Abstract
The elucidation of the mechanisms of transcriptional activation and repression in eukaryotic cells has shed light on the important role of acetylation-deacetylation of histones mediated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. Another group belonging to the large family of sirtuins (silent information regulators (SIRs)) has an (nicotinamide adenine dinucleotide) NAD+-dependent HDAC activity. Several inhibitors of HDACs (HDIs) have been shown to exert antitumor effects. Interestingly, some of the HDIs exerted a broad spectrum of antiprotozoal activity. The purpose of this review is to analyze some of the current data related to the deacetylase enzymes as a possible target for drug development in cancer and parasitic diseases with special reference to protozoan infections. Given the structural differences among members of this family of enzymes, development of specific inhibitors will not only allow selective therapeutic intervention, but may also provide a powerful tool for functional study of these enzymes.
INTRODUCTION
Regulation of cellular processes by reversible phosphorylation of key regulatory proteins is fundamental to a large number of cellular functions. However, during the past few years, results from basic research studies pointed to the importance of the acetylation and deacetylation reactions not only at the level of histone lysine residues but also on other cellular factors which together could affect gene expression regulation. In fact, although the pioneering studies focused on the role of histone acetylation in the control of gene transcription, recent investigations have pointed to the notion that reversible nonhistone proteins acetylation is an important posttranslational modification that regulates a wide range of cellular functions, including protein stability, protein-protein interactions, and the recognition of DNA by proteins [1]. Although this might be regarded as an oversimplification, the acetylation and deacetylation could be considered as a molecular “on-off” switch. Indeed, in the diverse molecular complexes involving the deacetylases, the reversible acetylation may have either positive or negative effect, depending on the gene involved [2].
STRUCTURAL AND FUNCTIONAL DIVERSITIES OF HISTONE DEACETYLASES
A major breakthrough in the characterization of histone deacetylases came with the cloning of HDAC1 encoding gene [3]. Further studies revealed the existence of a large family of proteins in higher eukaryotes similar to the yeast RPD3 protein, a known factor involved in gene transcription regulation [4], suggesting for the first time the link between the histone deacetylation and transcriptional control. This large family comprises HDAC1, HDAC2, HDAC3, and HDAC8 exhibiting high sequence identity and similar domain organization, and thus grouped as class I HDACs. Given that the yeast cells express another histone deacetylase, a complex with the active part carried by the HDA1 catalytic subunit [5] and the search for homologs in vertebrates allowed the identification of HAD1-like proteins, first in mouse: mHDA1 and mHDA2 [6], then in human: HDAC4, HDAC5, HDAC6, HDAC7, and HDAC9 [7–9]. Therefore, this family of enzymes was grouped as class II deacetylases. A third class of deacetylases (class III) comprises the SIR2 (silent information regulator)-like family of NAD-dependent deacetylases [1]. The SIR2 gene family members have been cloned from a variety of species ranging from bacteria to man indicating a high degree of conservation throughout evolution [10]. SIR2 was firstly described in budding yeasts to be involved with SIR3 and SIR4 proteins in transcriptional repression, or silencing, by modulating chromatin structure at mating-type loci (HML and HMR) and subtelomeric regions [11].
The protein encoded by the SIR2 gene belongs to a highly conserved family of closely related proteins in both prokaryotic and eukaryotic species named Hst proteins (Homologous of Sir two) or sirtuins. Based on their primary structure, the family could be divided into five classes [12]. The yeast Saccharomyces cerevisiae has, in addition to the founding member SIR2, four homologs (Hst 1–4) belonging to class I proteins. Eight sirtuins have been identified in human (SIRT1-8) [12, 13]. Human SIR-T1, SIR-T2, and SIR-T3 belong to class I, SIR-T4 is in class II, SIR-T5 is in class III, and SIR-T6 and SIR-T7 are in class IV sirtuins. SIR-T8 recently identified in thyroid carcinoma cell lines and tissues [13] shared 85% identity in the core sirtuin domain to the SIR-T7, and thus could be included in class IV. Several bacteria species have also sirtuins being all of class III. An intermediate class between classes II and III or classes I and IV is designed class U which comprises few gram-positive bacterial and Thermatoga maritima sirtuins [12].
Transcriptional repression is directly associated with the recruitment of multiprotein complexes containing histone deacetylases. The Yin Yan 1 (YY1), Mad/Max heterodimer, and the nuclear hormone receptors represent paradigms of transcriptional repressors [14]. The YY1 has been shown to interact with class I HDACs in vitro and in vivo. The HDAC1 and HDAC2 were immunoprecipitated with a mouse protein mSin3a which is known to bind to Mad. Other studies have shown that Sin3a-HDAC complex contains multiple components including histone-binding proteins. HDAC1 and HDAC2 were also found in association with multiprotein complexes known as NuRD/Mi2/NRD [15].
The HDAC3 has a distinguishable feature compared to HDAC1 and HDAC2 that is the shuttling between the nucleus and the cytoplasm [16]. The enzyme was not found in association with either Sin3/HDAC or NuDR/Mi2/NDR complexes; instead, its function seems to be linked to the activity of multiprotein complexes containing the nuclear corepressor (NCoR) [17]. Although some of the functions of class II deacetylases (HDAC4, HDAC5 and HDAC7) seem to be shared with class I members, they are not found in association with the two well-characterized deacetylase complexes: Sin3/HDAC and NuRD/Mi2/NRD. Furthermore, a particular characteristic of all members of class II HDACs is their nucleocytoplasmic shuttling.
Unlike the other class I and class II HDACs, sirtuins require NAD as a cofactor. The SIR2 protein of yeast is the founding member of a large family of NAD-dependent HDACs. In yeast, the SIR2 gene is involved in the regulation of transcriptional silencing, DNA damage responses, and lifespan extension mediated by caloric restriction [18, 19]. The mammalian SIRT1 gene encodes an NAD-dependent nuclear HDAC that is the closest structural ortholog of yeast SIR2 protein [12]. However, the mammalian SIRT1 has been shown to deacetylate a wide range of nonhistone substrates comprising TAF168, PCAF, p300, MyoD, p53, and Ku70 [20–23]. Furthermore, recent investigations have shown that SIRT1 mediates the deacetylation of RelA/p65, a subunit of the NF-κB transcription factor that regulates a wide range of cellular processes [24]. SIRTs 2 and 3 are cytoplasmic proteins. SIRT2 has been shown to play a role in tubulin deacetylation, whereas SIRT3 is a mitochondrial protein whose substrate (s) is not yet identified [25–27]. Therefore, it is possible that the actual extensive investigations focused on SIR2-related family of proteins will uncover new biochemical pathways, and thus new targets of these enzymes.
Histone deacetylases in cancer diseases
Specific events-driven changes that led to the cancer development and progression are interconnect complex molecular modifications including DNA methylation, histone acetylation, phosphorylation, ubiquitylation, and ADP ribosylation. The p53 protein is among the factors that play pivotal role in gene expression regulation and cell signaling pathways' network that govern the cell decision for life or death. Mutation of p53 correlates with many forms of cancer [28], and mice that lack p53 develop normally but rapidly succumb to cancers [29]. Moreover, low levels of p53 and increased production of Mdm2 protein, the negative regulator of p53, have been shown to be associated with increased cell growth and proliferation [30]. Furthermore, recent investigations have reported that overexpression of p53 in cisplatin-treated tumors might be associated with cell resistance to apoptosis [31]. In biopsies of involved psoriatic skin, decreased expression of p53 could be seen when compared to uninvolved skin areas, suggesting that the loss of growth-inhibitory activity due to downregulation of p53 is responsible for the keratinocyte hyperproliferation characteristic of the disease [32].
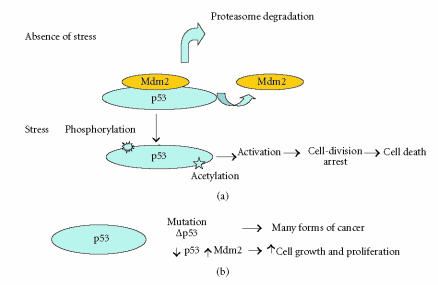
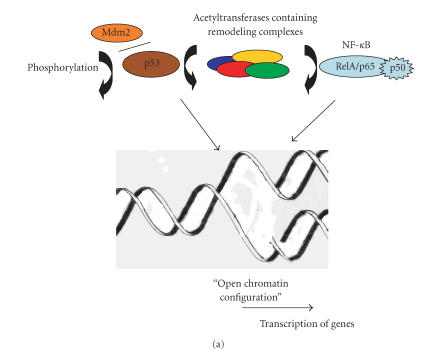
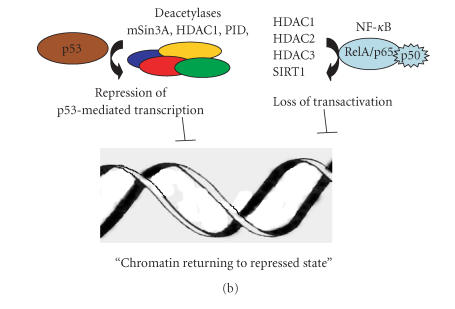
At the cellular level, in the absence of stress, p53 is not phosphorylated and linked to Mdm2 which directed the protein to the proteasome degradation pathway (Figure 1), a number of posttranslational modifications among which acetylation and deacetylation play a role in the activation of p53 [33]. Under stress conditions, its phosphorylation and release from Mdm2 and further acetylation led to its activation, inducing checkpoints in the cell-division cycle, permanent cell-division arrest (senescence), and cell death (Figure 1). The activity of acetylase remodeling complex-associated p53 may lead to “open chromatine” configuration and binding of transcriptional factors and mediators (Figure 2(a)). However, there are increasing lines of evidences indicating that deacetylases can contribute in the regulation of p53 activity. Indeed, the transcriptional repression of certain genes has been shown to involve p53 association with other molecular complexes containing mSin3A (a corepressor), a factor termed PID identical to metastasis-associated protein 2 (MTA2), and HDAC1 (Figure 2(b)). Furthermore, recognition of substrates other than histones by SIR2 family of proteins has broad implications in important biological processes. In fact, the ability of class III deacetylases to physically interact and modify the function and biological properties of proteins such as p53 [34, 35] and tubulin [36] has made them a paradigm for the investigation of new biochemical pathways involved in tumoregenesis. Thus, the selective binding of acetylases or HDAC might determine whether p53 will mediate the activation or repression of transcription (Figures 2(a) and 2(b)). Therefore, on account of their interaction with various molecular complexes involved in gene expression, HDACs have captured the attention of a large number or researchers.
Figure 1.
Schematic representation of some modifications affecting p53 leading to cell growth and proliferation or death. (a) Under stress conditions, phosphorylation of p53 and release from Mdm2 and further acetylation led to p53 activation; (b) mutations of p53 and the modifications in the levels of p53/Mdm2 might affect cell growth and proliferation.
Figure 2.
Roles of acetylation/deacetylation of p53 and NF-κB in transcriptional regulation. (a) Interaction of p53 with acetylases facilitates alteration of chromatine structure by histone acetylation and association of transcription complex machinery; p53 is itself subject to acetylation. NF-κB becomes attached to acetyltransferases containing complexes facilitating an “open chromatine” configuration and the binding of transcription factors and RNA polymerase II. (b) Deacetylases regulate p53 and NF-κB- mediated transcription, possibly by inducing a “repressed chromatin” configuration.
The potential implication of HDAC in tumorigenesis has been well documented [25]. A number of reports have shown HDACs to be overexpressed in various tumor cells [37, 38]. HDACs were also found to be overexpressed under certain circumstances such us hypoxia, hypoglycemia, and serum deprivation [39]. In the case of class III deacetylase members, there are several recent reports showing either up- or downregulation of SIRTs genes. For instance, isolation of an SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma and cell lines but not in adenomas has been recently reported [13]. Unexpectedly, in contrast to most SIR proteins which may be part of the telomere/telomerase functional complex, which is localized to the nucleolus [40], SIR-T8 is in the cytoplasm under the nuclear membrane. Moreover, in the case of human gliomas among differentially expressed genes evidenced by a proteomics-based approach, the SIRT2 gene was shown to be downregulated [26] suggesting that SIRT2 may serve as a novel molecular marker for gliomas. Interestingly, ectopic expression of SIRT2 gene induced cell-growth suppression in glioma cell lines having barely detectable endogenous SIRT2 levels. These observations allowed the authors to suggest SIRT2 being a tumor suppressor gene in glioma cells through tubulin acetylation and consequently microtubule organization.
Other important targets of HDACs are members of the nuclear factor Rel/NF-κB family known to be made of a plethora of transcriptional regulators sharing the so-called Rel homology domain [41]. In fact, NF-κB has been shown to be activated in the early malignant transformation of mammary cells of the breast [42]. Moreover, NF-κB is constitutively active in pancreatic adenocarcinoma in humans [43], in T leukemia lymphocytes [44], in human breast cancer [45], and in head and neck squamous carcinoma cell lines [46], therefore facilitating cell survival and tumor progression by activating biochemical pathways leading to the inhibition of apoptosis process. Once in the nucleus, the NF-κB associates with histone acetyltransferases containing complexes [47]. Acetylation of NF-κB RelA/p65 and p50 subunits affects both its DNA binding ability and transcriptional activity [48, 49] (Figure 2(a)). Deacetylation of NF-κB RelA/p65 subunit by HADC1, HDAC2 [50], or HDAC3 [51] induced increased IκBα association leading to loss of NF-κB transactivation capacity (Figure 2(b)). Moreover, recent investigations have shown that SIRT1, a member of class III deacetylases, represses NF-κB function by deacetylating the RelA/p65 lysine 310 [52], regulating therefore its antiapoptotic activity. Thus, HDACs seem to be key elements in the regulation of gene expression, differentiation and development, and the maintenance of homeostasis. Their association with a number of cellular oncogenes and tumor suppressor genes make them candidate targets for anticancer drugs and therapies.
Histone deacetylase inhibitors and cancer therapy
The HDAC inhibitors (HDIs) by releasing trancriptional repression-induced cell cycle arrest, differentiation, and/or apoptosis of different tumors are an interesting strategy for antitumor chemotherapy development [27]. The p53 may play an important role in HDIs-mediated growth inhibition. Indeed, it has been shown that although the sodium butyrate (NaB) induced the inhibition of DNA synthesis both in the presence or absence of p53, the levels of cell detah by apaoptosis are greatly reduced in the absence of p53 [53]. Nevertheless, there are other mechanisms not involving p53 by which HDIs function. For instance, in the absence of p53, the trichostatin A (TSA), a natural product isolated from Streptomyces hygroscopicus, used initially as an antifungal antibiotic, causes cell cycle arrest at the G2/M phase transition leading to cell death by apoptosis [54]. Furthermore, fusion proteins that utilize HDACs have been implicated in many acute leukemias [55]. In fact, in normal conditions and in the absence of retinoic acid (RA), retinoic acid receptor α (RARα) binds to DNA as a heterodimer with an RXR (retinoid-X receptor) in complex with transcriptional corepressor containing NcoR, Sin3A, and HDAC. The presence of RA induced the displacement of the corepressor complex which is replaced by a coactivator complex containing HAT (p300/CBP and PCAF). In certain acute promyelocytic leukemia (APL), a chromosomal translocation of t(11;17) fuses a large portion of the RARα to a coding sequence of a promyelocytic leukemia zinc finger (PLZF) protein rendering the RARα insensitive to physiological levels of RA leading to the blockage of hematopoietic cells differentiation and their proliferation without restraint [56]. The TSA is the most potent of the known HDACs class I and class II inhibitors that restore the capacity of leukemic cells carrying PLZF-RARα to respond to pharmacological doses of all-trans-retinoid acid [56–58]. Thus, HDIs by interfering with fusion protein-induced blockage of differentiation of hematopoietic cells relieve the repression of differentiation gene transcription allowing the cells to respond to RA. Based on the structure of TSA, a large number of synthetic drugs were designed. They possess a “cap” group, a central aliphatic chain, and a functional group that would chelate the zinc cation in the active site in HDAC. Various compounds have been tested in cell cultures and in preclinical murine models, several are currently under phase I/II clinical trials [59]. Hydroxamic acid derivatives represent efficient inhibitors of both HDAC class I and class II enzymes, with activities in vitro detected at nanomolar to micromolar concentrations. One of the potential problem is their reversible activity towards class I and class II enzymes. For instance, TSA, one of the first HDIs containing a hydroxamic functional group, inhibits the growth of pancreatic adenocarcinoma cells and induced ACHN renal carcinoma cell cycle arrest. Suberoylanilide hydroxamic acid and pyroxamide are compounds inducing growth arrest, differentiation and/or apoptosis in a wide range of tumor cells. A number of natural products such as apicidin, a cyclic tetrapeptide generated from the endophytic fungi, Fusarium pallidoroseum, are able to inhibit HDACs and the growth of tumor cells. In fact, a number of genes that are consistently upregulated have their promoters associated with acetylated histones. One such gene encodes the cyclin-dependent kinase (CDK) inhibitor p21WAF1, which inhibits cell cycle progression by blocking CDK activity and arrests the cell cycle in G1 phase. Most of the HDIs induce expression of p21, and apicidin had also similar effect being therefore able to block tumor cell proliferation [59]. Moreover, a compound, the FK228 a bicyclic depsipeptide derived from Chromobacterium violaceum, inhibits HDACs at nanomolar concentrations and exerts potent antitumor activity by inducing apoptosis cell death. Moreover, the FK228-induced HDAC inhibition led to increased expression of angiogenic-inhibitory factor encoding gene and suppression of gene encoding the angiogenic-stimulating factor interfering therefore with the neovascularization required for certain tumor growth.
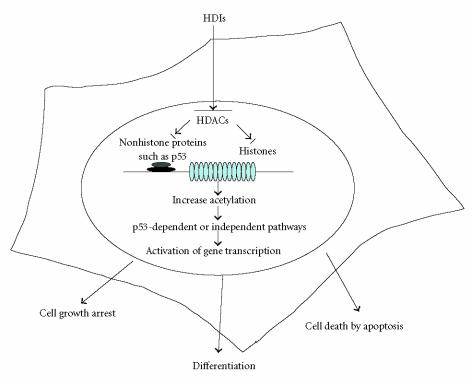
Thus, most of these HDIs act by interfering with cell cycle, differentiation, and apoptosis pathways depending on the nature of tumor cells (Figure 3). In fact, key regulators such as NF-κB as described above are target of HDACs. For instance, a link between inflammation and susceptibility to cancer has been already evidenced in population studies [60]. In precancerous cells, NF-κB by increasing the expression of proinflammatory and survival genes and inhibiting death-promoting pathway may enhance premaligant potential. HDIs by preventing NK-κB transactivation may enable the reexpression of silenced genes in tumor cells leading to the reversion of the malignant phenotype exerting therefore a therapeutic effect.
Figure 3.
A schematic model for HDIs mode of action. Complex molecular interactions involve acetylation and deacetylation events of histone in the vicinity of promoters and of nonhistone proteins, such as transcription factor p53. Acetylation and deacetylation affect cell cycle transition through regulation of proteins such as p21WAF1 and cyclin B1. The pathways could be dependent on or independent of p53.
It is also noteworthy that HDIs could potentiate the effect of antitumor drugs. Indeed, it has been shown that coincubation of human leukemia cells (K562) with a proteasome inhibitor, the bortezomid, and HDIs (suberoylanilide hydroxamid acid or sodium butyrate) resulted in increase of mitochondrial injury and apoptosis [61]. Moreover, pretreatment of human leukemic cells with MS-275, an orally active synthetic benzamidine derivative that belongs to HDIs, significantly enhanced the activity of the cytotoxic drug, Fludarabine, a purine analogue that has demonstrated significant activity in B-cell malignancies [62].
The enzymatic reaction of sirtuins involved nicotine adenine dinucleotide (β-NAD) and the N-acetylated substrate. The final reactions products comprise the deacetylated product, the nicotinamide (Nam), and 2′-O-acetyl-ADP-ribose. A feedback regulation of sirtuin activity is exerted by Nam. Indeed, Nam has been reported to be a physiological inhibitor of certain deacetylase SIR2 enzymes such as the yeast (ySIR2) and human SIRT1 [63]. Moreover, it is noteworthy that Nam was able to reduce inflammation in the case of patients suffering from osteoarthritis [64] and it is in trials as a therapy to prevent cancer recurrence and insulin-dependent (type I) diabetes [65].
Parasitic histone deacetylases
Although the cancer is not a single disease, the growth of the parasites in their host and that of cancer cells share at least one common feature that is their mutual capacity for rapid cell division. Moreover, we have recently attempted to delineate some similarities between the immune evasion strategies that parasites and tumors employ [66]. The protozoan parasites which cause important diseases in human and a number of other mammalian species have complex life cycles including different developmental stages. Most of these parasites are transmitted by insect vectors and invade a range of different tissues in their mammalian hosts. These series of transitions involve major modifications in morphology, gene expression, and cell cycle in order to survive environmental extremes such as toxic metabolites and acidic pH of the host cell phagolysosomes. Therefore, cells at different periods of the life cycle may differ significantly. Gene expression determines the state of differentiation and has a role in the regulation of activity of a given parasite form under a physiological state. The importance of HAT and HDACs in the life cycle of pathogenic protozoan parasites is only partially emerging. Thus, recent investigations have shown that like other eukaryotes, the protozoan parasites possess HATs and HDACs orthologues and the reversible acetylation seems to play a key role in gene transcriptional regulation and cell cycle progression.
Indeed, the African trypanosome, Trypanosoma brucei, which grows in the mammalian vasculature and causes sleeping sickness in humans and Nagana in cattle, expresses four HDACs ortologues named DAC1–4 [67]. The DAC1 and DAC2 share significant similarity with mammalian and yeast class I HDACs, whereas DAC3 and DAC4 are closely related to class II deacetylases. Despite the fact that none of the four genes were present among the > 5000 T brucei expressed sequence tags, RT-PCR analysis showed that the four genes were expressed in bloodstream parasites as well as in insect developmental stages. Functional analysis of DAC1–4 genes by reverse genetic approach showed that DAC1 and DAC3 appear to be essential, whereas DAC2 and DAC3 are not required for parasite viability. In a more recent study, an unusual member of the SIR2 family, the TbSIR2RP1, with both histone NAD-dependent deacetylase and ADP-ribosyltransferase activities, has been identified in T brucei [68]. The parasite resistance to DNA damage correlates with the level of TbSIR2RP1 expression. The authors suggest that histone modification (deacetylation/ribosylation) by TbSIR2RP1 is involved in DNA repair.
In the case of Plasmodium falciparum, a member of the apicomplexan protozoan family which causes malaria, one of the leading causes of loss of life worldwide accounting for over 1.5 million deaths annually, a gene encoding a protein sharing significant homology to yeast, human, and other eukaryotic HDACs, and thus termed PfHDAC1, has been characterized [69]. The PfHDAC1 mRNA transcript was found to be predominantly expressed in mature asexual blood stages and in gametocytes and the ≅ 50 kd PfHDAC1 protein localized in the P falciparum nucleus. Furthermore, an expressed sequence tag for a yeast RPD3 homologue has been identified in the genome of Cryptosporidium parvum, a protozoan parasite having biochemical and cell biological peculiarities but sharing a number of morphological and biochemical features with other apicomplexa [70]. Moreover, in recent investigations, a P falciparum protein homologous to the yeast SIR2 called PfSir2 was found to play a role in chromatin modification at telomeres [71]. Indeed, the data reported showed that PfSir2 protein localized to telomeric foci and the nucleolus and suggested that the protein generates a hypoacetylated state of chromosome ends, including subtelomeric var genes, that represses gene activity.
The Entamoeba histolytica, an intestinal parasite that infects about 50 million people worldwide, has also been shown to express both HAT and HDAC activities [72]. Parasite proteins ehGCN5 and ehMYST carrying a signature of acetytranferase domain (GNAT motif, ehGCN5) and a carboxy-proximal MOZ-SAS domain including a zinc finger (the signature of a MYST family) have been characterized. The HAT assays showed that the ehMYST protein produced in E coli was able to incorporate [3H]; from [3H] acetyl-CoA into calf thymus histones and standard assays revealed the presence of HDAC activity in E histolytica nuclear extracts. Moreover, a single putative gene encoding a member of class I family named ehHDAC, was characterized at the molecular level, although a functional native or recombinant protein has not been reported.
Leishmania spp, the causative agents of leishmaniasis, a group of several different diseases widely distributed in tropical and subtropical areas and also are commonly found in the Mediterranean basin, are kinetoplastid protozoan transmitted by a blood-feeding dipteran vector of the subfamily Phlebotominae. These parasites have complex life cycles that include different morphological forms. Within the insect vector, the parasite replicates as a noninfective promastigote, which transforms into infective metacyclic promastigote. In the mammalian host, the infective promastigotes invade the macrophages and differentiate into amastigotes, which are the proliferative forms within the vertebrate hosts.
In an earlier report, we described the molecular characterization of several L major proteins eluted from glutathione-agarose column [73]. Further studies allowed to show that one member of this group is among members of SIR2 family and thus was termed LmSIR2 [74]. A gene encoding homologous protein in L infantum, exhibiting NAD+-dependent deacetylase activity, was sequenced and termed LiSIR2 [75] and other related sequences can be found in the Leishmania genome database [L major sirtuin (CAB55543), L major cobB (LmjF34.2140)]. The expectation was to find the LmSIR2 associated with the nucleus, instead, the protein had a cytoplasmic localization [76]. However, further independent investigations revealed the presence of SIR2-related proteins in the cytoplasm of different species, and in the mitochondria [11]. Interestingly, L infantum amastigotes, the vertebrate stage of the parasite, carrying extracopies of LmSIR2 or L infantum SIR2 (LiSIR2) gene, when maintained under normal axenic culture conditions, showed striking increase in the survival due to an inherent resistance to apoptosis-like death, leading to a longer stationary phase of growth [75]. In a more recent study, it has been demonstrated that although L infantum tolerates deletion of one wild-type LiSIR2 allele, deletion of both genes is a lethal event, indicating that at least one active LiSIR2 gene is required for parasite survival. Monoallelic disruption of LiSIR2 (LiSIR2+/−) resulted in the obtention of L infantum lines with significantly reduced virulence in vitro and in vivo [77]. Although it remains to be demonstrated that a protein with a cytoplasmic localization could influence nuclear silencing events, it is reasonable to assume that inhibiting parasite SIR2 activity may represent a mean to interfere with parasite development.
Histone deacetylase inhibitors interfere with parasite development
Several studies using various pharmacological HDIs and different parasite species have concluded that HDACs play a critical role in the life cycle of theses organisms. It has been shown, for example, that apicidin exerted an antiparasitic activity against the apicomplexan family of protozoa, including P falciparum, C parvum, Toxoplasma gondii, Sarcocystis neurona, and Eimeria tenella [78]. The antiparasitic activity of apicidin appears to be attributable to inhibition of apicomplexa HDAC at nanomolar concentrations. Furthermore, recent investigations conducted on Neospora caninum, a protozoan parasite closely related to T gondii, that infects a wide range of vertebrate hosts (ie, cattle, sheep, goats, horses, and dogs) have shown that apicidin exerted antiparasitic activity through the inhibition of HDACs [79]. Another structurally unrelated compound, depucidin, a natural product isolated from Altermaria brassiciola, when used in an in vitro culture system at a defined concentration, inhibited significantly the intracellular multiplication of N caninum tachyzoites without exerting any cytotoxicity toward the host Vero cells. The inhibitory effect seems to be related to the parasite HDAC inhibition. Moreover, the amebic trophozoites ehHDAC activity could be completely inhibited in the presence of low concentration of the HDI inhibitor TSA, a feature shared by the classical HDAC enzymes.
In more recent studies, it has been reported that Sirtinol, a commercially available inhibitor of SIR2 deacetylases, induced the death of L infantum amastigotes, the vertebrate stage of the parasites; whereas, the promastigotes, the insect stage forms, were insensitive [80] . Interestingly, amastigotes which overexpress SIR2 were less sensitive to the drug. This might be related to stage-specific requirement for functional SIR2 enzyme. Indeed, molecular biological manipulation of L infantum SIR2 gene by homologous recombination allowed to show that single LiSIR2 gene disruption (LiSIR2+/−) does not affect the growth of parasite in the promastigote form, whereas axenic amastigotes display a marked reduction in their capacity to multiply in vitro inside macrophages and in vivo in Balb/c mice [77]. Furthermore, Nam, a physiological inhibitor of certain deacetylase SIR2 enzymes, has been shown to exert an antimicrobial and antiviral activities against Mycobacterium tuberculosis and human immunodeficiency virus, respectively [81]. In addition, recent investigations have also demonstrated that Nam could express antiparasitic activity in vitro on various Leishmania species [82]. However, although Nam could inhibit NAD-dependent deacetylase activity contained in the extracts from parasites overexpressing SIR2 enzyme, the mutant parasites were as sensitive to Nam activity as the parental wild-type clone, suggesting therefore that Nam may interfere with other parasite metabolic pathways in addition to those involving NAD-dependent SIR2 deacetylase activity.
Another aspect to be taken with care is the possible modulation of host cell transcription machinery by the parasite itself. One fascinating example is that of T cruzi and host cell NF-κB. The NF-κB plays an important role in the coordination of innate and acquired immune cellular responses during microbial infections. T cruzi has a marked tissue tropism with preference for infection of myocytes. It has been demonstrated that NF-κB activation plays a role in controlling intracellular infection in nonimmune cells [83]. Indeed, trypomastigotes, the vertebrate host infective stage of T cruzi, activate nuclear translocation and DNA binding of NF-κB p65 subunit and NF-κB-dependent gene expression in epithelial cells, endothelial cells, and fibroblasts. Inducible expression of the inhibitory mutant IκBαm leading to the inactivation of epithelial cell NF-κB induced increased parasite invasion. In contrast, T cruzi does not activate NF-κB in cells derived from skeletal, smooth, or cardiac muscle resulting in increased level of cell infection. Therefore, interfering with NF-κB activation through the use of indiscriminate HDIs may have dramatic consequences on the level of T cruzi invasion.
CONCLUDING REMARKS
The dynamic remodeling of chromatin is a complex process that governs gene expression relevant to cell division, differentiation, or death. Many molecular details are emerging and shed light on the key role of the deacetylase enzymes in the modification of chromatin structure leading to altered accessibility of certain genes. Given the degree of homology between members of this large family of enzymes and the putative redundancy of their functions, pharmacological inhibitors may have limited selectivity or specificity leading to a functional consequence which is the toxicity against normal tissues. However, studies of enzymatic activities of members of the same family, for instance human sirtuins, using histone H4 peptide, revealed low or undetectable activity [84]. This could reflect biochemical peculiarities (ie, substrate specificities, cofactors requirement) features that perhaps may be exploitable for drug design. The resolution of the catalytic core structures of a number of enzymes and their substrate-bound complex would allow the design of specific inhibitors with potential therapeutic efficacy in cancer and parasitic diseases.
ACKNOWLEDGMENTS
The first author is a Surgeon supported by the Service de Chirurgie Digestive et Générale, Hôpital Sainte Marguerite, Centre Universitaire de Marseille, France, and the second author is a Head of Research at Institut National de la Santé et de la Recherche Madicale (INSERM) and a Manager of IRD Research Unit no. 008.
References
- 1.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? The EMBO Journal. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun ZW, Hampsey M. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics. 1999;152(3):921–932. doi: 10.1093/genetics/152.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272(5260):408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 4.Leipe DD, Landsman D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Research. 1997;25(18):3693–3697. doi: 10.1093/nar/25.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(25):14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. Journal of Biological Chemistry. 1999;274(4):2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 7.Gray SG, Ekstrom TJ. The human histone deacetylase family. Experimental Cell Research. 2001;262(2):75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 8.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(9):4868–4873. [Google Scholar]
- 9.Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Development. 1995;9(23):2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 11.Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279(1):1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- 12.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochemical and Biophysical Research Communications. 2000;273(2):793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 13.de Nigris F, Cerutti J, Morelli C, et al. Isolation of a SIR-like gene, SIR-T8, that is overexpressed in thyroid carcinoma cell lines and tissues. British Journal of Cancer. 2002;86(6):917–923 . doi: 10.1038/sj.bjc.6600156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. Journal of Cellular Physiology. 2000;184(1):1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99(5):447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 16.Takami Y, Nakayama T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. Journal of Biological Chemistry. 2000;275(21):16191–16201. doi: 10.1074/jbc.M908066199. [DOI] [PubMed] [Google Scholar]
- 17.Underhill C, Qutob MS, Yee S-P, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. Journal of Biological Chemistry. 2000;275(51):40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 18.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes & Development. 2000;14(9):1021–1026. [PubMed] [Google Scholar]
- 19.Lin S-J, Kaeberlein M, Andalis AA, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418(6895):344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 20.Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. Journal of Leukocyte Biology. 2004;75(6):939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 21.Fulco M, Schiltz RL, Iezzi S, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Molecular Cell. 2003;12(1):51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 22.Cohen HY, Lavu S, Bitterman KJ, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Molecular Cell. 2004;13(5):627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 23.Motta MC, Divecha N, Lemieux M, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 24.Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO Journal. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristeleit R, Stimson L, Workman P, Aherne W. Histone modification enzymes: novel targets for cancer drugs. Expert Opinion on Emerging Drugs. 2004;9(1):135–154. doi: 10.1517/eoed.9.1.135.32947. [DOI] [PubMed] [Google Scholar]
- 26.Hiratsuka M, Inoue T, Toda T, et al. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochemical and Biophysical Research Communications. 2003;309(3):558–566. doi: 10.1016/j.bbrc.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Pandolfi PP. Histone deacetylases and transcriptional therapy with their inhibitors. Cancer Chemotherapy and Pharmacology. 2001;48(suppl 1):S17–S19. doi: 10.1007/s002800100322. [DOI] [PubMed] [Google Scholar]
- 28.Kaghad M, Bonnet H, Yang A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90(4):809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 29.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 30.Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407(6805):777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama K, Takebayashi Y, Nakayama S, et al. Prognostic value of overexpression of p53 in human ovarian carcinoma patients receiving cisplatin. Cancer Letters. 2003;192(2):227–235. doi: 10.1016/s0304-3835(02)00686-9. [DOI] [PubMed] [Google Scholar]
- 32.Michel G, Auer H, Kemény L, Böcking A, Ruzicka T. Antioncogene P53 and mitogenic cytokine interleukin-8 aberrantly expressed in psoriatic skin are inversely regulated by the antipsoriatic drug tacrolimus (FK506) Biochemical Pharmacology. 1996;51(10):1315–1320. doi: 10.1016/0006-2952(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 33.Prives C, Manley JL. Why is p53 acetylated? Cell. 2001;107(7):815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 34.Vaziri H, Dessain SK, Eaton EN, et al. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 35.Luo J, Nikolaev AY, Imai S-I, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 36.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Molecular Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 37.Hu E, Chen Z, Fredrickson T, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. Journal of Biological Chemistry. 2000;275(20):15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 38.Yang W-M, Yao Y-L, Sun J-M, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. Journal of Biological Chemistry. 1997;272(44):28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 39.Kim MS, Kwon HJ, Lee YM, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nature Medicine. 2001;7(4):437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 40.Morales CP, Holt SE, Ouellette M, et al. Absence of cancerassociated changes in human fibroblasts immortalized with telomerase. Nature Genetics. 1999;21(1):115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- 41.Karin M, Cao Y, Greten FR, Li Z-W. NF-κB in cancer: from innocent bystander to major culprit. Nature Reviews Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 42.Kim DW, Sovak MA, Zanieski G, et al. Activation of NF-κB/Rel occurs early during neoplastic transformation of mammary cells. Carcinogenesis. 2000;21(5):871–879. doi: 10.1093/carcin/21.5.871. [DOI] [PubMed] [Google Scholar]
- 43.Wang W, Abbruzzese JL, Evans DB, Chiao PJ. Overexpression of urokinase-type plasminogen activator in pancreatic adenocarcinoma is regulated by constitutively activated RelA. Oncogene. 1999;18(32):4554–4563. doi: 10.1038/sj.onc.1202833. [DOI] [PubMed] [Google Scholar]
- 44.Mori N, Nunokawa Y, Yamada Y, Ikeda S, Tomonaga M, Yamamoto N. Expression of human inducible nitric oxide synthase gene in T-cell lines infected with human T-cell leukemia virus type-I and primary adult T-cell leukemia cells. Blood. 1999;94(8):2862–2870. [PubMed] [Google Scholar]
- 45.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-κB subunits in human breast cancer: potential roles for NF-κB2/p52 and for Bcl-3. Oncogene. 2000;19(9):1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 46.Ondrey FG, Dong G, Sunwoo J, et al. Constitutive activation of transcription factors NF-κB, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express proinflammatory and pro-angiogenic cytokines. Molecular Carcinogenesis. 1999;26(2):119–129. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 47.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/ p300 or HDAC-1. Molecular Cell. 2002;9(3):625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 48.Chen L-F, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. The EMBO Journal. 2002;21(23):6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiernan R, Brès V, Ng RWM, et al. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. Journal of Biological Chemistry. 2003;278(4):2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 50.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Molecular and Cellular Biology. 2001;21(20):7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L-F, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293(5535):1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 52.Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO Journal. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joseph J, Wajapeyee N, Somasundaram K. Role of p53 status in chemosensitivity determination of cancer cells against histone deacetylase inhibitor sodium butyrate. International Journal of Cancer. 2005;115(1):11–18. doi: 10.1002/ijc.20842. [DOI] [PubMed] [Google Scholar]
- 54.Hirose T, Sowa Y, Takahashi S, et al. p53-independent induction of Gadd45 by histone deacetylase inhibitor: coordinate regulation by transcription factors Oct-1 and NF-Y. Oncogene. 2003;22(49):7762–7773. doi: 10.1038/sj.onc.1207091. [DOI] [PubMed] [Google Scholar]
- 55.Fenrick R, Hiebert SW. Role of histone deacetylases in acute leukemia. Journal of Cellular Biochemistry. Supplement. 1998;72(S30-31):194–202. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<194::AID-JCB24>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 56.Grignani F, De Matteis S, Nervi C, et al. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391(6669):815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 57.He L-Z, Guidez F, Tribioli C, et al. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nature Genetics. 1998;18(2):126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 58.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391(6669):811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 59.Acharya MR, Sparreboom A, Venitz J, Figg WD. Rational development of histone deacetylase inhibitors as anticancer agents: a review. Molecular Pharmacology. 2005;68(4):917–932. doi: 10.1124/mol.105.014167. [DOI] [PubMed] [Google Scholar]
- 60.Balkwill F, Coussens LM. Cancer: an inflammatory link. Nature. 2004;431(7007):405–406. doi: 10.1038/431405a. [DOI] [PubMed] [Google Scholar]
- 61.Yu C, Rahmani M, Conrad D, Subler M, Dent P, Grant S. The proteasome inhibitor bortezomib interacts synergistically with histone deacetylase inhibitors to induce apoptosis in Bcr/Abl+ cells sensitive and resistant to STI571. Blood. 2003;102(10):3765–3774. doi: 10.1182/blood-2003-03-0737. [DOI] [PubMed] [Google Scholar]
- 62.Maggio SC, Rosato RR, Kramer LB, et al. The histone deacetylase inhibitor MS-275 interacts synergistically with fludarabine to induce apoptosis in human leukemia cells. Cancer Research. 2004;64(7):2590–2600. doi: 10.1158/0008-5472.can-03-2631. [DOI] [PubMed] [Google Scholar]
- 63.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. Journal of Biological Chemistry. 2002;277(47):45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 64.Jonas WB, Rapoza CP, Blair WF. The effect of niacinamide on osteoarthritis: a pilot study. Inflammation Research. 1996;45(7):330–334. doi: 10.1007/BF02252945. [DOI] [PubMed] [Google Scholar]
- 65.Kaanders JHAM, Pop LAM, Marres HAM, et al. ARCON: experience in 215 patients with advanced head-and-neck cancer. International Journal of Radiation Oncology, Biology, Physics. 2002;52(3):769–778. doi: 10.1016/s0360-3016(01)02678-5. [DOI] [PubMed] [Google Scholar]
- 66.Ouaissi A, Ouaissi M. Molecular basis of Trypanosoma cruzi and Leishmania interaction with their host(s): exploitation of immune and defense mechanisms by the parasite leading to persistence and chronicity, features reminiscent of immune system evasion strategies in cancer diseases. Archivum Immunologiae et Therapiae Experimentalis. 2005;53(2):102–114. [PubMed] [Google Scholar]
- 67.Ingram AK, Horn D. Histone deacetylases in Trypanosoma brucei two are essential and another is required for normal cell cycle progression. Molecular Microbiology. 2002;45(1):89–97. doi: 10.1046/j.1365-2958.2002.03018.x. [DOI] [PubMed] [Google Scholar]
- 68.García-Salcedo JA, Gijón P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. The EMBO Journal. 2003;22(21):5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joshi MB, Lin DT, Chiang PH, et al. Molecular cloning and nuclear localization of a histone deacetylase homologue in Plasmodium falciparum. Molecular and Biochemical Parasitology. 1999;99(1):11–19. doi: 10.1016/s0166-6851(98)00177-7. [DOI] [PubMed] [Google Scholar]
- 70.Strong WB, Nelson RG. Preliminary profile of the Cryptosporidium parvum genome: an expressed sequence tag and genome survey sequence analysis. Molecular and Biochemical Parasitology. 2000;107(1):1–32. doi: 10.1016/s0166-6851(99)00225-x. [DOI] [PubMed] [Google Scholar]
- 71.Freitas LH, Jr, Hernandez-Rivas R, Ralph SA, et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121(1):25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 72.Ramakrishnan G, Gilchrist CA, Musa H, et al. Histone acetyltransferases and deacetylase in Entamoeba histolytica. Molecular and Biochemical Parasitology. 2004;138(2):205–216. doi: 10.1016/j.molbiopara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 73.Yahiaoui B, Loyens M, Taibi A, Schöneck R, Dubremetz JF, Ouaissi A. Characterization of a Leishmania antigen associated with cytoplasmic vesicles resembling endosomal-like structure. Parasitology. 1993;107(pt 5):497–507. doi: 10.1017/s0031182000068074. [DOI] [PubMed] [Google Scholar]
- 74.Yahiaoui B, Taibi A, Ouaissi A. A Leishmania major protein with extensive homology to silent information regulator 2 of Saccharomyces cerevisiae. Gene. 1996;169(1):115–118. doi: 10.1016/0378-1119(95)00785-7. [DOI] [PubMed] [Google Scholar]
- 75.Vergnes B, Sereno D, Madjidian-Sereno N, Lemesre J-L, Ouaissi A. Cytoplasmic SIR2 homologue overexpression promotes survival of Leishmania parasites by preventing programmed cell death. Gene. 2002;296(1-2):139–150. doi: 10.1016/s0378-1119(02)00842-9. [DOI] [PubMed] [Google Scholar]
- 76.Zemzoumi K, Sereno D, Francois C, Guilvard E, Lemesre JL, Ouaissi A. Leishmania major: cell type dependent distribution of a 43 kDa antigen related to silent information regulatory-2 protein family. Biology of the Cell. 1998;90(3):239–245. doi: 10.1016/s0248-4900(98)80020-8. [DOI] [PubMed] [Google Scholar]
- 77.Vergnes B, Sereno D, Tavares J, et al. Targeted disruption of cytosolic SIR2 deacetylase discloses its essential role in Leishmania survival and proliferation. Gene. 2005;363:85–96. doi: 10.1016/j.gene.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 78.Darkin-Rattray SJ, Gurnett AM, Myers RW, et al. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(23):13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwon HJ, Kim J-H, Kim M, Lee J-K, Hwang W-S, Kim D-Y. Anti-parasitic activity of depudecin on Neospora caninum via the inhibition of histone deacetylase. Veterinary Parasitology. 2003;112(4):269–276. doi: 10.1016/s0304-4017(03)00035-9. [DOI] [PubMed] [Google Scholar]
- 80.Vergnes B, Vanhille L, Ouaissi A, Sereno D. Stage-specific antileishmanial activity of an inhibitor of SIR2 histone deacetylase. Acta Tropica. 2005;94(2):107–115. doi: 10.1016/j.actatropica.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 81.Murray MF. Nicotinamide: an oral antimicrobial agent with activity against both Mycobacterium tuberculosis and human immunodeficiency virus. Clinical Infectious Diseases. 2003;36(4):453–460. doi: 10.1086/367544. [DOI] [PubMed] [Google Scholar]
- 82.Sereno D, Monte Alegre A, Silvestre R, Vergnes B, Ouaissi A. In vitro antileishmanial activity of Nicotinamide. Antimicrobial Agents and Chemotherapy. 2005;49(2):808–812. doi: 10.1128/AAC.49.2.808-812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hall BS, Tam W, Sen R, Pereira MEA. Cell-specific activation of nuclear factor-κB by the parasite Trypanosoma cruzi promotes resistance to intracellular infection. Molecular Biology of the Cell. 2000;11(1):153–160. doi: 10.1091/mbc.11.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Molecular Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]