Abstract
Meiotic crossovers detected by pedigree analysis in the mouse MHC cluster into hotspots. To explore the properties of hotspots, we subjected the class II Eβ gene to high-resolution sperm crossover analysis. We confirm the presence of a highly localized hotspot 1.0–1.6 kb wide in the second intron of Eβ and show that it is flanked by DNA which is almost completely recombinationally inert. Mice heterozygous for haplotype s and another MHC haplotype show major haplotype-dependant variation in crossover rate but always the same hotspot, even in crosses including the highly diverged p haplotype. Crossovers in reciprocal orientations occur at similar rates but show different distributions across the hotspot, with the position of centre points in the two orientations shifted on average by 400 bp. This asymmetry results in crossover products showing biased gene conversion in favour of hotspot markers from the non-initiating haplotype, and supports the double-strand break repair model of recombination, with haplotype s as the most efficient crossover initiator. The detailed behaviour of the Eβ hotspot, including evidence for highly localized recombination initiation, is strikingly similar to human hotspots.
Keywords: gene conversion/meiotic drive/meiotic recombination hotspot/mouse MHC Eβ gene
Introduction
There is growing evidence that meiotic crossovers are not distributed randomly along mammalian chromosomes. Rather, mammalian genomes appear, at least to some extent, to be a mosaic of recombinationally suppressed regions separated by intervals that undergo very elevated and targeted crossover activity (Pittman and Schimenti, 1998; Petes, 2001; Yu et al., 2001). Recently, high-resolution sperm typing has shown that crossovers in the human MHC tend to cluster into very narrow (1–2 kb) recombination hotspots separated by long stretches of DNA exhibiting strong linkage disequilibrium (Jeffreys et al., 2000, 2001). Similarly, meiotic recombination events in the mouse MHC also show strong clustering. Eight mouse hotspots have been identified so far by breakpoint mapping in recombinant inbred lines and in pedigrees. These hotspots vary in the degree to which they have been characterized, from the Eα-C4 region at 300 kb in length (Lafuse et al., 1989), down to the Eα hotspot ∼0.5 kb wide (Khambata et al., 1996).
Mouse studies have also shown that hotspot location and/or intensity can be influenced by MHC haplotype, as well as genetic background (strain of mouse) and sex (Steinmetz et al., 1987; Shiroishi et al., 1991, 1993, 1995). For example, the very active hotspot in the class II Eβ gene, first characterized by Steinmetz et al. (1982), is ∼4 kb wide. Its activity varies between hybrid strains, and appears to be completely suppressed in heterozygotes carrying the p haplotype, in which crossovers occur instead preferentially at the Eα gene (Lafuse and David, 1986; Zimmerer and Passmore, 1991). Heine et al. (1994) found that these Eα haplotype p recombinants only result when p is from the intra-H2 recombinant strain B10F(13R), suggesting that distal modifiers of crossover can influence hotspot activity. Similarly, crossover rates at the Psmb9 (formerly known as Lmp2) hotspot are governed not only by haplotype and genetic background, but also by the sex of the animal, with high female rates occurring only in some strains (Shiroishi et al., 1993, 1995).
While crossover breakpoint mapping in mice has provided valuable information about meiotic recombin ation, the use of recombinant inbred lines and back-crossed offspring to identify MHC-recombinant progeny is extremely laborious. The limited numbers of crossovers that can be detected prevent the detailed definition of hotspot morphology and of the recombination processes operating within hotspots. Similarly, little is known about the recombinational activity of DNA outside these mouse hotspots. These limitations can be overcome by recovering crossover products directly from sperm DNA (Jeffreys et al., 1998, 2001), as shown by Guillon and De Massy (2002), who mapped 69 sperm recombinants at the Psmb9 locus to show that crossover density was maximal in an interval of 210 bp and decreased roughly symmetrically on both sides, in a manner similar to human recombination hotspots. We now extend this approach to analyse recombination at the mouse Eβ locus and to investigate the influence of haplotype on male recombination rates and on the distribution and reciprocity of crossover events within the hotspot. We show that the Eβ hotspot shares remarkably similar properties with its counterparts in the human MHC, implying similar recombination processes in these two very different species.
Results
Detection of meiotic crossovers in sperm DNA
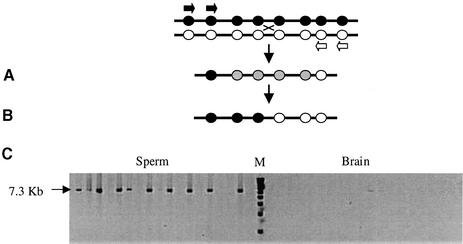
Re-sequencing across the Eβ gene did not reveal any previously unidentified polymorphisms across the crossover hotspot region in the second intron (see Materials and methods). Mice homozygous for MHC haplotypes b, d, k or p were crossed with haplotype s homozygotes. Sperm DNA from a mature F1 hybrid from each cross was prepared and Eβ-recombinant DNA molecules recovered from multiple pools of sperm DNA by nested repulsion-phase allele-specific PCR directed to heterozygous single nucleotide polymorphism (SNP) sites, as shown in Figure 1. Nested PCR gave sufficient allele specificity to allow the selective amplification of crossover molecules even from large pools of sperm DNA (up to 30 000 amplifiable molecules of each progenitor haplotype per PCR). Parallel analysis of brain DNA yielded much lower levels of PCR products, establishing that the sperm crossovers recovered were genuine products of meiotic recombination (Jeffreys et al., 2000) (Figure 1). Cross overs in reciprocal orientation were analysed either by using haplotype s 5′ allele-specific primers plus non-s 3′ primers (crossovers in orientation A) or 5′ non-s primers plus 3′ s haplotype primers (orientation B). We analysed a total of 6.5 × 106 amplifiable molecules from the four crosses and isolated 516 crossover molecules.
Fig. 1. Recovery of sperm crossovers. (A) Batches of sperm DNA from a hybrid mouse with heterozygosities (black and white circles) across the test region are amplified using two rounds of allele-specific PCR in repulsion phase (primers shown as black and white arrows) to selectively amplify recombinant molecules. Grey circles represent alleles of unknown status. (B) SNP typing is used to localize the breakpoint interval for each recombinant molecule amplified. (C) Examples of crossover molecules recovered at the Eβ locus from s × k sperm DNA at 280 molecules per reaction, electrophoresed next to a 1 kb marker (Gibco) (M), and alongside 840 molecules per reaction of brain DNA.
Sperm crossover rates in the Eβ region varied significantly between different crosses (Fisher’s exact test, P <0.001 for each pair-wise comparison; Table I), but not between individuals from the same cross (data not shown). The s × k heterozygotes showed the highest crossover rate (1.6 × 10–3 per sperm) and s × p showing the lowest (0.021 × 10–3). These rate differences were consistent both for orientation A crossovers and for orientation B exchanges (Table I), and are therefore reproducible. This establishes that recombination rates decrease in the order k > d > b >> p in hybrids carrying haplotype s.
Table I. Summary of sperm crossover data at the mouse MHC Eβ gene for separate orientation A and B crossovers as well as for combined A + B exchanges.
| Cross | Orientation | No. sperm typed | No. crossoversrecovered | Rate (×10–3) (95% CI) | Centre (bp)a | Shift (bp)b | 95% width (kb)c | Kolmogorov–Smirnov Pd |
|---|---|---|---|---|---|---|---|---|
| s × b | A | 400 000 | 66 | 0.16 (0.13–0.21) | 7070 | 0.7 | ||
| B | 240 000 | 63 | 0.26 (0.20–0.33) | 7400 | 330 | 1.3 | <0.001 | |
| A + B | 640 000 | 129 | 0.20 | 7190 | 1.0 | |||
| s × d | A | 120 000 | 79 | 0.66 (0.53–0.83) | 7050 | 1.3 | ||
| B | 240 000 | 73 | 0.30 (0.24–0.38) | 7350 | 300 | 0.8 | <0.001 | |
| A + B | 360 000 | 152 | 0.42 | 7220 | 1.2 | |||
| s × k | A | 34 000 | 61 | 1.79 (1.40–2.35) | 7190 | 0.9 | ||
| B | 44 000 | 63 | 1.43 (1.11–1.84) | 7410 | 220 | 1.4 | 0.037 | |
| A + B | 78 000 | 124 | 1.59 | 7300 | 1.3 | |||
| s × p | A | 2 300 000 | 50 | 0.022 (0.016–0.028) | 7050 | 0.9 | ||
| B | 3 100 000 | 61 | 0.020 (0.015–0.025) | 7590 | 540 | 1.5 | <0.001 | |
| A + B | 5 400 000 | 111 | 0.021 | 7280 | 1.6 | |||
| All four | A | 2 900 000 | 256 | – | 7070 | 1.0 | ||
| B | 3 600 000 | 260 | – | 7470 | 400 | 1.2 | <0.001 | |
| A + B | 6 500 000 | 516 | – | 7260 | 1.4 |
aBased on position relative to DDBJ/EMBL/GenBank accession No. AF050157, clone BAC 135G15.
bDisplacement of the centres of orientation B versus orientation A distributions.
cWidth within which 95% of crossovers occur, assuming that they are normally distributed across the hotspot.
dComparing orientation A and B crossover distributions.
Allele-specific oligonucleotide mapping of crossover breakpoints
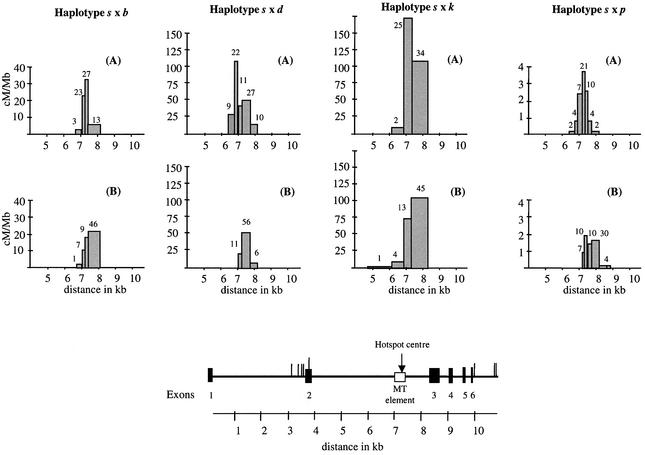
The selected polymorphisms (mainly SNPs) used to map crossover breakpoints by hybridizing amplified recombinant molecules with allele-specific oligonucleotides (ASOs) (Wood et al., 1985) are shown in Table II. All 516 sperm crossovers were simple (Figure 1), with breakpoints mapping to a single inter-marker interval. There were no instances of more complex exchanges involving switching back and forth between haplotypes near the site of exchange. Crossover breakpoints were non-randomly distributed across the Eβ region in all hybrids (Figure 2). Nearly all crossovers (99%) occurred within the second intron of the Eβ gene and were highly concentrated in the 3′ half, demonstrating the presence of a highly localized hotspot. Crossover frequencies in each interval were used to determine the recombination activity per unit length in cM/Mb (Figure 2). Each hybrid showed an approximately normal distribution of combined orientation A + B crossover breakpoints across the hotspot (Figure 3A). All four heterozygotes showed the centre of the hotspot at a very similar location (Table I), within an MT element (Figure 2). The width of the hotspot was also similar in all crosses (1.0–1.6 kb), although the peak activity varied dramatically from 3.5 cM/Mb in the s × p hybrid to 170 cM/Mb in s × k. The same hotspot therefore functions in male recombination in all heterozygotes, despite the substantial rate variation. Only one crossover, located upstream of position 6114 bp, was found outside the hotspot. The mean recombination activity over all crosses in DNA flanking the hotspot is therefore extremely low, at 0.005 cM/Mb (95% CI 0.0002–0.02 cM/Mb) and ∼100-fold below the mouse genome average rate of 0.5 cM/Mb.
Table II. List of ASO sequences.
| Marker name | Location (bp)a | ASO sequence |
||||
|---|---|---|---|---|---|---|
| Haplotype s | Haplotype b | Haplotype d | Haplotype k | Haplotype p | ||
| 0 | 4089 | ccttggtaccattaactg | ctttggtgccattaactg | |||
| 0.04 | 4115 | ctcactggcccgagcaaa | ctcactgacccgagcaaa | |||
| 0.39 | 4453 | taagagcactcttgggcc | taagagcgctcttgggcc | taagagcgctcttgggcc | ||
| 0.65 | 4723 | tggcaatcgtcagggtga | tggcaatggtcagggtga | tggcaatggtcagggtga | tggcaatggtcagggtga | |
| 1.4-1 | 5480 | ccagtctccgctgtcaat | ccagtctccactgtcaat | |||
| 1.4-2 | 5482 | ccagtctccgctgtcaat | ccagtctccgcagtcaat | |||
| 2.05 | 6114 | tgtgtttttttttactg | tgtgtttttttt-actg | |||
| 2.4 | 6472 | ggtgcctagatccacatg | ggtgcctggatccacatg | ggtgcctggatccacatg | ||
| 2.45 | 6524 | gctgtagatgaagccaga | gctgtaggtgaagccaga | |||
| 2.64 | 6734, 6739 | acccgggggtgggctgcc | acccgagggtaggctgcc | |||
| 2.7-1 | 6765 | gcctggggtagtactgtgg | gcctgggtagtactgtgg | gcctggg-tagtactgtgg | ||
| 2.7-2 | 6768, 6775 | gcctggggtagtactgtgg | gcctggggtagaactgtgt | gcctggggtagaactgtgt | ||
| 2.82 | 6901 | gtgtgtcgggccctggag | gtgtgtcaggccctggag | gtgtgtcaggccctggag | ||
| 2.85 | 6930, 6933 | gggaccgaagtggatttc | gggaccaaaatggatttc | |||
| 2.93 | 7016 | gcttagctcacacttctt | gcttagcttacacttctt | |||
| 3.1 | 7158 | tatagagtccaggacaac | tatagagcccaggacaac | tatagagcccaggacaac | tatagagcccaggacaac | tatagagcccaggacaac |
| 3.14 | 7206 | gagtccccccccatcaat | gagtccctccccccccca | |||
| 3.2 | 7247 | cctaaaggcttacctaca | cctaaagacttacctaca | |||
| 3.3 | 7324 | gtgtcagatcaacataaa | gtgtcagttcaacataaa | |||
| 3.35 | 7393, 7399 | gtgtgggaccacaacccc | gtatgggatcacaacccc | |||
| 3.5 | 7575 | gttatgacttgttgccta | gttatgatttgttgccta | |||
| 3.58 | 7609 | tggcctggcttcttcagc | tggcctgacttcttcagc | |||
| 4 | 8113 | ggatccattctggatgga | ggatccagtctggatgga | ggatccagtctggatgga | ggatccagtctggatgga | ggatccagtctggatgga |
| 4.48 | 8538, 8542 | gaatgcccgcccgggtgg | gaatgcctgcctgggtgg | |||
| 4.58 | 8638 | cttacagtgtaggtgcgt | cttacagggtaggtgcgt | cttacagggtaggtgcgt | ||
| 4.8-1 | 8821, 8825 | ggtcatgactgaactcag | ggtcatggctgacctcag | ggtcatggctgacctcag | ||
| 4.8-2 | 8823 | tgactgaactcagggaca | tgaccgacctcagggaca | |||
| 5.4 | 9429 | gccatgggcactgccggt | gccatggtcactgccggt | |||
| 5.79 | 9855 | tgtaaggagaggcctgaa | tgtaaggtgaggcctgaa | |||
| 5.9 | 9969 | gaaggagttcccccccgg | gaaggaattcccccccgt | |||
Nucleotide differences are shown in bold.
aBased on position relative to DDBJ/EMBL/GenBank accession No. AF050157, clone BAC 135G15.
Fig. 2. Mouse Eβ sperm crossover distributions for orientation A (s/non-s) and orientation B (non-s/s) recombinants. The haplotypes in each mouse analysed are given at top. The numbers of recombinants recovered in each interval, shown above the histograms, were used to calculate the crossover frequency per unit length of DNA for each interval in cM/Mb. Note the considerable differences in recombination scaling due to substantial variation in crossover activity across the hybrids tested. The Eβ gene is shown beneath the histograms. The locations of the allele-specific primers used to recover crossovers are shown as vertical lines and the locations of the hotspot and MT element are indicated. Locations are relative to clone BAC 135G15 (DDBJ/EMBL/GenBank accession No. AF050157).
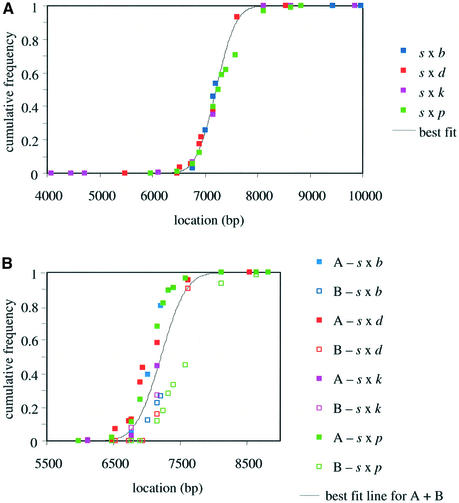
Fig. 3. Distribution of sperm crossovers across the Eβ hotspot. (A) Cumulative frequency distributions of sperm crossovers in the four hybrids analysed, for the combined orientation A + B recombinants. The least-squares best-fit cumulative frequency distribution assumes that crossovers are normally distributed across the hotspot (centre at 7260 bp, standard deviation 350 bp). (B) Cumulative frequency distributions of separate orientation A (filled squares) and B (open squares) crossovers. The best-fit line for the combined A + B distributions is indicated. The B distributions are shifted 3′ relative to A distributions in all crosses.
Reciprocal crossover asymmetry and transmission distortion
Orientation A and B crossovers were detected in each cross at similar frequencies, consistent with these sperm recombinants being the products of reciprocal crossover. Minor differences in A versus B rates within a cross (at most only 2.2-fold for s × d; Table I) show no consistent bias towards A or B exchanges, and could easily have arisen from variation in PCR efficiency between different primer sets. While sperm crossovers appear to be reciprocal in rate, the separate A and B crossovers showed different distributions, with orientation A (s→non-s) crossovers consistently displaced 5′-ward relative to orientation B (non-s→s) exchanges in all four hybrids (Table I; Figures 2 and 3B). This displacement varied from 220 bp for s × k to 540 bp for s × p, and averaged ∼400 bp over all four heterozygotes.
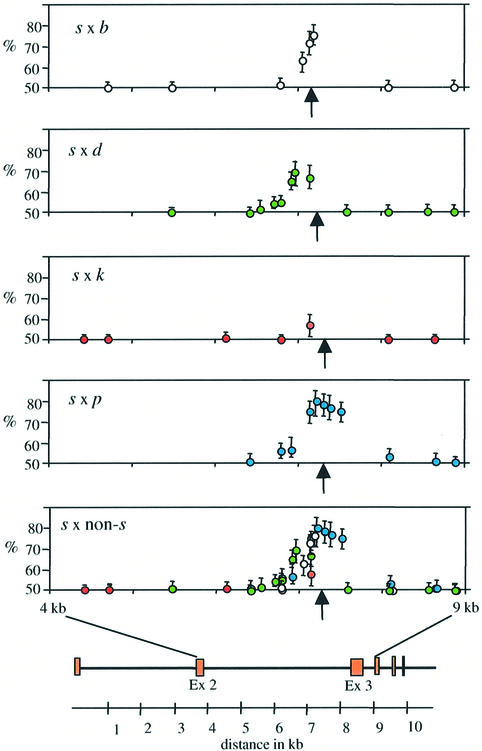
This reciprocal crossover asymmetry results in transmission ratio distortion of alleles into crossover products. Without asymmetry, s and non-s alleles should show a 50:50 Mendelian ratio in combined A and B crossovers. However, crossover asymmetry results in highly significant over-transmission of non-s alleles within the hotspot (Figure 4). The most extreme distortion was seen for marker 3.2 at 7247 bp, very close to the centre of the hotspot, which in s × p animals show a transmission ratio of 81:19 for non-s:s alleles in A + B crossover products. This distortion also affects neighbouring markers, with the strength of distortion decreasing with distance from the centre of the hotspot. Superimposition of data from all four hybrids (Figure 4, bottom) showed that the transmission distortion profiles were similar in all animals. However, the maximum degree of distortion varied between crosses in the order p > (b, d) > k; this order is the inverse of the order of crossover rates in these heterozygotes (Table I).
Fig. 4. Transmission distortion in Eβ crossover products in each heterozygote. The proportions of non-s alleles in equal numbers of orientation A and B sperm crossovers are shown for each marker, along with 95% CIs. The bottom graph shows all hybrids superimposed. The positions of the markers with respect to the Eβ gene are shown at bottom, and the centre of the hotspot is indicated with arrows.
Discussion
Male recombination at the Eβ hotspot
This work describes the application of high-resolution sperm typing to analyse meiotic recombination in the class II Eβ gene in four mouse hybrid crosses. Our data confirm the presence of a recombination hotspot in the second intron of the gene, centred on an MT element and only 1.0–1.6 kb wide, and within the interval determined by prior mapping of recombinant inbred lines (Steinmetz et al., 1982, 1987; Bryda et al., 1992). This hotspot is flanked by DNA that is almost totally suppressed for recombination. Male crossovers are symmetrically distributed across the hotspot and occur at a rate strongly influenced by MHC haplotype. Sperm recombination rates for the s × b, s × d and s × k crosses (0.2–1.6 × 10–3 per sperm) are comparable to crossover rates determined from pedigrees (1.0 × 10–3 across the Eβ region; Passmore et al., 1987; Shiroishi et al., 1995), establishing that sperm analysis yields reliable quantitative data on male recombination.
Sequence determinants of recombination rates
It has previously been suggested that recombination rates across mouse MHC hotspots, as in other species, can be reduced by sequence divergence between the inter acting haplotypes (Sant’Angelo et al., 1992; Yoshino et al., 1995). Datta et al. (1997) found that even a single mismatch over a 350 bp sequence between other wise identical sequences in yeast can inhibit mitotic recombination up to 4-fold; this anti-recombination was exerted through the action of mismatch repair proteins. Our data are consistent with the hypothesis that sequence divergence results in decreased recombination efficiency. The degree of divergence between the s haplotype and the other haplotypes over the central 200–400 bp of the hotspot is least for haplotype k (0.25%) and by far the greatest for haplotype p (2.5%) (Table III). The order of divergence from haplotype s over the centre of the hotspot (k < d < b << p) inversely correlates with the relative rates of recombination in the heterozygotes (k > d > b >> p), consistent with the hypothesis that recombination resulting in crossover is favoured when nucleotide identity is at its greatest at the Eβ hotspot (Bryda et al., 1992; Sant’Angelo et al., 1992). However, it is possible that more distal flanking polymorphisms, as well as genetic background, could influence hotspot activity. For example, cis-acting elements have been shown to be involved in controlling recombination rates and sex-specificity at the mouse Psmb9 hotspot (Shiroishi et al., 1991; Heine et al., 1994).
Table III. Number of sequence differences between haplotype s and each of the non-s haplotypes, around the centre of the Eβ hotspot or over the whole second intron.
| Distance | Haplotype b | Haplotype d | Haplotype k | Haplotype p |
|---|---|---|---|---|
| ±100 bp | 2 | 1 | 0 | 4 |
| ±200 bp | 3 | 2 | 1 | 10 |
| Entire 2nd intron | 12 | 86 | 8 | Highly divergeda |
aSee section on haplotype p in the Discussion.
It has also been suggested that recombination may be influenced by an imperfect four-base repeat sequence (AGGC)n located just upstream of the third exon (8130 bp) and outside the hotspot (Kobori et al., 1986). This sequence shows similarity to the lambda phage chi sequence and 80% sequence identity to the human minisatellite core sequence (Jeffreys et al., 1985), and has been postulated to provide a recombination signal with the number of repeats governing the activity of the hotspot (Zimmerer and Passmore, 1991). Haplotype s has 11 copies of this repeat sequence, compared with 18 in haplotype d, 10 in b and k, and six in haplotype p. There is no obvious correlation between relative recombination rates (Table I) and either the absolute number of repeats or the difference in repeat copy number compared with haplotype s. There is therefore no clear evidence that this tetramer plays a role in regulating recombination activity.
Reciprocal crossover asymmetry and meiotic drive
Previous evidence of reciprocal crossover asymmetry between the k and b haplotypes at the Eβ hotspot was based on the mapping of only seven recombinants and was therefore very speculative (Zimmerer and Passmore, 1991). The large numbers of sperm crossovers analysed in the present study confirm that asymmetry in the location of reciprocal A and B crossover breakpoints does occur in all hybrids carrying haplotype s. It remains to be seen whether other heterozygous crosses will show a similar phenomenon. The asymmetry results in non-s alleles being preferentially over-transmitted to crossover products in all heterozygotes, with transmission distortion being strongest for markers closest to the centre of the hotspot (Figure 4). This phenomenon is remarkably similar to the asymmetry seen in one of the human MHC crossover hotspots (Jeffreys and Neumann, 2002). The degree of transmission distortion in human sperm crossover products (up to 87:13) is very similar to that seen in the Eβ hotspot (up to 89:11), as is the degree of displacement of orientation A and B crossovers (on average 400 bp in Eβ, compared with 430 bp in the human hotspot).
Reciprocal crossover asymmetry and transmission distortion can be readily explained by the double-strand break (DSB) repair model of recombination (Szostak et al., 1983; Pâques and Haber, 1999; Jeffreys and Neumann, 2002). This model is supported by previous data at human hotspots (Jeffreys et al., 2000, 2001; Jeffreys and Neumann, 2002) and at the Psmb9 hotspot in mice (Guillon and De Massy, 2002) which show that crossovers are approximately symmetrical and normally distributed across a hotspot, compatible with the presence of initiation site(s) located at or close to the centre of the hotspot. Initiation creates a DSB, which then undergoes resection to create 3′ overhanging segments. The 3′ overhangs invade the undamaged chromosome where mismatch removal from the invading strand may occur (Alani et al., 1994), and repair is carried out using information from the undamaged (donor) chromosome (Jeffreys and Neumann, 2002). This process results in sites of crossover resolution occurring at distances from the initiation site determined, in part at least, by the extent of resection, and contributes to the observed width of hotspots.
If crossovers are initiated on both chromosomes with equal frequency, then breakpoints will be identically distributed in orientation A and B crossovers. However, if initiation rates vary between haplotypes, then asymmetry and transmission distortion will arise, increasing in strength with the degree of initiation suppression on one of the haplotypes (Jeffreys and Neumann, 2002). If a hybrid carries a high and low efficiency haplotype, then the majority of crossovers will arise from initiations on the active haplotype. DSB repair will therefore result in the replacement of markers from the initiating chromosome with those from the non-initiating donor chromosome, resulting in biased gene conversion in favour of the donor chromosome. The consistent over-transmission of non-s alleles in all heterozygous mice therefore suggests that haplotype s is most active in initiation. The relative strengths of transmission distortion in different crosses (Table I; Figure 4) further suggest that the initiation efficiency on different haplotypes decreases in the order s > k > (b, d) > p. The extreme over-transmission of the non-s allele (up to 89:11 in the s × p cross), combined with the inverse correlation between recombination rate and the strength of over-transmission, provides strong evidence in favour of this initiation model. Alternative models involving biased mismatch repair of heteroduplex DNA following strand invasion and subsequent branch migration are less plausible (for a fuller discussion see Jeffreys and Neumann, 2002).
There are, however, differences in the properties of reciprocal crossover asymmetry in the Eβ hotspot and in the human MHC hotspot. In the latter, the trigger for asymmetry has been traced to a single SNP at the centre of the hotspot that, when heterozygous, appears to be sufficient to cause asymmetry, presumably by directly influencing the efficiency of DSB formation (Jeffreys and Neumann, 2002). One SNP heterozygosity inside the Eβ hotspot is shared by all four mouse hybrids (marker 3.1, located 100 bp 3′ of the average hotspot centre, with allele C shared by haplotypes b, d, k and p, and the alternative allele T on haplotype s), and the C allele is over-transmitted in every cross. However, without recombinant haplotype information, it is impossible to determine whether this SNP is triggering asymmetry. Given the variation in transmission distortion between different crosses, it is unlikely that this one SNP is exclusively responsible for asymmetry. Other hotspot SNPs might influence initiation in different heterozygotes, although this seems unlikely since first, they would always have to favour the s haplotype, and secondly, most SNPs within human MHC hotspots do not cause asymmetry (Jeffreys and Neumann, 2002). Instead, it seems likely that more distal elements influence the efficiency of recombination initiation in the Eβ hotspot.
The second difference between the mouse and human hotspots is revealed by recombination rates. In the human hotspot, heterozygosity for the suppressing variant causes asymmetry but has little influence on crossover rate (in theory, the recombination rate in a heterozygote with one totally suppressed chromosome should be 50% of that in a person homozygous for the initiation-proficient variant) (Jeffreys and Neumann, 2002). In mice, recombination rates can be strongly suppressed in hybrids, particularly in s × p heterozygotes. This implies that in such hybrids, initiating events may be suppressed in trans on the s haplotype or, if they occur, fail to form a productive recombination complex. Perhaps sequence mismatches between haplotypes result in single strands from the s haplotype, produced by gap resection, that either fail to invade the other chromosome or are rejected on invasion. Subsequent DSB recombination repair may then occur preferentially from the sister chromatid, resulting in a sister chromatid exchange. Alternatively, mismatched invasions might be preferentially resolved to yield gene conversions rather than crossovers. The decrease of sperm crossover rates with increasing divergence from the s haplotype is consistent with these suggestions.
Exchanges involving haplotype p
Recombination has never before been detected between p and the other haplotypes at the Eβ hotspot (Lafuse and David, 1986; Shiroishi et al., 1993). The present data indicate that this hotspot does function in s × p heterozygotes, but at a very low efficiency, apparently via recombination initiation occurring preferentially, if not exclusively, on the s haplotype. Yoshino et al. (1995) suggested that insertions and deletions (in their case at a tandem repeat locus at the Psmb9 hotspot) may play a significant role in influencing recombination. In this respect, haplotype p is unique in containing a 597 bp ancestral DNA segment in the 5′ half of the second intron, outside the hotspot, that is replaced by a 1634 bp retroposon element in all other haplotypes (Zimmerer and Passmore, 1991). It is not clear how this insertion/deletion might influence Eβ recombination. It has been suggested that this retroposon element creates a recombination initiation activator on non-p haplotypes (Zimmerer and Passmore, 1991). This would be very difficult to test, given that it would require analysis of p/non-p recombinant haplotypes either carrying or devoid of the retroposon.
Similarities between mammalian recombination hotspots
Several mammalian hotspots have been characterized using pedigree approaches, and more recently by high resolution sperm typing. The Eβ hotspot shares remarkably similar features with human hotspots (Jeffreys et al., 1998, 2000, 2001), including the same width, the extreme suppression of recombination in the immediate flanking DNA, the simple structure of the vast majority of sperm crossovers and the evidence from crossover asymmetry for highly localized sites of initiation at the centres of hotspots. These shared features suggest that very similar recombination mechanisms operate in humans and in mice. Over-transmission of markers from recombination-suppressed haplotypes into crossover progeny also seems to be a common phenomenon in hotspots, having now been seen in one of six human MHC hotspots (Jeffreys and Neumann, 2002), in a human minisatellite-associated hotspot (Jeffreys et al., 1998) and in the mouse Eβ hotspot. The resulting meiotic drive of markers from recombin ationally suppressed chromosomes, which can include markers that actually suppress recombination, can be sufficiently strong to influence the population frequencies of alleles and to favour the eventual extinction of hotspot activity (Boulton et al., 1997; Jeffreys and Neumann, 2002). The prevalence and evolutionary significance of this recombination-based meiotic drive will require further high-resolution studies of meiotic crossover in a range of species.
Materials and methods
Mouse strains and breeding
Mice from strains A.SW, B10.S/SgmedJ (Eβ haplotype s), C57BL/10J (Eβ haplotype b), B10.D2 (Eβ haplotype d), B10.A/SgSnJ (Eβ haplotype k) and P/J (Eβ haplotype p) were purchased from the Jackson Laboratory (Bar Harbor, ME) (haplotype and strain information available on the Jackson Laboratory website: www.jax.org). All relevant sequence information has been published previously (Saito et al., 1983; Widera and Flavell, 1984; Kobori et al., 1986; Padgett et al., 1991; Zimmerer and Passmore, 1991; Bryda et al., 1992). Mice with MHC haplotypes b, d, k and p were crossed with haplotype s mice. Caudal epidydimus and brains were collected from 8- to 10-week-old mature offspring. All crosses were generated at the University of Leicester under guidance issued by the MRC in ‘Responsibility in the use of animals for medical research’ and Home Office project license No. PPL 80/1353.
Genotyping and SNP verification
We used PCR primers designed from the sequenced clone BAC 135G15 (DDBJ/EMBL/GenBank accession No. AF050157) to identify and verify SNPs in the Eβ gene of all strains in the study. We re-sequenced using BigDye Terminators (ABI) on an ABI 377 Automated Sequencer and identified SNPs using ABI AutoAssembler software. Selected SNPs were used to map recombination breakpoints in subsequent sperm analyses. The ASO probes used for genotyping are shown in Table II.
DNA extraction and quantification
Sperm DNA was prepared using procedures similar to those described by Jeffreys et al. (1994). To minimize the risk of contamination, all manipulations were carried out in a laminar flow hood. Caudal epidydimus from one mouse was chopped in 1 ml of phosphate-buffered saline (138 mM NaCl, 2.7 mM KCl, 5.4 mM Na2HPO4, 0.2 mM KH2PO4), filtered and centrifuged at 10 000 g. The cell pellet was resuspended in 1× SSC and somatic cells were lysed at room temperature by the addition of SDS to 0.15%. The lysate was centrifuged and the resulting sperm pellet resuspended in 1 ml 0.2× SSC, 0.1% SDS, 1 M 2-mercaptoethanol and digested with 200 µg/ml proteinase K (Sigma) for 1 h at 37°C. DNA was recovered by phenol/chloroform extraction and ethanol precipitation. DNA from brain was prepared by tissue homogenization in 1× SSC, followed by SDS lysis, phenol/chloroform extraction and ethanol precipitation. All DNAs were dissolved in 5 mM Tris–HCl (pH 7.5). Approximately 20 µg of DNA was digested with enzymes (HpaI, BssHII or XhoI) that cleave outside the test interval analysed, to render genomic DNA fully soluble prior to dilution. DNA concentrations were determined by UV spectrometry and by agarose gel electrophoresis against λ phage DNA.
Sperm crossover analyses
We designed allele-specific primers 17–20 nt long with at least 50% GC content for SNP sites 5′ and 3′ of the putative Eβ hotspot to amplify across a maximum of 7.7 kb. 5′ primers were located between 3227 and 4301 bp on the reference clone sequence, and 3′ primers between 9855 and 10952 bp. The 5′ allele-specific primers, named on the basis of distance from the first ASO typed (assigned as 0.0), were:
–1.0p, 5′-GGT GGG GAG GAT CTG AAA G-3′, used for haplotypes d and p;
–0.6g, 5′-TAA CTG TCC AGC CTG GGG-3′, used for haplotypes b, d and k;
–0.56A, 5′-TGC CCT GTT AGT TGT GGA-3′, used for haplotypes b and k;
–0.56T, 5′-TGC CCC GTT AGT TGT GGT-3′, used for haplotype s;
–0.57T, 5′-TTG GAG CTG GGG CAT GT-3′, used for haplotype s;
–0.5C, 5′-GGG TCT GGT CGG TCA TTC-3′, used for haplotype s;
–0.5T, 5′-CTT GGG TCT GGT CAT TT-3′, used for haplotype p;
–0.2C, 5′-GGA GCA AAA GCG GGC CGC-3′, used for haplotype s;
0.2F, 5′-CCT ACC TAC ACG GTG TGC GG-3′, used for haplotype s.
The 3′ primers were:
5.8T, 5′-CTG ACT GCT TCA GGC CTC T-3′, used for haplotype s;
5.8g, 5′-CTG ACT GCT TCA GGC CTC G-3′, used for haplotypes p and d;
6.75C, 5′-GGC CCA CAG CAA CAG CTT C-3′, used for haplotype s;
6.75T, 5′-GGC CCA CAG CAA CAT CTT T-3′, used for haplotypes b and k;
6.85C, 5′-CTC TTC CCA TCC TGC TGC-3′, used for haplotypes b, d, k and p;
6.85T, 5′-CTC TTC CCA TCC TGC TGT-3′, used for haplotype s.
The optimal annealing temperature for each primer was determined by amplification of inbred parental DNA samples from each strain, using the allele-specific primer in combination with an internal universal primer that can amplify from all haplotypes. Primers were selected that were both very efficient and highly specific for the allele of interest. Primers specific to the 5′ end of haplotype s were used in combination with primers specific to the 3′ end of the non-s haplotype to amplify orientation A crossovers, and vice versa for orientation B.
We amplified recombinant molecules using procedures similar to those described previously (Jeffreys et al., 2000, 2001). We amplified multiple batches of restriction-digested and diluted DNA samples, from sperm and brain of a single mouse per cross, each containing on average between 0.4 and 2 amplifiable crossover molecules per reaction. We tested two heterozygous mice for the b and p crosses, and one mouse each for the other two crosses. Since data for mice from the same cross were not significantly different (data not shown), such data were pooled. Brain DNA was used as a negative control for PCR artefacts and/or bleed-through amplification of one or other progenitor haplotype. DNA inputs depended on crossover rates and were determined in pilot experiments using 300–30 000 amplifiable molecules of each haplotype per reaction; we assume a single molecule PCR efficiency of 85% (Jeffreys et al., 2000). Poisson analysis of the number of positive and negative reactions (Jeffreys et al., 2000, 2001) was used to estimate the number of amplifiable crossover molecules. In the full crossover analyses, inputs varied from 216 ng sperm DNA per reaction (30 000 molecules) for s × p hybrids down to 2 ng DNA per reaction (280 molecules per reaction) for s × k crosses. DNA was amplified in two rounds of nested repulsion-phase PCR to selectively amplify recombinant molecules (Jeffreys et al., 1998, 2000) (Figure 1). Touch-down PCRs were used [typically 96°C for 1 min, followed by 26 rounds of 96°C for 20 s, 60–56°C for 30 s (step-down) and 63°C for 7–8 min (depending on length of amplicon)] with two allele-specific primers in repulsion phase. Primary PCR products were digested with S1 nuclease to remove single-stranded DNA and PCR artefacts (Jeffreys et al., 1998) and re-amplified with a second round of repulsion-phase allele-specific PCR as above. Secondary PCR products were electrophoresed on an agarose gel and visualized with ethidium bromide and UV light. Positive reactions were re-amplified using internal universal primers. PCR products were dot blotted and SNPs typed by ASO hybridization (Wood et al., 1985) as modified by Jeffreys et al. (2000) using the probes listed in Table II. Poisson estimations of the number of amplifiable recombinant molecules, to correct for instances of multiple recombinants recovered in a single positive reaction, were performed as described previously (Jeffreys et al., 1998).
Acknowledgments
Acknowledgements
We thank Dr John Stead, Dr Celia May, Liisa Kauppi and four anonymous reviewers for helpful comments, and Rita Neumann for help with ASO typing. This work was supported by grants to A.J.J. from the Royal Society and the Wellcome Trust (ref. 058084/Z/99/Z).
References
- Alani E., Reenan,R.A. and Kolodner,R.D. (1994) Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics, 137, 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton A., Myers,R.S. and Redfield,R.J. (1997) The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl Acad. Sci. USA, 94, 8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryda E.C., DePari,J.A., Sant’Angelo,D.B., Murphy,D.B. and Passmore,H.C. (1992) Multiple sites of crossing over within the Eb recombinational hotspot in the mouse. Mamm. Genome, 2, 123–129. [DOI] [PubMed] [Google Scholar]
- Datta A., Hendrix,M., Lipsitch,M. and Jinks-Robertson,S. (1997) Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc. Natl Acad. Sci. USA, 94, 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon H. and de Massy,B. (2002) An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat. Genet., 32, 296–299. [DOI] [PubMed] [Google Scholar]
- Heine D., Khambata,S., Wydner,K.S. and Passmore,H.C. (1994) Analysis of recombinational hotspots associated with the p haplotype of the mouse MHC. Genomics, 23, 168–177. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J. and Neumann,R. (2002) Reciprocal crossover asymmetry and meiotic drive in a human recombination hotspot. Nat. Genet., 31, 267–271. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Wilson,V. and Thein,S.L. (1985) Hypervariable ‘minisatellite’ regions in human DNA. Nature, 314, 67–73. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Tamaki,K., MacLeod,A., Monckton,D.G., Neil,D.L. and Armour,J.A.L. (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet., 6, 136–145. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Murray,J. and Neumann,R. (1998) High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell, 2, 267–273. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Ritchie,A. and Neumann,R. (2000) High-resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum. Mol. Genet., 9, 725–733. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J., Kauppi,L. and Neumann,R. (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet., 29, 217–222. [DOI] [PubMed] [Google Scholar]
- Khambata S., Mody,J., Modzelewski,A., Heine,D. and Passmore,H.C. (1996) Ea recombinational hotspot in the mouse major histocompatibility complex maps to the fourth intron of the Ea gene. Genome Res., 6, 195–201. [DOI] [PubMed] [Google Scholar]
- Kobori J.A., Strauss,E., Minard,K. and Hood,L. (1986) Molecular analysis of the hotspot of recombination in the murine major histocompatibility complex. Science, 234, 173–179. [DOI] [PubMed] [Google Scholar]
- Lafuse W.P. and David,C.S. (1986) Recombination hotspots within the I region of the mouse H-2 complex map to the Eβ and Eα genes. Immunogenetics, 24, 352–360. [DOI] [PubMed] [Google Scholar]
- Lafuse W.P., Lee,S.T., Castle,L. and David,C.S. (1989) Restriction fragment analysis of H-2 recombinant mouse strains with crossovers between Eα and C4 genes. Immunogenetics, 30, 387–389. [DOI] [PubMed] [Google Scholar]
- Padgett K.A., Shreffler,D.C. and Saha,B.K. (1991) Molecular mapping of murine I region recombinants. III. Crossing over at two discrete sites within the beta 1–beta 2 intron of the E beta gene. J. Immunol., 147, 2764–2770. [PubMed] [Google Scholar]
- Pâques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore H.C., Kobori,J.A., Zimmerer,E.J., Spinella,D.G. and Hood,L. (1987) Molecular characterization of meiotic recombination within the major histocompatibility complex of the mouse: mapping of crossover sites within the I region. Biochem. Genet., 25, 513–526. [DOI] [PubMed] [Google Scholar]
- Petes T.D. (2001) Meiotic recombination hotspots and cold spots. Nat. Rev. Genet., 2, 360–369. [DOI] [PubMed] [Google Scholar]
- Pittman D.L. and Schimenti,J.C. (1998) Recombination in the mammalian germ line. Curr. Top. Dev. Biol., 37, 1–35. [DOI] [PubMed] [Google Scholar]
- Saito H., Maki,R.A., Clayton,L.K. and Tonegawa,S. (1983) Complete primary structures of the Eβ chain and gene of the mouse major histocompatibility complex. Proc. Natl Acad. Sci. USA, 80, 5520–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Angelo D.B., Lafuse,W.P. and Passmore,H.C. (1992) Evidence that nucleotide sequence identity is a requirement for meiotic crossing over within the mouse Eb recombinational hotspot. Genomics, 13, 1334–1336. [DOI] [PubMed] [Google Scholar]
- Shiroishi T., Sagai,T., Hanzawa,N., Gotoh,H. and Moriwaka,K. (1991) Genetic control of sex-dependent meiotic recombination in the major histocompatibility complex of the mouse. EMBO J., 10, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroishi T., Sagai,T. and Moriwaki,K. (1993) Hotspots of meiotic recombination in the mouse major histocompatibility complex. Genetica, 88, 187–196. [DOI] [PubMed] [Google Scholar]
- Shiroishi T., Koide,T., Yoshino,M., Sagai,T. and Moriwaki,K. (1995) Hotspots of homologous recombination in mouse meiosis. Adv. Biophys., 31, 119–132. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Minard,K., Horvath,S., McNicholas,J., Srelinger,J., Wake,C., Long,E., Mach,B. and Hood,L. (1982) A molecular map of the immune response region from the major histocompatibility complex of the mouse. Nature, 300, 35–42. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Uematsu,Y. and Fischer Lindahl,K. (1987) Hotspots of homologous recombination in mammalian genomes. Trends Genet., 3, 7–10. [Google Scholar]
- Szostak J.W., Orr-Weaver,T.L., Tothstein,R.J. and Stahl,F.W. (1983) The double-strand-break repair model for recombination. Cell, 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Widera G. and Flavell,R.A. (1984) The nucleotide sequence of the murine I–E β b immune response gene: evidence for gene conversion events in class II genes of the major histocompatibility complex. EMBO J., 3, 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W.I., Gitschier,J., Lasky,L.A. and Lawn,R.M. (1985) Base composition-independent hybridization in tetramethyleammonium chloride: a method for oligonucleotide screening of highly complex gene libraries. Proc. Natl Acad. Sci. USA, 82, 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino M., Sagai,T., Fischer Lindahl,K., Toyoda,Y., Moriwaki,K. and Shiroishi,T. (1995) Allele-dependent recombination frequency: homology requirement in meiotic recombination at the hotspot in the mouse major histocompatibility complex. Genomics, 27, 298–305. [DOI] [PubMed] [Google Scholar]
- Yu A. et al. (2001) Comparisons of human genetic and sequence-based physical maps. Nature, 409, 951–953. [DOI] [PubMed] [Google Scholar]
- Zimmerer E.J. and Passmore,H.C. (1991) Structural and genetic properties of the Eb recombinational hotspot in the mouse. Immunogenetics, 33, 132–140. [DOI] [PubMed] [Google Scholar]