ice to separate the DNA strands prior to incubation with plant mitochondria, incorporation into the organelles was abolished (Figure 2A), indicating that mitochondrial import is restricted to double-strand DNA. Replacing the maize mitochondrial 2.3 kb linear plasmid by linearized pBluescript, a bacterial vector (3.0 kb), or by a 2.1 kb linear DNA fragment corresponding to the coding sequence of the Arabidopsis thaliana threonyl-tRNA synthetase cDNA (previously cloned in our laboratory; Souciet et al., 1999), showed that mitochondrial DNA import is not specific either for plant plasmid sequences or for plasmid sequences at all (Figure 2B). Assays run with linearized pBluescript containing the 2.3 kb maize plasmid sequence as an insert (final size 5.3 kb) indicated that the efficiency of mitochondrial incorporation seems to decrease when increasing the length of the substrate DNA (Figure 2B). Finally, labeled circular pBluescript was poorly recovered in the mitochondrial fraction following DNA import assays. As a whole, it appears that DNA uptake into plant mitochondria does not depend on the sequence content, but is most efficient for linear fragments up to a few kilobase pairs.
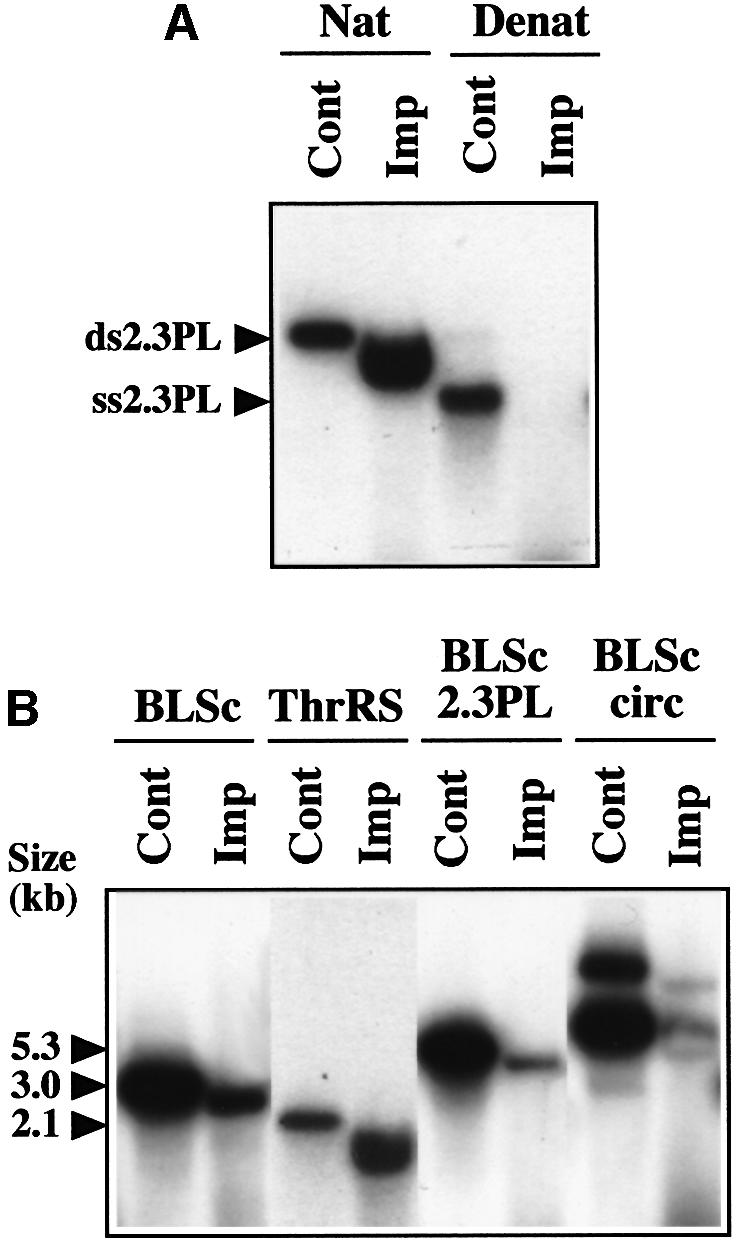
Fig. 2. Mitochondrial uptake of different DNA substrates. (A) Single-strand DNA is not taken up by plant mitochondria. The labeled linear maize 2.3 kb plasmid was incubated in its native form (Nat) or heat- denatured form (Denat) in the presence of potato mitochondria (Imp). Incubation time for uptake was 30 min. Aliquots of the initial native or denatured DNA probe were also loaded on the gel (Cont), showing the migration of the double-strand (ds2.3PL) or single-strand (ss2.3PL) plasmid. The migration shift of the incorporated DNA versus the initial probe was due to the presence of the bulk of the mitochondrial nucleic acids, as shown by mixing a probe aliquot with a nucleic acid sample obtained from an import assay run without substrate. (B) DNA uptake is not sequence specific. Labeled linear DNA corresponding to the pBluescript vector (BLSc), the A.thaliana threonyl-tRNA synthetase (ThrRS), the maize 2.3 kb plasmid cloned in pBluescript (BLSc 2.3PL), as well as labeled circular pBluescript DNA (BLSc circ), were incubated in the presence of potato mitochondria (Imp). Incubation time for uptake was 30 min. Aliquots of the initial DNA probes were also loaded on the gel (Cont). The size and migration of the probes are indicated.
The imported DNA is transcribed and used as a template for DNA synthesis in mitochondria
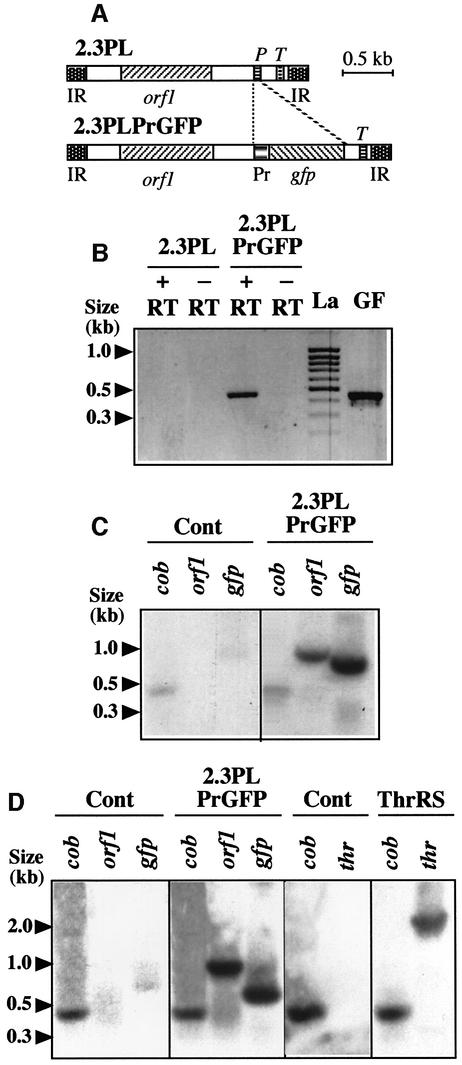
The broad specificity of mitochondrial DNA incorporation allowed the development of expression tests, both to add more information to the body of evidence supporting the existence of the DNA import process and to establish whether the experimental system can be suitable for molecular analyses. For this purpose, DNA construct pCK/GFP/PRmt was prepared (for details, see the Materials and methods) as a basis to yield, by PCR amplification, a 2.3 kb maize mitochondrial plasmid containing the GFP gene from the jellyfish Aequoria victoria as a reporter unrelated to plant mitochondria. The GFP gene replaced the tRNAPro gene present in the wild-type 2.3 kb maize plasmid (Leon et al., 1989). It was placed under the control of the previously characterized promoter sequence of the potato mitochondrial 18S ribosomal RNA gene (Giese et al., 1996).
Unlabeled linear 2.3 kb plasmid containing the GFP gene under the control of the plant mitochondrial promoter sequence (Figure 3A) was incubated with isolated S.tuberosum mitochondria in standard DNA import conditions. Following import and DNase digestion, mitochondria were further incubated for different times in a transcription medium designed to keep the organelles intact (Farré and Araya, 2001). Mitochondrial RNA was finally prepared and used for RT–PCR amplification with primers derived from the GFP sequence. A specific reverse transcriptase-dependent product of the appropriate size was amplified with the RNA extracted from mitochondria pre-incubated in the presence of the 2.3 kb plasmid containing the GFP gene and the mitochondrial promoter (Figure 3B). No RT–PCR product was detected when the wild-type 2.3 kb maize plasmid was used during the import step preceding transcription (Figure 3B). This implied that the GFP-containing 2.3 kb plasmid was imported and the reporter gene transcribed inside mitochondria. For further proof, [α-32P]UTP was substituted for UTP during the transcription step following import, so as to yield labeled transcripts which were hybridized to Southern blots carrying GFP-encoding DNA. The mitochondrial cob gene served as an internal control. Confirming the RT–PCR results, a strong hybridization signal was obtained with the labeled RNA extracted from mitochondria pre-incubated in the presence of the 2.3 kb plasmid containing the GFP gene (Figure 3C), whereas no signal was observed when the import step preceding transcription was run without exogenous DNA (Figure 3C). Interestingly, a clear signal was also detected when the labeled transcripts extracted from mitochondria pre-incubated in the presence of the 2.3 kb plasmid/GFP construct were hybridized against the DNA corresponding to the 885 nucleotide (nt) ORF1 present on this plasmid (Leon et al., 1992), whereas RNAs from the control mitochondria still showed no hybridization (Figure 3C). Hence, two distinct coding sequences with independent promoters could be expressed in mitochondria upon incorporation of exogenous DNA, bringing further evidence for the DNA import process and for the capability of isolated mitochondria to engage transcription of the introduced DNA.
Fig. 3. The incorporated DNA is transcribed and is a template for DNA synthesis in plant mitochondria. (A) Organization of the construct used as an import substrate for transcription and DNA synthesis studies. The tRNAPro gene present in the wild-type 2.3 kb maize plasmid (2.3PL) was replaced by the GFP gene (gfp) driven by the promoter of the potato mitochondrial 18S ribosomal RNA gene (Pr) in plasmid 2.3PLPrGFP. (B) GFP gene transcription in plant mitochondria as shown by RT–PCR. Potato mitochondria were incubated with PCR-amplified unlabeled 2.3PL or 2.3PLPrGFP DNA. Following DNA uptake, transcription was run for 2 h. Mitochondrial nucleic acids were isolated, treated with DNase and used for RT–PCR (+RT) with primers specific for the gfp sequence; amplification controls were run in identical conditions only omitting the reverse transcriptase enzyme in the corresponding medium (–RT). The expected DNA fragment (nucleotides 98–539 of the GFP gene) was also PCR-amplified directly from the initial construct (GF). (C) ORF1 and GFP gene transcription in plant mitochondria as shown by Southern hybridization. Potato mitochondria were incubated in the absence (Cont) or presence (2.3PLPrGFP) of PCR-amplified unlabeled 2.3PLPrGFP DNA. Following DNA uptake, transcription was run for 3 h in the presence of [α-32P]UTP. Mitochondrial nucleic acids were isolated from the different samples and identical amounts of radioactive transcripts were hybridized to Southern blots carrying DNA probes for the potato mitochondrial cob gene (cob), the ORF1 of the maize 2.3 kb mitochondrial plasmid (orf1) and the GFP gene (gfp). (D) DNA synthesis in plant mitochondria as shown by Southern hybridization. Potato mitochondria were incubated in the absence of exogenous DNA (Cont) or in the presence of PCR-amplified unlabeled 2.3PLPrGFP DNA (2.3PLPrGFP) or A.thaliana threonyl-tRNA synthetase encoding DNA (ThrRS). Following uptake, DNA synthesis was run for 1.5 h in the presence of [α-32P]dCTP. Mitochondrial nucleic acids were isolated from the different samples and identical amounts of radioactive DNA were hybridized to Southern blots carrying the (cob), (orf1) and (gfp) probes as in panel (C), or a threonyl-tRNA synthetase probe (thr). Fragment sizes determined with a DNA ladder [(La) in panel (B)] are indicated.
Similar experiments were developed to test whether the mitochondrially incorporated 2.3 kb plasmid/GFP could be a template for DNA synthesis. Following import and DNase digestion, mitochondria were further incubated in essentially the same medium as for transcription, but [α-32P]dCTP and unlabeled dNTPs were substituted for [α-32P]UTP and unlabeled NTPs. The radioactive DNA generated was hybridized to Southern blots carrying cob, maize plasmid orf1 and gfp sequences. A strong hybridization to the orf1 and gfp probes was obtained with the labeled DNA extracted from mitochondria pre-incubated in the presence of the 2.3 kb plasmid containing the GFP gene, whereas no hybridization to these probes was observed when the import step was run without added DNA (Figure 3D). However, the generation of radioactive DNA was likely to reflect some repair processes, rather than replication of the 2.3 kb plasmid/GFP construct, because the coding sequence of the A.thaliana threonyl-tRNA synthetase (see above), a DNA fragment presumably deprived of replication information, was also a good template for DNA synthesis upon uptake into mitochondria (Figure 3D).
Mitochondrial import of DNA is an active process
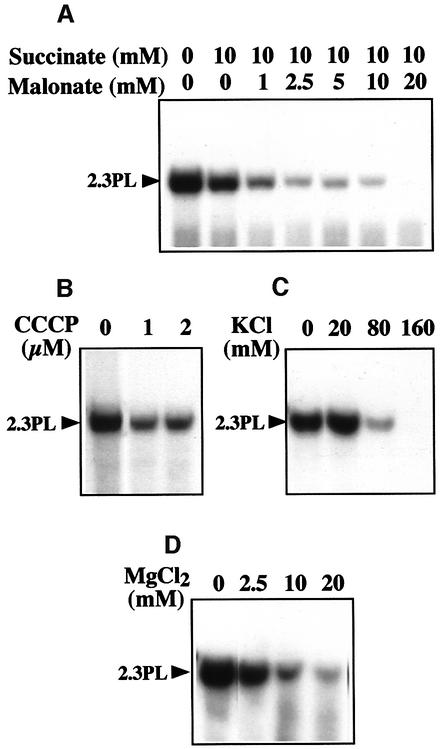
To gather some hints about the mechanisms allowing DNA import into plant mitochondria, further functional tests were carried out. Malonate inhibits, in a competitive manner, the oxidation of succinate via complex II, leading to a decrease in electron transport. Increasing amounts of malonate thus gradually decrease the ΔΨ membrane potential (Millar et al., 1995). Titrating out the mitochondrial ΔΨ potential by increasing concentrations of malonate in the presence of succinate, progressively inhibited the incorporation of the labeled 2.3 kb maize plasmid into plant mitochondria (Figure 4A). The protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) allows proton diffusion across the inner membrane and collapses the proton-motive force (ΔpH, ΔΨ). CCCP also impaired the uptake of the 2.3 kb plasmid (Figure 4B). DNA import therefore appears to be an active process that requires both components of the mitochondrial inner membrane electrochemical potential. The inhibition of DNA uptake in the presence of increasing KCl concentrations in the import medium (Figure 4C) can tentatively be interpreted in terms of ΔΨ decrease or pH shift (Garlid and Paucek, 2001). Mg2+-mediated inhibition (Figure 4D) is likely to reflect an interaction with the structure of the DNA, as the same effect was obtained with spermidine.
Fig. 4. Mitochondrial uptake of DNA is an active process. (A) DNA uptake requires the ΔΨ potential. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of a constant concentration of succinate and increasing concentrations of malonate. (B) DNA uptake requires the proton motive force. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of increasing concentrations of the protonophore CCCP. (C and D) DNA uptake is inhibited by KCl and MgCl2. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of increasing concentrations of KCl (C) or MgCl2 (D). Migration of the incorporated plasmid (2.3PL) is indicated.
Mitochondrial import of DNA potentially involves the VDAC and the ANT
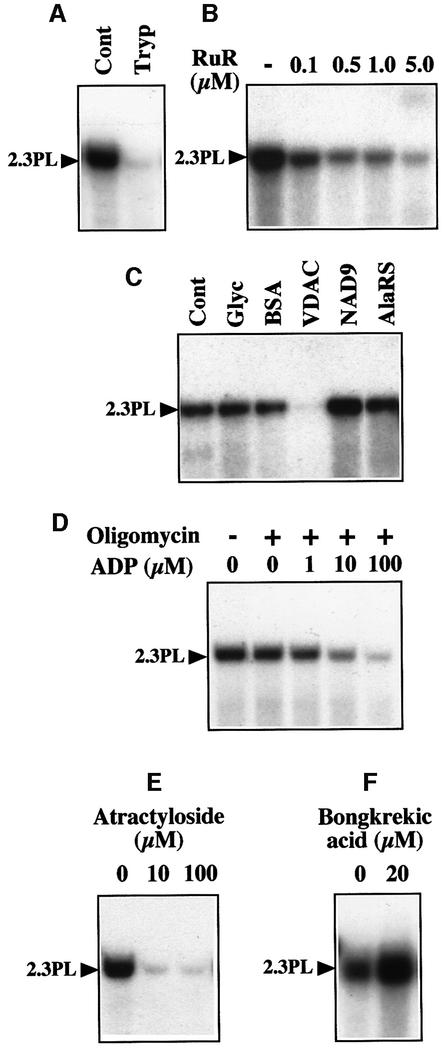
Trypsin pretreatment of the mitochondria prior to the import assay completely abolished DNA uptake (Figure 5A), implying that surface accessible outer-membrane proteins are involved. The two main constitutive pores known in the mitochondrial outer membrane are the translocase of the outer-membrane (TOM) complex and the VDAC (also called porin). The TOM complex is responsible for the transport of nuclear-encoded preproteins (Werhahn et al., 2001) and thus was not an obvious candidate for DNA translocation. We chose to test a possible involvement of the VDAC in mitochondrial DNA uptake. Ruthenium Red was shown previously to induce closure of the VDAC from rat liver mitochondria (Gincel et al., 2001). Addition of increasing concentrations of Ruthenium Red progressively inhibited DNA uptake by plant mitochondria (Figure 5B). Furthermore, incorporation of the 2.3 kb plasmid was abolished when mitochondria were pretreated with an antiserum against potato mitochondrial VDAC (Heins et al., 1994) prior to the import assay (Figure 5C), whereas no inhibition was observed with mock-treated mitochondria and with mitochondria pretreated with an antiserum against the wheat (Triticum aestivum) subunit 9 (NAD9) of the inner-membrane complex I (NADH dehydrogenase) (Lamattina et al., 1993) or against the A.thaliana cytosolic/mitochondrial alanyl-tRNA synthetase (Mireau et al., 1996). The DNA probe was stable when incubated with the antibodies in the absence of mitochondria, showing that there was no DNase activity in the antisera and implying that the lack of incorporation into mitochondria pretreated with the VDAC antiserum was not due to hydrolysis of the probe during the import step. As the VDAC controls the outer membrane permeability to ADP and ATP (Rostovtseva and Colombini, 1997), ADP-stimulated respiration rates in the presence of succinate were tentatively measured in parallel with DNA uptake to assess antibody inhibition. A 20–30% decrease in the respiratory control ratio was observed with mitochondria pretreated with the VDAC antiserum, as compared with mock-treated mitochondria or mitochondria pretreated with the NAD9 antiserum, which was taken as an indication for impairment of ADP import due to the binding of antibodies to the VDAC.
Fig. 5. Mitochondrial uptake of DNA is likely to involve VDAC/ANT complexes. (A) DNA uptake requires surface-accessible protein(s). The labeled maize 2.3 kb plasmid was incubated in the presence of mock-pretreated (Cont) or trypsin-pretreated (Tryp) potato mitochondria. (B) DNA uptake is inhibited by Ruthenium Red. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of increasing concentrations of Ruthenium Red (RuR). (C) DNA uptake is inhibited by antibodies against the VDAC. Incorporation of labeled maize 2.3 kb plasmid was tested in the presence of mock-treated mitochondria (Cont), mitochondria pretreated with BSA, or mitochondria pretreated with a polyclonal antiserum against the VDAC, NAD9 or alanyl-tRNA synthetase (AlaRS). As the antisera used contained 50% (v/v) glycerol, a control assay was also run in the presence of the relevant amount of glycerol (Glyc). (D) DNA uptake is inhibited by ADP. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of increasing concentrations of ADP and a constant concentration (10 µg/ml) of oligomycin (+). (E and F) DNA uptake is inhibited by atractyloside, but enhanced by bongkrekic acid. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of atractyloside (E) or bongkrekic acid (F). Migration of the incorporated plasmid (2.3PL) is indicated.
The ANT is the major channel in the mitochondrial inner membrane. It imports ADP and exports ATP. In the presence of oligomycin, an inhibitor of the ATP synthase, ADP cannot be converted into ATP and therefore binds to the ANT, but does not translocate into the mitochondrial matrix, thereby blocking the channel. In such conditions, DNA uptake into plant mitochondria was clearly inhibited, even at low ADP concentrations (Figure 5D). Similarly, atractyloside, an ANT inhibitor acting on the intermembrane space side, abolished DNA uptake already at a 10 µM concentration (Figure 5E). On the contrary, bongkrekic acid, a matrix-side ANT inhibitor, stimulated DNA import into plant mitochondria (Figure 5F). In control oxygen electrode assays run in parallel, bongkrekic acid completely inhibited ADP-stimulated respiration depending on the ANT activity (‘state III’ respiration) in potato mitochondria.
Mitochondrial import of DNA does not occur through permeability transition
The above results strongly suggest that plant mitochondrial DNA uptake involves the VDAC and the ANT, which are actually the core components of the mitochondrial PTPC as defined in animal cells (Zamzami and Kroemer, 2001). In current models of the animal mitochondrial PTPC, hexokinase is associated to the VDAC on the cytosolic side. Hexokinase binding to mitochondria has also been reported in plants (Wilson, 1997). Addition of yeast hexokinase in DNA import assays with plant mitochondria led to a strong inhibition of the incorporation. These observations altogether, raised the question as to whether DNA import is related to MPT.
Permeability transition leads to mitochondrial swelling and ultimately to the rupture of the outer membrane (Zamzami and Kroemer, 2001). Neither incubation in the import medium nor addition of DNA triggered any swelling of the mitochondria, as tested by absorption kinetics at 546 nm, indicating that there was no permeabilization of the membranes. DNA import was inhibited by atractyloside and stimulated by bongkrekic acid (see above), which is just the opposite of the effects these effectors have on MPT. Cyclosporin A had no influence on the incorporation of the 2.3 kb plasmid into plant mitochondria, although it is a strong inhibitor of the animal PTPC. MPT is mainly triggered by high Ca2+ and oxidative stress, including thiol oxidation (Vieira et al., 2000; Martinou and Green, 2001). Our experimental data show that DNA uptake by plant mitochondria does not require addition of Ca2+. When increasing amounts of CaCl2 were added to the import assays, the process was gradually inhibited (Figure 6A). Similarly, oxidative stress provided by hydroxyl radical (HO·) generation in the presence of H2O2 and FeSO4 (Fenton’s reagent) strongly impaired mitochondrial DNA uptake (Figure 6B). The same effect was observed upon thiol oxidation mediated by diamide (Figure 6C). As a whole, plant mitochondrial DNA import does not seem to be related to known mitochondrial membrane permeabilization processes.
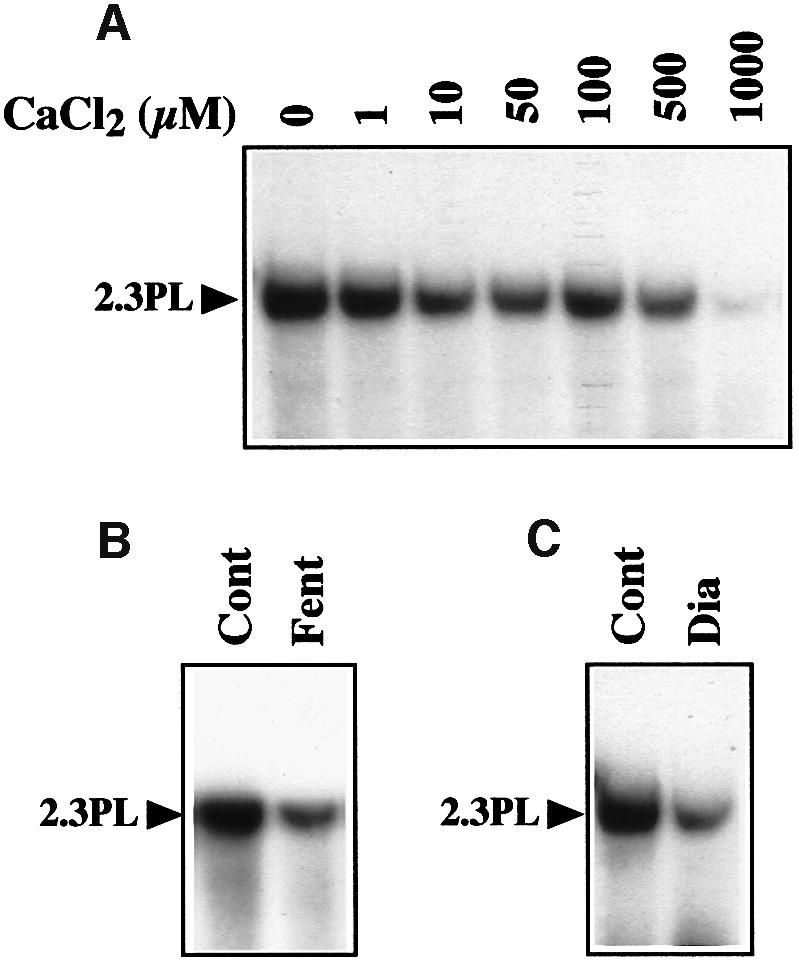
Fig. 6. Uptake of DNA is not related to MPT. (A) DNA uptake is inhibited by CaCl2. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the presence of increasing concentrations of CaCl2. (B) DNA uptake is inhibited in oxidative conditions. Incorporation of labeled maize 2.3 kb plasmid was tested with mock-treated mitochondria (Cont) and with mitochondria pretreated with H2O2 and FeSO4 (Fenton’s reagent, Fent). (C) DNA uptake is inhibited by thiol oxidation. Mitochondrial incorporation of labeled maize 2.3 kb plasmid was tested in the absence (Cont) and presence (Dia) of 3 mM diamide. Migration of the incorporated plasmid (2.3PL) is indicated.
Discussion
Transport of DNA or RNA across membrane barriers is a key challenge in a number of physiological and pathological cell processes. The involvement of proteic channels in these mechanisms receives growing evidence both in prokaryotes and in eukaryotes. Bacterial transformation, conjugation and phage infection recrute constitutive channels or trigger formation of specific pores for DNA transfer (Dreiseikelmann, 1994; Dubnau, 1999). The Escherichia coli outer membrane protein FhuA is among the best documented examples and was shown to have a dual function of transporter and phage DNA translocation channel (Bonhivers et al., 1998). Bacillus subtilis high conductance ion channels confer DNA permeability to planar bilayers in a reconstituted system (Szabo et al., 1997). DNA polymers can be electrophoretically driven through 2 nm nanopores made by a bacterial exotoxin with membrane-damaging function, the Staphylococcus aureus α-hemolysin, in a lipid bilayer (Meller et al., 2000). Translocation of the transferred DNA through the plasma membrane during Agrobacterium tumefaciens transformation of plant cells occurs through a channel formed by the bacterial virulence protein VirE2, which also participates in coating of the T-DNA during its transfer to the nucleus (Dumas et al., 2001). The present work brings strong lines of evidence for the existence of a novel organellar transport process, i.e. active import of large-size double-strand DNA into plant mitochondria. The mechanisms underlying this process may rely on alternate functional capabilities of the metabolite/nucleotide translocators VDAC and ANT. Our data suggest that VDAC/ANT-based multifunctional protein complexes similar to the PTPC of animal mitochondria (Zamzami and Kroemer, 2001) are formed at contact sites between the outer and inner membranes in plant mitochondria and are able, in appropriate conditions, to promote active translocation of DNA.
Considering its pore size capacity and its properties in reconstituted systems, the VDAC (the most abundant protein in the mitochondrial outer membrane in all eukaryotes) is a quite plausible candidate for such an alternate function. It mediates the exchange of metabolites, and especially of nucleotides (Colombini et al., 1996). VDAC pores can reach a diameter of 3–4 nm (Mannella et al., 1992), which is compatible with the passage of DNA molecules according to estimations of the double helix minimal diameter. Moreover, purified bovine VDAC allows electrophoretically driven double-strand DNA translocation across a planar membrane (Szabo et al., 1998). Although the way the ANT (the major channel in the mitochondrial inner membrane) could function in translocating DNA seems less obvious to understand, such a possibility is also supported by existing data. The regular role of the ANT is to translocate nucleotides, as it ensures exchange of extra-mitochondrial ADP on intra-mitochondrial ATP. A sequence motif similar to a DNA recognition element of the estrogen receptor was detected in the ANT, as well as in the uncoupling protein (UCP; a nucleotide regulated proton uniporter) of animal mitochondria (Bouillaud et al., 1994). This motif is mostly conserved in the plant ANT (Winning et al., 1991; Schuster et al., 1993), in particular, the three key amino acids. Permeability analyses of bilayer and proteoliposome systems showed that the ANT may also form a pore (Vieira et al., 2000). However, given its sensitivity to an accumulation of external ADP, to atractyloside and to bongkrekic acid, DNA uptake seems to remain related to the regular conformation properties of the ANT involved in the ADP/ATP exchange. The ANT binding center can be open to the external face (‘c’ for ‘cytosolic’ conformation) or to the internal face (‘m’ for ‘matrix’ conformation) of the inner membrane (Klingenberg and Nelson, 1994). It seems that arresting the ANT ‘c’ conformation with atractyloside or ADP blocks DNA transport, whereas ‘m’ conformation arrest with bongkrekic acid shows some enhancing effect.
Mitochondrial permeability currently receives much attention in the mammalian field, mainly due to its role in apoptosis and cancer (Martinou and Green, 2001). MPT is the most documented among the models that have been developed (Zamzami and Kroemer, 2001). MPT was initially defined as a Ca2+-dependent, non-specific permeabilization of the inner membrane of animal mitochondria, inhibited by submicromolar concentrations of cyclosporin A. It is driven by the permeability transition pore involving the VDAC and the ANT as the major proteins. The occurrence of MPT has recently been characterized for plant (i.e. potato) mitochondria (Arpagaus et al., 2002). The MPT induced in isolated potato mitochondria can be insensitive to cyclosporin A (Fortes et al., 2001). The data we collected show that, although it is likely to be promoted by the VDAC and the ANT, plant mitochondrial DNA uptake does not seem to be related to MPT or non-specific permeabilization processes. Rather, the characteristics emerging from our studies (membrane potential requirement, sensitivity to a number of mitochondrial effectors, involvement of representative channel proteins) suggest that DNA import into plant mitochondria is a physiological phenomenon. Although its occurrence in vivo remains to be established, it is tempting to speculate that this process has some significance in genetic fluxes or fulfills some yet unknown function. As stressed in the Introduction, the expansion of plant mitochondrial genomes through integration of extra sequences versus the minimal mitochondrial DNA of mammals or Plasmodium is striking in the light of the unique evolutionary origin of mitochondria in all eukaryotes (Gray et al., 2001). DNA uptake can also be regarded as a potential way of genetic information exchange between mitochondria, contributing to the genetic stability and the spread of useful mutations in the plant mitochondrial population, as well as to the distribution of mitochondrial plasmids. The presence of viral sequences in the organellar genomes and plasmids raises the idea that plant mitochondria might contribute to sink processes for invading pathogen genetic material. Plants often restrict pathogen growth through rapid programmed cell death (PCD) in and around the infection site. Mitochondria are involved in the control of PCD, but many of the cell-death regulators that have been characterized in animal systems seem to be absent in plants (Lam et al., 2001). Uptake of pathogen DNA might be a candidate as one of the stress signals promoting mitochondrial damage, thereby initiating cell death.
Earlier attempts at introducing DNA sequences into mitochondria relied on chemically synthesized chimeras between mitochondrial preproteins or targeting peptides and oligonucleotides. A short piece of single-strand DNA covalently linked to a mitochondrial precursor protein could be imported into yeast mitochondria (Vestweber and Schatz, 1989). As a strategy towards gene therapy of diseases due to mutations in the mitochondrial DNA, a chimera composed of a targeting peptide, and a 322 bp double-strand passenger DNA was successfully incorporated into rat liver mitochondria (Seibel et al., 1995). Peptide-nucleic acid oligomers, synthetic DNA analogs tentatively designed for therapeutic purposes, were imported into both isolated mitochondria and mitochondria within human cultured cells when conjugated to a lipophilic phosphonium cation (Muratovska et al., 2001). Large-size DNA was also forced into mammalian (Collombet et al., 1997), trypanosomatid (Estevez et al., 1999) and plant (Farré and Araya, 2001) mitochondria by electroporation. Further studies on the DNA uptake mechanism characterized in this work could result in the tentative design of plant mitochondrial transformation strategies based on natural DNA import, allowing new developments in plant research and biotechnology.
Materials and methods
Isolation and functional tests of plant mitochondria
Mitochondria were isolated from potato (S.tuberosum) tubers mostly according to Neuburger et al. (1982). Tubers were homogenized in a juice extractor and the suspension mixed with one half volume of three times concentrated extraction buffer [90 mM sodium pyrophosphate, 900 mM sucrose, 6 mM EDTA, 0.9% (w/v) BSA, 2.4% (w/v) polyvinylpyrrolidone 25, 9 mM cysteine, 15 mM glycine, 6 mM β-mercaptoethanol, pH 7.5]. Following filtration through a nylon net (100 µm mesh), crude mitochondria were recovered by two cycles of low speed (1500 g) and high speed (16 000 g) centrifugation with intermediate resuspension in washing buffer [10 mM potassium phosphate, 300 mM sucrose, 1 mM EDTA, 0.1% (w/v) BSA, 5 mM glycine, pH 7.5]. The resuspended second pellet was loaded on a 30% (v/v) Percoll solution in washing buffer and centrifuged for 1 h at 40 000 g. Mitochondria were recovered in the bottom third of the Percoll gradient established during centrifugation and washed twice.
Outer membrane integrity was estimated by testing the ability of the isolated mitochondria to reduce exogenously added cytochrome c, as reflected by an increase of the absorption at 550 nm (Douce et al., 1972). For control assays, complete breakage of the outer membrane was obtained upon a 30 s osmotic shock in 5 mM potassium phosphate (pH 7.5). The mitochondrial respiratory control ratio, i.e. the ratio between the respiration rate (measured as oxygen consumption) of the mitochondria upon addition of 100 µM ADP (‘active state’ or state III) and the respiration rate in the absence of added ADP or when the added ADP was entirely converted to ATP (‘resting state’ or state IV), was determined with an oxygraph chamber using 10 mM succinate as a respiration substrate (Douce, 1985). Organelle swelling was analyzed by following the absorption of the suspension at 546 nm during 30 min. Control kinetics were run in 20 mM Tris–HCl pH 7.5 and 100 mM NH4NO3 (Panov et al., 1980).
Labeled DNA substrates for mitochondrial import
The main substrate DNA used for mitochondrial import assays was the 2.3 kb linear plasmid from maize (Z.mays; Leon et al., 1989; DDBJ/EMBL/GeneBank accession No. X13704). Mitochondria were isolated from 10-day-old, dark-grown maize seedlings following the same procedures as for potato tubers. Total DNA extracted from maize mitochondria served as a template for PCR amplification of the 2.3 kb linear plasmid using a primer corresponding to the 5′ end of the inverted repeat present at both termini. To obtain the full-length radioactive linear plasmid, 50 ng of unlabeled PCR product and the same primer were used for a single PCR cycle in which a 10 min elongation in an unlabeled dCTP-deprived reaction medium containing 100 µCi of [α-32P]dCTP (3000 Ci/mmole) per 50 µl was followed by the addition of 0.2 mM unlabeled dCTP and a further 5 min elongation. The same PCR procedures were developed to label ApaI-linearized pBluescript KS(+) plasmid (Stratagene) and to prepare a 2.1 kb labeled DNA fragment corresponding to the coding sequence of A.thaliana threonyl-tRNA synthetase (Souciet et al., 1999; accession No. Y14329) deprived of translation initiation and termination sequences. To obtain labeled circular DNA, the 2.3 kb maize plasmid amplified as above was cloned into the pBluescript KS (+) vector. Circular plasmid DNA was subsequently extracted from recombinant bacteria cultured in the presence of 100 µCi [32P]orthophosphate (285 Ci/mg) per ml of medium. Linear labeled pBluescript and pBluescript containing the 2.3 kb maize plasmid as an insert were also obtained upon EcoRI digestion of the corresponding labeled circular DNAs.
Gene construct for mitochondrial DNA import and transcription
The 1–1806 nt sequence of the 2.3 kb maize linear plasmid was amplified by PCR from the complete clone in pBluescript KS (+) and cloned into the XhoI and EcoRI sites of the pUC and pRTL2-derived plasmid pCK-GFP3 (Menand et al., 1998) upstream of the tobacco etch virus (TEV) translation leader (TL) and the GFP gene from the jellyfish A.victoria. The 1881–2312 nt sequence of the 2.3 kb maize linear plasmid was amplified by PCR and cloned into the BamHI and XbaI sites downstream of the GFP gene in the pCK-GFP3 plasmid already containing the 1–1806 maize sequence, yielding plasmid pCK/GFP/MLP in which the TL/GFP sequences were replacing the tRNAPro gene within the 2.3 kb maize plasmid. To exchange the TEV TL with plant mitochondrial sequences, the 140 nt region containing the promoter sequences upstream of the mature 5′ end of the 18S ribosomal RNA (Giese et al., 1996; accession No. X98800) was amplified by PCR from potato (S.tuberosum) mitochondrial DNA. In addition to the sequence complementary to the 3′ end of the potato mitochondrial 18S rRNA promoter, the reverse primer contained 18 nt complementary to the region immediately upstream of the initiation codon of the ATP1 subunit in the tobacco (Nicotiana tabacum) mitochondrial genome (Bergman et al., 2000; accession No. AF121903). The resulting PCR product was cloned into the EcoRI and NcoI sites of plasmid pCK/GFP/MLP after removal of the TEV TL sequence with the same restriction endonucleases, yielding construct pCK/GFP/PRmt. Finally, the unlabeled DNA substrate actually used for mitochondrial import was PCR-amplified from construct pCK/GFP/PRmt with the primer corresponding to the 5′ end of the inverted repeat present at both termini of the maize 2.3 kb plasmid.
Mitochondrial import assays
Standard mitochondrial import of DNA was carried out in 40 mM potassium phosphate and 0.4 M sucrose pH 7.0 (import buffer). The samples (200 µl) containing 1–5 ng of 32P-labeled DNA (100 000–200 000 c.p.m.) and an amount of purified mitochondria corresponding to 300 µg of proteins were incubated at 25°C for 30 min under mild shaking. Following addition of 200 µg of DNase I and 10 mM MgCl2, the incubation was continued for 20 min in the same conditions. Mitochondria were subsequently washed three times by resuspension in 1 ml of 10 mM potassium phosphate, 300 mM sucrose, 10 mM EDTA, 10 mM EGTA, 0.1% (w/v) BSA, 5 mM glycine pH 7.5, and centrifuged for 5 min at 10 000 g. In some experiments, the organelles were re-isolated on a Percoll gradient. The final pellets were extracted with one volume of 10 mM Tris–HCl, 1 mM EDTA, 1% (w/v) SDS pH 7.5 and one volume of phenol. The nucleic acids recovered in the aqueous phase were ethanol-precipitated, fractionated by electrophoresis on a 1% (w/v) agarose gel and transferred onto a nylon membrane (Hybond N+, Amersham Biosciences) for autoradiography.
To break the outer membrane by osmotic shock after DNA import, mitochondria were centrifuged (5 min at 10 000 g), resuspended in 1 ml of 5 mM potassium phosphate pH 7.5, incubated on ice for 1 min and centrifuged again before resuspension in import buffer. For mock treatment, washing buffer (see isolation of mitochondria) was used instead of 5 mM potassium phosphate. To generate oxidative stress, mitochondria were pre-incubated for 30 min at room temperature in import buffer containing 50 µM H2O2 and 50 µM FeSO4, washed once with import buffer and used in DNA uptake assays. To test the implication of surface-accessible proteins, mitochondria were pretreated with 50 µg/ml trypsin for 10 min on ice. Following dilution with washing buffer, addition of 1 mg/ml soybean trypsin inhibitor and further incubation for 12 min on ice, organelles were re-isolated and washed twice in the presence of 1 mg/ml soybean trypsin inhibitor prior to use in import assays. Mock-treated mitochondria went through the same steps omitting the addition of trypsin. For antibody inhibition, mitochondria (60 µg protein) were pre-incubated for 30 min on ice and 30 min at room temperature in 100 µl washing buffer completed with 5 µl α2-macroglobulin (2.5 µg/µl) and 20 µl polyclonal antiserum (150–300 µg protein) containing 50% (v/v) glycerol. The organelles were subsequently centrifuged and resuspended in import buffer. Mock treatment was without or with addition of 20 µl of 50% (v/v) glycerol.
Mitochondrial transcription of imported DNA sequences and DNA synthesis
To analyze the expression of the imported DNA, mitochondrial import was carried out with 200 ng of unlabeled DNA amplified by PCR (see above). After incubation for 30 min at 25°C, mitochondria were submitted to DNase digestion as above, washed twice with 1 ml of import buffer and resuspended in 200 µl transcription medium according to Farré and Araya (2001; 330 mM sucrose, 90 mM KCl, 10 mM MgCl2, 12 mM tricine, 5 mM KH2PO4, 1.2 mM EGTA, 1 mM GTP, 2 mM DTT, 2 mM ADP, 10 mM sodium succinate, 0.15 mM CTP, 0.15 mM UTP pH 7.2). Upon further incubation for 1–16 h at 25°C under mild shaking, mitochondria were pelleted and extracted with one volume of 10 mM Tris–HCl, 1 mM EDTA, 1% (w/v) SDS pH 7.5 and one volume of phenol. The nucleic acids recovered in the aqueous phase were ethanol-precipitated and submitted to RNase-free DNase I digestion twice for 1 h in the presence of 2 U of enzyme (Promega), with an intermediate ethanol precipitation. After phenol extraction and reprecipitation, the RNA was used as a template for RT–PCR.
Alternatively, unlabeled UTP was replaced by [α-32P]UTP (800 Ci/mmole) in the above transcription medium (80 µCi/200 µl medium) to generate radioactive transcripts. Following DNA import and transcription, nucleic acids were extracted as above and unincorporated [α-32P]UTP was eliminated by gel-filtration through a Sephadex G-50 spin column. Equal amounts of radioactivity were taken for hybridization to Southern blots carrying 1 µg per lane of PCR-generated DNA corresponding to the potato mitochondrial apocytochrome b (cob gene), the ORF1 of the 2.3 kb maize plasmid and the GFP. The cob probe was amplified from potato mitochondrial DNA using primers originally designed according to the A.thaliana cob sequence (accession No. Y08501; nucleotides 60235–61416). Hybridizations were at 42°C in 5× SSPE, 50% (v/v) deionized formamide, 5× Denhardt solution, 1% (w/v) SDS and 20 µg/ml salmon sperm DNA.
The same procedures were developed to analyze DNA synthesis after mitochondrial uptake, substituting dATP, dGTP, dTTP (50 µM each), dCTP (2 µM) and [α-32P]dCTP (3000 Ci/mmole, 50 µCi/200 µl medium) for NTPs in the above transcription medium.
Acknowledgments
Acknowledgements
We thank L.Delage, J.L.Evrard, D.Gagliardi, L.Maréchal-Drouard, A.Panov, V.Tarasenko and J.H.Weil for helpful discussions and suggestions during the course of these studies. We are indebted to V.Podsosonny for his interest and insight. We are grateful to K.Newton for the gift of a clone containing the 2.3 kb maize plasmid, to H.P.Braun for the gift of an antiserum against potato mitochondrial VDAC and to G.Souciet for the ThrRS cDNA clone. M.K. was funded by a fellowship from the French Government. This work was supported by a cooperation agreement between the French Centre National de la Recherche Scientifique (CNRS) and the Russian Academy of Sciences (RAS).
References
- Arpagaus S., Rawyler,A.J. and Braendle,R.A. (2002) Occurrence and characteristics of the mitochondrial permeability transition in plants. J. Biol. Chem., 277, 1780–1787. [DOI] [PubMed] [Google Scholar]
- Bergman P., Edqvist,J., Farbos,I. and Glimelius,K. (2000) Male-sterile tobacco displays abnormal mitochondrial atp1 transcript accumulation and reduced floral ATP/ADP ratio. Plant Mol. Biol., 42, 531–544. [DOI] [PubMed] [Google Scholar]
- Bonhivers M., Plancon,L., Ghazi,A., Boulanger,P., le Maire,M., Lambert,O., Rigaud,J.L. and Letellier,L. (1998) FhuA, an Escherichia coli outer membrane protein with a dual function of transporter and channel which mediates the transport of phage DNA. Biochimie, 80, 363–369. [DOI] [PubMed] [Google Scholar]
- Bouillaud F., Arechaga,I., Petit,P.X., Raimbault,S., Levi-Meyrueis,C., Casteilla,L., Laurent,M., Rial,E. and Ricquier,D. (1994) A sequence related to a DNA recognition element is essential for the inhibition by nucleotides of proton transport through the mitochondrial uncoupling protein. EMBO J., 13, 1990–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.G. and Zhang,M. (1995) Mitochondrial plasmids: DNA and RNA. In Levings,C.S.,III and Vasil,I.K. (eds), The Molecular Biology of Plant Mitochondria. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 61–91.
- Collombet J.M., Wheeler,V.C., Vogel,F. and Coutelle,C. (1997) Introduction of plasmid DNA into isolated mitochondria by electroporation. A novel approach toward gene correction for mitochondrial disorders. J. Biol. Chem., 272, 5342–5347. [DOI] [PubMed] [Google Scholar]
- Colombini M., Blachly-Dyson,E. and Forte,M. (1996) VDAC, a channel in the outer mitochondrial membrane. Ion Channels, 4, 169–202. [DOI] [PubMed] [Google Scholar]
- Douce R. (1985) The function of plant mitochondrial matrix. In Mitochondria in Higher Plants. Structure, Function and Biogenesis. American Society of Plant Physiologists Monograph Series, Academic Press, Orlando, FL, pp. 155–156.
- Douce R. and Neuburger,M. (1989) The uniqueness of plant mitochondria. Annu. Rev. Plant Physiol. Plant Mol. Biol., 40, 371–414. [Google Scholar]
- Douce R., Christensen,E.L. and Bonner,W.D.J. (1972) Preparation of intact plant mitochondria. Biochim. Biophys. Acta, 275, 148–160. [DOI] [PubMed] [Google Scholar]
- Dreiseikelmann B. (1994) Translocation of DNA across bacterial membranes. Microbiol. Rev., 58, 293–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. (1999) DNA uptake in bacteria. Annu. Rev. Microbiol., 53, 217–244. [DOI] [PubMed] [Google Scholar]
- Dumas F., Duckely,M., Pelczar,P., Van Gelder,P. and Hohn,B. (2001) An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc. Natl Acad. Sci. USA, 98, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez A.M., Thiemann,O.H., Alfonzo,J.D. and Simpson,L. (1999) T7 RNA polymerase-driven transcription in mitochondria of Leishmania tarentolae and Trypanosoma brucei. Mol. Biochem. Parasitol., 103, 251–259. [DOI] [PubMed] [Google Scholar]
- Farré J.C. and Araya,A. (2001) Gene expression in isolated plant mitochondria: high fidelity of transcription, splicing and editing of a transgene product in electroporated organelles. Nucleic Acids Res., 29, 2484–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes F., Castilho,R.F., Catisti,R., Carnieri,E.G. and Vercesi,A.E. (2001) Ca2+ induces a cyclosporin A-insensitive permeability transition pore in isolated potato tuber mitochondria mediated by reactive oxygen species. J. Bioenerg. Biomembr., 33, 43–51. [DOI] [PubMed] [Google Scholar]
- Garlid K.D. and Paucek,P. (2001) The mitochondrial potassium cycle. IUBMB Life, 52, 153–158. [DOI] [PubMed] [Google Scholar]
- Giese A., Thalheim,C., Brennicke,A. and Binder,S. (1996) Correlation of nonanucleotide motifs with transcript initiation of 18S rRNA genes in mitochondria of pea, potato and Arabidopsis. Mol. Gen. Genet., 252, 429–436. [DOI] [PubMed] [Google Scholar]
- Gincel D., Zaid,H. and Shoshan-Barmatz,V. (2001) Calcium binding and translocation by the voltage-dependent anion channel: a possible regulatory mechanism in mitochondrial function. Biochem. J., 358, 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover K.E., Spencer,D.F. and Gray,M.W. (2001) Identification and structural characterization of nucleus-encoded transfer RNAs imported into wheat mitochondria. J. Biol. Chem., 276, 639–648. [DOI] [PubMed] [Google Scholar]
- Gray M.W., Burger,G. and Lang,B.F. (2001) The origin and early evolution of mitochondria. Genome Biol., 2, REVIEWS1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins L., Mentzel,H., Schmid,A., Benz,R. and Schmitz,U.K. (1994) Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J. Biol. Chem., 269, 26402–26410. [PubMed] [Google Scholar]
- Klingenberg M. and Nelson,D.R. (1994) Structure-function relationships of the ADP/ATP carrier. Biochim. Biophys. Acta, 1187, 241–244. [DOI] [PubMed] [Google Scholar]
- Kubo T., Nishizawa,S., Sugawara,A., Itchoda,N., Estiati,A. and Mikami,T. (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res., 28, 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E., Kato,N. and Lawton,M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature, 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Lamattina L., Gonzalez,D., Gualberto,J. and Grienenberger,J.M. (1993) Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem., 217, 831–838. [DOI] [PubMed] [Google Scholar]
- Leon P., Walbot,V. and Bedinger,P. (1989) Molecular analysis of the linear 2.3 kb plasmid of maize mitochondria: apparent capture of tRNA genes. Nucleic Acids Res., 17, 4089–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon P., O’Brien-Vedder,C. and Walbot,V. (1992) Expression of ORF1 of the linear 2.3 kb plasmid of maize mitochondria: product localization and similarities to the 130 kDa protein encoded by the S2 episome. Curr. Genet., 22, 61–67. [DOI] [PubMed] [Google Scholar]
- Mannella C.A., Forte,M. and Colombini,M. (1992) Toward the molecular structure of the mitochondrial channel, VDAC. J. Bioenerg. Biomembr., 24, 7–19. [DOI] [PubMed] [Google Scholar]
- Maréchal-Drouard L., Weil,J.H. and Dietrich,A. (1993) Transfer RNAs and transfer RNA genes in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol., 44, 13–32. [Google Scholar]
- Marienfeld J., Unseld,M. and Brennicke,A. (1999) The mitochondrial genome of Arabidopsis is composed of both native and immigrant information. Trends Plant Sci., 4, 495–502. [DOI] [PubMed] [Google Scholar]
- Martinou J.C. and Green,D.R. (2001) Breaking the mitochondrial barrier. Nat. Rev. Mol. Cell Biol., 2, 63–67. [DOI] [PubMed] [Google Scholar]
- Meller A., Nivon,L., Brandin,E., Golovchenko,J. and Branton,D. (2000) Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl Acad. Sci. USA, 97, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Maréchal-Drouard,L., Sakamoto,W., Dietrich,A. and Wintz,H. (1998) A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 95, 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Atkin,O.K., Lambers,H., Wiskich,J.T. and Day,D.A. (1995) A critique of the use of inhibitors to estimate partitioning of electrons between mitochondrial respiratory pathways in plants. Physiol. Plant, 95, 523–532. [Google Scholar]
- Mireau H., Lancelin,D. and Small,I.D. (1996) The same Arabidopsis gene encodes both cytosolic and mitochondrial alanyl-tRNA synthetases. Plant Cell, 8, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratovska A., Lightowlers,R.N., Taylor,R.W., Turnbull,D.M., Smith,R.A., Wilce,J.A., Martin,S.W. and Murphy,M.P. (2001) Targeting peptide nucleic acid (PNA) oligomers to mitochondria within cells by conjugation to lipophilic cations: implications for mitochondrial DNA replication, expression and disease. Nucleic Acids Res., 29, 1852–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuburger M., Journet,E.P., Bligny,R., Carde,J.P. and Douce,R. (1982) Purification of plant mitochondria by isopycnic centrifugation in density gradients of Percoll. Arch. Biochem. Biophys., 217, 312–323. [DOI] [PubMed] [Google Scholar]
- Panov A., Filippova,S. and Lyakhovich,V. (1980) Adenine nucleotide translocase as a site of regulation by ADP of the rat liver mitochondria permeability to H+ and K+ ions. Arch. Biochem. Biophys., 199, 420–426. [DOI] [PubMed] [Google Scholar]
- Rostovtseva T. and Colombini,M. (1997) VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys. J., 72, 1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster W., Kloska,S. and Brennicke,A. (1993) An adenine nucleotide translocator gene from Arabidopsis thaliana. Biochim. Biophys. Acta, 1172, 205–208. [DOI] [PubMed] [Google Scholar]
- Seibel P., Trappe,J., Villani,G., Klopstock,T., Papa,S. and Reichmann,H. (1995) Transfection of mitochondria: strategy towards a gene therapy of mitochondrial DNA diseases. Nucleic Acids Res., 23, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souciet G., Menand,B., Ovesna,J., Cosset,A., Dietrich,A. and Wintz,H. (1999) Characterization of two bifunctional Arabidopsis thaliana genes coding for mitochondrial and cytosolic forms of valyl-tRNA synthetase and threonyl-tRNA synthetase by alternative use of two in-frame AUGs. Eur. J. Biochem., 266, 1–8. [DOI] [PubMed] [Google Scholar]
- Szabo I., Bathori,G., Tombola,F., Brini,M., Coppola,A. and Zoratti,M. (1997) DNA translocation across planar bilayers containing Bacillus subtilis ion channels. J. Biol. Chem., 272, 25275–25282. [DOI] [PubMed] [Google Scholar]
- Szabo I., Bathori,G., Tombola,F., Coppola,A., Schmehl,I., Brini,M., Ghazi,A., De Pinto,V. and Zoratti,M. (1998) Double-stranded DNA can be translocated across a planar membrane containing purified mitochondrial porin. FASEB J., 12, 495–502. [DOI] [PubMed] [Google Scholar]
- Turmel M., Otis,C. and Lemieux,C. (2002) The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol. Biol. Evol., 19, 24–38. [DOI] [PubMed] [Google Scholar]
- Vestweber D. and Schatz,G. (1989) DNA–protein conjugates can enter mitochondria via the protein import pathway. Nature, 338, 170–172. [DOI] [PubMed] [Google Scholar]
- Vieira H.L., Haouzi,D., El Hamel,C., Jacotot,E., Belzacq,A.S., Brenner,C. and Kroemer,G. (2000) Permeabilization of the mitochondrial inner membrane during apoptosis: impact of the adenine nucleotide translocator. Cell Death Differ., 7, 1146–1154. [DOI] [PubMed] [Google Scholar]
- Werhahn W., Niemeyer,A., Jansch,L., Kruft,V.V., Schmitz,U.K. and Braun,H.P. (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis. Identification of multiple forms of TOM20. Plant Physiol., 125, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.E. (1997) Homologous and heterologous interactions between hexokinase and mitochondrial porin: evolutionary implications. J. Bioenerg. Biomembr., 29, 97–102. [DOI] [PubMed] [Google Scholar]
- Winning B.M., Day,C.D., Sarah,C.J. and Leaver,C.J. (1991) Nucleotide sequence of two cDNAs encoding the adenine nucleotide translocator from Zea mays L. Plant Mol. Biol., 17, 305–307. [DOI] [PubMed] [Google Scholar]
- Zamzami N. and Kroemer,G. (2001) The mitochondrion in apoptosis: how Pandora’s box opens. Nat. Rev. Mol. Cell Biol., 2, 67–71. [DOI] [PubMed] [Google Scholar]