Abstract
Dlx3 is a homeodomain transcription factor and a member of the vertebrate Distal-less family. Targeted deletion of the mouse Dlx3 gene results in embryonic death between day 9.5 and day 10 because of placental defects that alter the development of the labyrinthine layer. In situ hybridization reveals that the Dlx3 gene is initially expressed in ectoplacental cone cells and chorionic plate, and later in the labyrinthine trophoblast of the chorioallantoic placenta, where major defects are observed in the Dlx3 −/− embryos. The expression of structural genes, such as 4311 and PL-1, which were used as markers to follow the fate of different derivatives of the placenta, was not affected in the Dlx3-null embryos. However, by day 10.5 of development, expression of the paired-like homeodomain gene Esx1 was strongly down-regulated in affected placenta tissue, suggesting that Dlx3 is required for the maintenance of Esx1 expression, normal placental morphogenesis, and embryonic survival.
The development of the placenta begins with the formation of the specialized epithelial trophoblasts that will give rise to the ectoplacental cone and chorionic ectoderm. The remainder of the cells in the embryo segregate and constitute the inner cell mass that will form the embryo proper. During implantation in mice, trophoblasts begin to attach to the receptive uterine epithelium, after which the trophectoderm proliferates to form the ectoplacental cone, and later the spongiotrophoblast layer. The outermost trophoblasts of the ectoplacental cone differentiate into secondary trophoblast giant cells, which lie in the periphery of the placenta, forming the interface with maternal cells in the decidua (1, 2). The expression of placental lactogen (PL) is regulated during trophoblast giant cell differentiation (3). The 4311 gene is specific for a group of cells of the ectoplacental cone, suggesting there may be a compartmentalization within the trophoblast cells, with 4311 marking the cells that will form the densely packed spongiotrophoblast layer, but not the trophoblast giant cells or the labyrinthine layer of the placenta (4).
Several transcription factors have been reported to be expressed in the chorioallantoic placenta, suggesting a possible role in the differentiation of this tissue; some examples are the zinc finger factor Rex-1 (5), a member of the GATA family, GATA-3 (6), and members of the helix–loop–helix family of transcription factors Mash-2 (7) and Hxt (8). Mash-2 is an important regulator of trophoblast proliferation; its expression diminishes as mouse trophoblasts differentiate into giant cells, and the targeted deletion of the Mash-2 gene results in an increase in the number of giant cells at the expense of the proliferative population, resulting in a diminished spongiotrophoblast layer (7). Transcription factors of the Ets winged helix–turn–helix family have been detected in trophoblastic tissues by day 6.5. In Ets2 mutant embryos, both migration and differentiation of trophoblast cells are defective, resulting in embryonic death before day 8.5 of gestation (9). For both Mash-2 and Ets2 mutants, the defects were rescued by tetraploid cell aggregation experiments, reinforcing the conclusion that there is an essential role for these genes during extraembryonic tissue development (7, 9).
Members of the homeodomain-containing transcription factors have also been reported in the developing placenta: Pem (10, 11), and more recently MOX2, MSX2, DLX4 (12, 13), and HB24 (12, 14), which were isolated from a third-trimester human placental library. The paired-like homeobox gene Esx1 has been shown to be expressed in the labyrinthine layer (15). The specific role for these homeodomain proteins in placental development and function is under current investigation, but it has been suggested that DLX4 may be important for trophoblast invasion (13) and HB24 may play a role in trophoblast differentiation (14).
Dlx3 is part of the Distal-less (16) family of non-Antennapedia homeobox genes, which comprises six members in mice and humans (13, 17–20) that are organized as three convergently transcribed pairs, each closely linked to three of the four mammalian Hox clusters. In the human genome, DLX3 is paired with DLX4 (also reported as DLX7 and DLX8) on chromosome 17 (location 17q21.3; refs. 13, 19, and 20), telomeric to the HOXB cluster (19). Dlx3 is expressed throughout development in a series of structures derived from epithelial mesenchymal interaction such as the teeth, hair follicles, and limb buds (21). Dlx3 is also expressed in the differentiated strata of vertebrate epidermis, and transgenic ectopic expression of this gene in mouse epidermal basal cells leads to severe disruption of this tissue as well as cessation of proliferation and precocious activation of late differentiation markers, such as loricrin and profilaggrin in the basal layer (22). Recently, the genetic abnormality in the hereditary disease tricho-dento-osseous (TDO) syndrome was identified as a four-nucleotide deletion in the human DLX3 gene, immediately downstream from the homeodomain coding region (23). This syndrome is inherited as an autosomal dominant trait, and it includes abnormalities in the teeth, hair, and facial bones. To study the function of Dlx3 during development we generated Dlx3-null mutant mice by gene targeting.
Here we report additional sites of expression of the Dlx3 gene: during placentation in the mouse, Dlx3 is expressed in the ectoplacental cone cells and chorionic plate, and later in placental development in the labyrinthine layer. We also show that targeted deletion of Dlx3 results in embryonic developmental arrest around day 9.5–10, associated with a gross failure of the placenta to undergo proper morphogenesis.
MATERIALS AND METHODS
Construction of Targeting Vector.
The targeting vector KO/Dlx3 (Fig. 1) contains 5.2 kb of Dlx3 genomic sequence from a 129/Sv strain in the pPNT vector (24). This vector was modified by converting the NotI site into an SfiI site, which was used to linearize the vector before electroporation. The 1.2-kb 5′ genomic flank was subcloned into the XhoI site in the same vector. The 4-kb 3′ homologous flank was subcloned as an XbaI fragment into the XbaI site in pPNT.
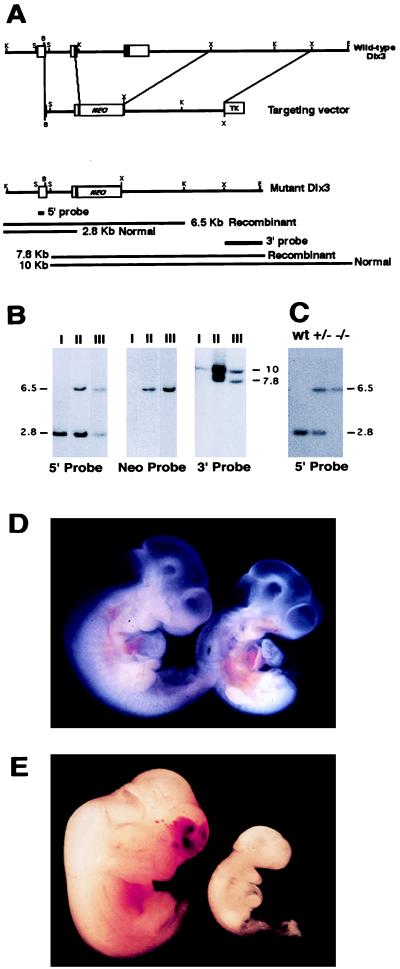
Figure 1.
Targeted disruption of Dlx3. (A) Restriction map of the Dlx3 gene, the targeting vector, and the predicted restriction map after the homologous recombination event. The probes used and the predicted length of restriction fragments in the Southern blot analysis are shown. B, BamHI; K, KpnI; E, EcoRI; S, SmaI; X, XbaI; NEO, neomycin phosphotransferase gene; TK, thymidine kinase gene. (B) Genomic analysis of wild-type-derived ES cell DNA (lanes I) and two independently targeted ES cell clones (lanes II and III). Recombination was detected at the 5′ end by digesting ES cell DNA with KpnI/Asp718 and hybridizing the Southern blot with 5′ probe, and at the 3′ end by digesting DNA with EcoRI and SmaI and hybridizing with 3′ probe. A single integration event was confirmed by hybridizing KpnI/Asp718-digested DNA with probe Neo (Center). Numbers at sides of the figure indicate fragment size in kb. (C) Genotyping of tail-derived DNA revealing the expected pattern for wild-type (wt), heterozygous (+/−), and homozygous Dlx3-null (−/−) embryonic day 9.5 (E9.5) embryos. (D) Phenotype of wild-type (left) and Dlx3-null (right) E9.5 embryos genotyped by Southern blotting. (E) Phenotype of wild-type (left) and regressed Dlx3 −/− (right) E12.5 embryos genotyped by Southern blotting.
Gene Targeting and Generation of Mouse Mutants.
Tissue culture of embryonic stem (ES) cells and conditions for electroporation of the targeting construct have been described (25). Successful integration events in the R1 line of ES cells (a gift from A. Nagy, Mount Sinai Hospital, Toronto) were isolated by simultaneous selection with G418 (350 μg/ml) and ganciclovir (2 μM). Homologous recombination events were identified by Southern blotting and PCR of DNA purified by proteinase K digestion followed by extraction with phenol/chloroform. The hybridization probes and the predicted sizes of fragments generated by the endogenous and targeted alleles in Southern blotting are shown in Fig. 1B. Recombination was detected at the 5′ end by digesting the DNA with Asp718 and hybridizing the blot with the 5′ probe (280-bp SmaI–BamHI fragment). The normal allele generated a 2.8-kb band and the disrupted allele a 6.5-kb band. Correct recombination at the 3′ end was demonstrated by double digesting DNA with EcoRI and SmaI and hybridizing with the 3′ probe (1.6-kb XbaI–EcoRI fragment). The fragment for the normal allele was 7.8 kb and that for the disrupted allele was 10 kb. Gene-specific integration of the targeting construct resulted in the elimination of all protein coding sequences C-terminal to the ninth amino acid of the centrally located homeodomain, a region postulated to bind the DNA and necessary to exert the transcriptional activity (26), and the complete C terminus of the protein. The presence of a single integration event was confirmed by probing the blot of EcoRI–SmaI-digested genomic DNA with a 0.6-kb neomycin phosphotransferase gene probe (Neo). For PCR, two oligonucleotides were used to determine heterozygosity, InF (CAGGCTGAGACTAAACTGGTGTTTTTCCCCTA) and Neo5′R (CGGCCGGAGAACCTGCGTGCAATCCATCTTG) in 50-μl reaction mixtures containing 2.25 mM MgCl2, 200 μM each deoxynucleotide triphosphate, and 1 unit of Taq/Pwo DNA polymerase mix (Boehringer Mannheim). The cycling conditions were: 94°C for 5 min followed by 36 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. The InF oligonucleotide is specific for intronic sequence of Dlx3 and Neo5′R is specific for the Neo gene; amplification with these two oligonucleotides generated a 635-bp fragment. Blastocyst injections with two independent targeted ES cell lines were as described in ref. 27. Mice heterozygous for the Dlx3 gene deletion (Dlx3 +/−) were generated by mating chimeras with CF1 mice. Dlx3 −/− mutant embryos were generated by heterozygous intermatings.
RNA Blotting.
Total RNA was isolated from E16.5 placenta, neonatal epidermis, and adult brain by a guanidinium thiocyanate/phenol method (28). The RNA samples (10 μg) were separated in a 1.2% agarose/methymercury hydroxide gel, transferred to a nylon membrane, and hybridized according to ref. 29. The blot was hybridized with a 1.1-kb mouse Dlx3 cDNA probe (21) labeled with 32P by random priming (Stratagene) and washed at high stringency (0.1× SSC/0.1% SDS at 65°C).
In Situ Hybridization.
Entire conceptus and placentas were isolated from staged pregnancies and were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Sections of 9 μm were stained with hematoxylin and eosin for histology or prepared for in situ hybridization. Antisense RNA probes were generated for PL-1 (3), 4311 (4), Mash-2 (7), Esx1 (15), Dlx3 (21), and mouse germ cell nuclear factor (mGCNF) gene (30). Hybridization was performed according to ref. 31.
RESULTS
Targeted Mutagenesis of Dlx3.
Dlx3-deficient mice were derived from ES cells in which one Dlx3 allele was disrupted (Fig. 1A) as described in Materials and Methods. Two independently isolated ES cell lines carrying the mutation (Fig. 1B) were used to generate chimeric mice that transmitted the mutated allele to their offspring. Founder chimeras were mated, and the progeny were analyzed to determine heterozygosity. Matings of heterozygous pairs did not generate any live births of Dlx3 −/− mice, nor were any null embryos found at a stage later than E12.5. However, at E9.5 the distribution of +/+, +/−, and −/− genotypes (Fig. 1C) showed approximately Mendelian ratios (data not shown). Morphological defects were already evident in E9.5 −/− embryos, such as swelling of the pericardium indicating a disturbance of fluid balance across the yolk sac and an accumulation of blood in the embryo (Fig. 1D). At this stage in normal embryogenesis, the Dlx3 gene has been reported to be expressed only in the first and second branchial arches (21). No gross abnormality was observed in this tissue by visual inspection of the E9.5 mutant embryos, which is reasonable because the expression of Dlx3 has only initiated at this stage. By E12.5, only two Dlx3 −/− embryos could be genotyped, and these were severely delayed in development compared with wild type (Fig. 1E). At this stage, mostly empty residual decidual swellings were found, and the genotype for these could not be determined. Thus, homozygous Dlx3 mutation leads to developmental arrest shortly after day 9.5. Identical results were obtained with derivatives of the two targeted ES cell lines.
Expression of Dlx3 in Early Development.
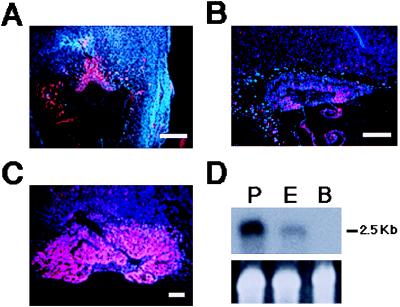
Analysis of Dlx3 expression showed that this gene is transcribed in the ectoplacental cone and chorionic plate at day 8.5 (Fig. 2A), in a region that overlaps with the domain of Esx1 (15) and Mash-2 (7) expression. This region gives rise to both the labyrinthine and spongiotrophoblast trophectoderm layers of the placenta. The expression of Dlx3 is maintained through early placental development, with abundant transcripts in the labyrinthine layer at day 10.5 (Fig. 2 B and C). No Dlx3 RNA was detected in placental sections of the Dlx3-null embryos. Northern blot analysis of RNA from E16.5 placenta (Fig. 2D) revealed a single mRNA of 2.5 kb, which was expressed at a higher level than that previously reported in neonatal epidermis, a major site of Dlx3 gene activity (21, 22). No Dlx3 RNA was detected in adult brain as reported previously (21).
Figure 2.
Expression of Dlx3. In situ hybridization with antisense Dlx3 of sagittal sections of wild-type conceptus within the uterus at E8.5 (A; scale bar = 400 μm) and placentas from wild-type embryos at 9.5 (B; scale bar = 300 μm) and 10.5 (C; scale bar = 500 μm) days of development. (D) Northern blot of 10 μg each of RNA from E16.5 placenta (P), neonatal epidermis (E), and adult brain (B) hybridized with a Dlx3 cDNA probe. (Upper) Hybridization with Dlx3 probe; number at the right indicates RNA size. (Lower) Ethidium bromide staining of 18S ribosomal RNA.
Defects in Extraembryonic Tissues Associated with the Dlx3-Null Genotype.
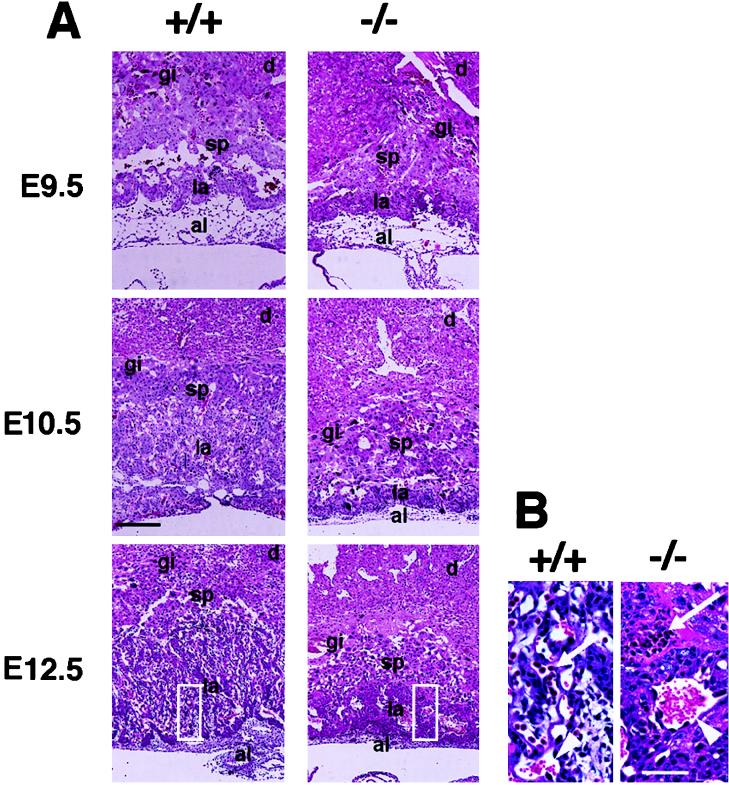
Histological analysis of sagittal sections of placentas from Dlx3 +/+ and −/− embryos at E9.5, E10.5, and E12.5 showed that by day 12.5 the spongiotrophoblast and labyrinthine layers are greatly reduced in size and very compact in the mutant compared with the wild type (Fig. 3A). In addition, the parietal and visceral yolk sacs were not as tightly associated with the placenta in the Dlx3-null embryos (data not shown). During formation of the chorioallantoic placenta, the chorion collapses onto the ectoplacental plate as the allantois fuses with the chorion. Invasion of the chorionic plate by the allantoic vessels results in the formation of the loose network of vascular cells that comprises the labyrinthine layer. In the Dlx3 −/− embryos, the characteristic labyrinths are absent from this layer. The decidual tissue was more dense than in the wild type, but this is probably secondary to the placental defects, since no Dlx3 expression in the decidua was detected.
Figure 3.
(A) Trophoblast tissues from Dlx3 +/+ (Left) and −/− (Right) littermates. Histology of sagittal sections of +/+ and −/− placentas at E9.5, E10.5, and E12.5 stained with hematoxylin and eosin. al, allantois; la, labyrinthine layer; sp, spongiotrophoblast layer, gi, giant trophoblast layer; d, decidua. (Scale bar = 250 μm.) (B) Higher magnification of boxed areas in A for panels +/+ and −/− at E12.5. Arrowhead indicates maternal enucleated blood cell and arrow indicates fetal blood cell. A few blood vessels are apparent in the compacted labyrinthine and spongiotrophoblast layers in the −/− E12.5 placenta. (Scale bar = 40 μm.)
By E12.5 there are significant defects evident in the placental vascular system. In the Dlx3 −/− labyrinthine layer, there are very few fetal red blood cells (RBCs), which can be distinguished from the maternal RBCs by the presence of a nucleus (Fig. 3B). Most of these fetal RBCs were found in the periphery, and those located more internally within the labyrinthine layer tended to have fragmented nuclear morphology. Vascularization was also less extensive in the Dlx3 −/− labyrinthine layer, resulting in the compact morphology shown in Fig. 3. This decreased vascularization could be due to a lack of proper invasion by the mesodermal vasculature of the allantois or a failure of the allantoic cells to survive once they have entered the extraembryonic ectoderm.
Expression of Trophoblast Markers in Dlx3 −/− Embryos.
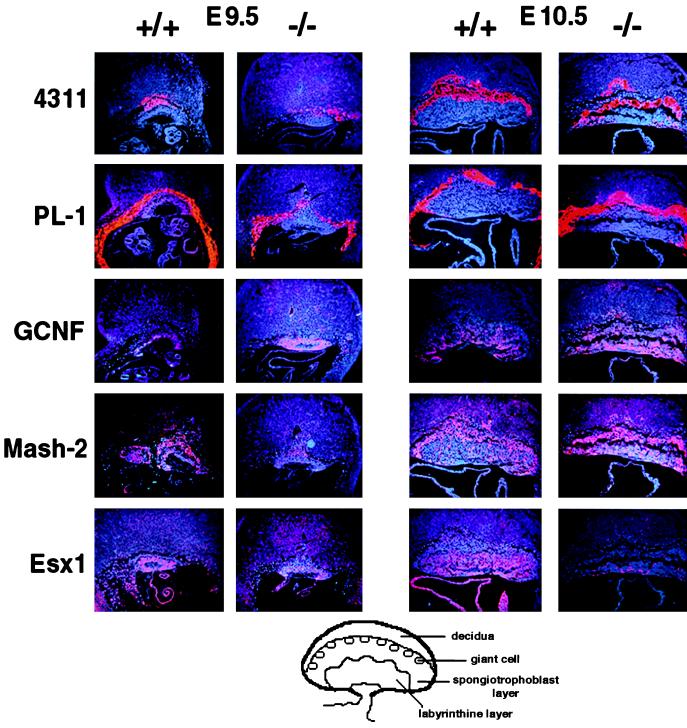
In situ hybridization analysis (Fig. 4) of sections throughout the entire conceptus at stage E9.5 showed no dramatic difference in the expression of the spongiotrophoblast-specific marker 4311 (4) or the giant trophoblast cell marker PL-1 (3) in the Dlx3-null compared with wild-type embryos. Thus, although the placental morphology was highly abnormal in Dlx3 −/− embryos, cytodifferentiation of giant cell and spongiotrophoblast placental cell types and expression of their specific markers was not prevented. The orphan receptor mGCNF (30) was detected in a subset of cells in the labyrinthine layer of wild-type placentas at E10.5, with the highest number of mGCNF-expressing cells proximal to the allantois. mGCNF was not expressed in the spongiotrophoblast cells of the Dlx3 +/+ placenta. In placentas of Dlx3-null mice, mGCNF expression appeared up-regulated in the labyrinthine layer, and expressing cells were also detected in the spongiotrophoblast layer. The transcription factor Mash-2 is highly expressed in the ectoplacental cone, the chorion, and its derivatives in the placenta (7) in regions coincident with the domain of expression of Dlx3. At E10.5, the expression of Mash-2 in the labyrinthine layer of Dlx3-null placentas persisted compared with wild type, and it was stronger at this stage in the spongiotrophoblast layer. A dramatic change was observed for the homeobox-containing gene Esx1, normally expressed in the chorion and labyrinthine trophoblast of the chorioallantoic placenta (15). At E9.5, Esx1 expression was reduced in the Dlx3 −/− placentas compared with wild type, and by E10.5 it was strongly down-regulated in the labyrinthine layer (Fig. 4). These results indicate that Dlx3 is essential for the proper regulation of genes expressed during chorioallantoic placenta development.
Figure 4.
Expression of cell-type-specific markers analyzed by in situ hybridization of sagittal sections of conceptus at E9.5, and placentas from Dlx3 +/+ and −/− embryos at E10.5. Hybridizations were performed with antisense probes for 4311 (spongiotrophoblast cells and their precursors in the ectoplacental cone); PL-1 (trophoblast giant cell); mGCNF (labyrinthine layer); Mash-2 (spongiotrophoblast and labyrinthine layers and chorion); and Esx1 (endoderm layer of the visceral yolk sac and labyrinthine trophoblast of the chorioallantoic placenta). (×10.) At the bottom of the figure there is a schematic representation of E10.5 placenta indicating the different cell types.
DISCUSSION
The mammalian embryo is critically dependent on the establishment of a chorioallantoic placenta, which functions as the primary means of nutrient and gas exchange (2). We have generated Dlx3 −/− embryos, which die from placental failure around E9.5–10. The null embryos first show slight morphological defects by day 9.5, when an enlarged pericardium and accumulation of blood in the embryo are visible. By day 10.5, the defects are detectable in both the embryo proper, which is being resorbed, and in the placenta, where the labyrinthine layer shows a compacted morphology, and the highly vascularized organization characteristic of this layer is absent. The spatial and temporal distribution of Dlx3 in the placenta correlates with the observed phenotype. Most Dlx3 −/− embryos die later than mice that are homozygous nulls for other regulatory genes expressed in the chorion, such as Mash-2 and ERR-β (7, 32). These two mutants show increased numbers of giant cells, suggesting an early misallocation or differential proliferation of trophoblast cells, neither of which occurs in the Dlx3 −/− placentas. The Dlx3 −/− embryos also survive longer than mice null for DNA methyltransferase (27), in which the allantois does not fuse at all with the chorion. In the Dlx3 −/− embryos, the fusion of the allantois with the chorion occurs, suggesting that some fetomaternal exchange takes place, but the highly vascularized phenotype characteristic of the labyrinthine layer does not develop, indicating deficient invasion or survival of the allantoic cells in the chorionic environment.
Cell type-specific markers expressed during placenta maturation, such as PL-1 in giant cells and 4311 in ectoplacental cone cells destined to form the densely packed spongiotrophoblast layer, were not affected in the Dlx3 −/− embryos. The apparent up-regulation of mGCNF expression in the labyrinthine layer of the Dlx3 −/− placenta compared with wild type could be because of (i) compaction of the labyrinthine layer, (ii) increased expression of mGCNF, or (iii) deletion of mGCNF-nonexpressing cells in the Dlx3 −/− labyrinthine layer. In the context of the spongiotrophoblast layer, the same possibilities could also explain the increased expression of Mash-2. On the other hand, the homeobox-containing gene Esx1, normally expressed in the visceral yolk sac, the ectoderm of the chorion, and the labyrinthine layer, was strongly down-regulated. This down-regulation could be because of a secondary effect of the disrupted placental patterning or to the requirement of Dlx3 for the maintenance of Esx1 expression, as part of a mechanism essential for proper labyrinthine morphogenesis.
Dlx3 clearly is a distinct member of the Dlx family; it differs from other members in that it is not detected in the central nervous system (21), and although all the Dlx genes are expressed in the first and second branchial arches, Dlx3 is the latest and most distally expressed member (21, 33). From the six reported murine Dlx genes, three have been analyzed by targeted disruption. Dlx1 −/− died within one month of birth, probably because of a dysfunction of the enteric nervous system, and Dlx2 −/− neonates died within a few hours after birth. Mice with combined elimination of both genes presented a phenotype much like Dlx2 −/− mice, characterized by the failure to differentiate a specific subset of neurons in the forebrain and craniofacial abnormalities, including the absence of maxillary molars (33, 34). Disruption of the Dlx5 gene results in abnormal olfactory bulb development and perinatal lethality (35). It has been postulated that there is a redundancy in function for the Dlx loci (Dlx1/2, 5/6) on the basis of the overlapping patterns of expression and the results from ablation experiments for Dlx1/2 genes (34). The human DLX4 gene (13, 19, 20), which is closely linked to DLX3, is also expressed in the placenta (12, 13). The extent of overlap in expression and function of the DLX3/4 genes remains to be determined. However, in view of the results presented here, it is clear that the mouse Dlx3 plays a unique and important role in placenta morphogenesis and has a function that cannot be provided by other Dlx genes.
Mice heterozygous for the Dlx3 disruption presented in this paper do not show any of the abnormalities characteristic of the tricho-dento-osseous syndrome, suggesting that the dominant pattern of inheritance of the syndrome is due to formation of nonfunctional complexes involving the frame-shifted/truncated DLX3 protein, as opposed to haploinsufficiency.
In conclusion, Dlx3 is required at postimplantation stages for the generation of a functional chorioallantoic placenta. The death of Dlx3 −/− embryos is due to placenta malfunction, specifically in failure to establish proper circulation. It will be of clinical importance to determine how the human DLX3 is expressed during pregnancy and what are the correlations of pathological conditions with mutations in this gene.
Acknowledgments
We thank Sing-Ping Huang for the cultured feeder cells and other members of the Laboratory of Mammalian Genes and Development for technical advice; Kelly Cornelis and Daniel Sussman for technical support; and Li Qui for help with the in situ hybridization analysis. We also thank A. Nagy for the ES cells; R. Behringer, J. Rossant, and A. Cooney for the Esx1, Mash-2, and mGCNF probes, respectively; and R. Farnsworth for helpful comments on the manuscript. We thank Dr. P. Steinert for his support.
ABBREVIATIONS
- PL
placental lactogen
- ES cells
embryonic stem cells
- En
embryonic day n
- mGCNF
mouse germ cell nuclear factor
References
- 1.Kaufman M H. In: Biology of Trophoblast. Loke Y W, White A, editors. Amsterdam: Elsevier; 1983. pp. 23–68. [Google Scholar]
- 2.Cross J C, Werb Z, Fisher S J. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 3.Colosi P, Swiergiel J J, Wilder E L, Oviedo A, Linzer D I H. Mol Endocrinol. 1987;2:579–586. doi: 10.1210/mend-2-6-579. [DOI] [PubMed] [Google Scholar]
- 4.Lescisin K R, Varmuza S, Rossant J. Genes Dev. 1988;2:1639–1646. doi: 10.1101/gad.2.12a.1639. [DOI] [PubMed] [Google Scholar]
- 5.Rogers M B, Hosler B A, Gudas L J. Development (Cambridge, UK) 1991;113:815–824. doi: 10.1242/dev.113.3.815. [DOI] [PubMed] [Google Scholar]
- 6.Ng Y K, George K M, Engel J D, Linzer D I H. Development (Cambridge, UK) 1994;120:3257–3266. doi: 10.1242/dev.120.11.3257. [DOI] [PubMed] [Google Scholar]
- 7.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner A L. Nature (London) 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 8.Cross J C, Flannery M L, Blanar M A, Steingrimsson E, Jenkins N A, Copeland N G, Rutter W J, Werb Z. Development (Cambridge, UK) 1995;121:2513–2523. doi: 10.1242/dev.121.8.2513. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto H, Flannery M L, Kupriyanov S, Pearce J, McKercher S R, Henkel G W, Maki R A, Werb Z, Oshima R G. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson M F, Kleeman J, Richards J, MacLeod C L. Dev Biol. 1990;141:451–455. doi: 10.1016/0012-1606(90)90400-d. [DOI] [PubMed] [Google Scholar]
- 11.Lin T P, Chen I, MacLeod C. Dev Biol. 1995;170:756. [Google Scholar]
- 12.Quinn L M, Johnson B V, Nicholl J, Sutherland G R, Kalionis B. Gene. 1997;187:55–61. doi: 10.1016/s0378-1119(96)00706-8. [DOI] [PubMed] [Google Scholar]
- 13.Quinn L M, Latham S E, Kalionis B. Placenta. 1998;19:87–93. doi: 10.1016/s0143-4004(98)90103-5. [DOI] [PubMed] [Google Scholar]
- 14.Quinn L M, Latham S E, Kalionis B. Reprod Fertil Dev. 1997;9:617–623. doi: 10.1071/r97025. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Lemaire P, Behringer R R. Dev Biol. 1997;188:85–95. doi: 10.1006/dbio.1997.8640. [DOI] [PubMed] [Google Scholar]
- 16.Cohen S M, Brönner G, Kütter F, Jürgens G, Jäckle H. Nature (London) 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- 17.Robinson G W, Wray S, Mahon K A. New Biol. 1991;3:1183–1194. [PubMed] [Google Scholar]
- 18.Simeone A, Acampora D, Pannese M, D’Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K, Boncinelli E. Proc Natl Acad Sci USA. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura S, Stock D W, Wynder K L, Bolekens J A, Takeshita K, Nagai B M, Chiba S, Kitamura T, Freeland T M, Zhao Z, et al. Genomics. 1996;38:314–324. doi: 10.1006/geno.1996.0634. [DOI] [PubMed] [Google Scholar]
- 20.Morasso M I, Yonescu R, Griffin C A, Sargent T D. Mamm Genome. 1997;8:302–303. doi: 10.1007/s003359900427. [DOI] [PubMed] [Google Scholar]
- 21.Robinson G W, Mahon K A. Mech Dev. 1994;48:199–215. doi: 10.1016/0925-4773(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 22.Morasso M I, Markova N G, Sargent T D. J Cell Biol. 1996;135:1879–1887. doi: 10.1083/jcb.135.6.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price J A, Bowden D W, Wright J T, Pettenati M J, Hart T C. Human Mol Genet. 1998;7:563–569. doi: 10.1093/hmg/7.3.563. [DOI] [PubMed] [Google Scholar]
- 24.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 25.Love P E, Tremblay M L, Westphal H. Proc Natl Acad Sci USA. 1992;89:9929–9933. doi: 10.1073/pnas.89.20.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 27.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 28.Sargent T D, Jamrich M, Dawid I B. Dev Biol. 1986;114:238–246. doi: 10.1016/0012-1606(86)90399-4. [DOI] [PubMed] [Google Scholar]
- 29.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susens U, Aguiluz J B, Evans R M, Borgmeyer U. Dev Neurosci. 1997;19:410–420. doi: 10.1159/000111238. [DOI] [PubMed] [Google Scholar]
- 31.Mackem S, Mahon K A. Development (Cambridge, UK) 1991;112:791–806. doi: 10.1242/dev.112.3.791. [DOI] [PubMed] [Google Scholar]
- 32.Luo J, Sladek R, Bader J, Matthyssen A, Rossant J, Giguere V. Nature (London) 1997;388:778–783. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- 33.Qui M, Bulfone A, Ghattas I, Meneses J, Christensen L, Sharpe P T, Presley R, Pedersen R A, Rubenstein J L R. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 34.Anderson S A, Qui M, Bulfone A, Eisenstat D D, Meneses J, Pedersen R, Rubenstein J L R. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 35.Long J E, Depew M, Liu J K, Meneses J J, Pedersen R A, Rubenstein J L R. Dev Biol. 1998;198:195. [Google Scholar]