The nucleosome core particle is the fundamental unit of chromatin structure in all eukaryotes. It comprises eight core histones (two each of H2A, H2B, H3, and H4) around which are wrapped 146 bp of DNA, and its structure has been defined at 2.8 Å resolution by x-ray crystallography (1). Since it was first described, it has been assumed that the function of the core particle is to package DNA into the interphase cell nucleus and metaphase chromosomes. Wrapping DNA around the histone octamer in left-handed supercoils results in an approximately sevenfold reduction in its length. Although this is only a small fraction of the several-thousand-fold length reduction required for compaction into metaphase chromosomes, it is presumably an essential first step that enables higher-order structures to assemble.
The nucleosome’s rather mundane packaging job became more interesting with the realization that compaction of DNA into chromatin is a crucial element in eukaryotic gene regulation (2). Appropriately located nucleosomes can hinder assembly of transcription initiation complexes and polymerase progression, and over recent years, whole families of chromatin-remodeling enzymes have been identified whose primary purpose is to reorganize histone-DNA interactions so as to facilitate (or repress) transcription (3). These enzymes disrupt the nucleosome in an ATP-dependent manner that often results in movement of the histone core relative to the DNA, thereby exposing recognition sequences for DNA-binding activators or repressors. However, even these insights still left the nucleosome as a passive inhibitor of transcription, something to be moved aside so transcription could proceed.
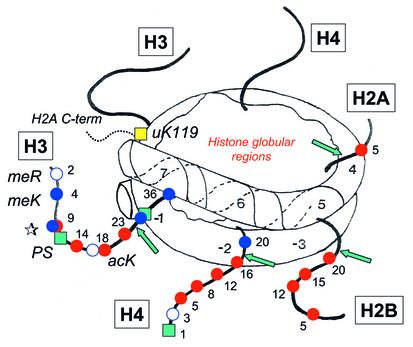
The final transformation of the nucleosome into a truly exciting and colorful character has come through studies on the effects and properties of other enzymes that act on chromatin. It has been known for many years that the histone N-terminal tails are exposed on the surface of the nucleosome and that selected amino acid residues are subject to a variety of enzyme-catalyzed, posttranslational modifications. These include acetylation of lysines, phosphorylation of serines, and methylation of lysines and arginines. The locations of the histone N-terminal tails in the nucleosome and the residues that can be modified are shown in Figure 1. The function of these modifications, and indeed of the tails themselves, is the focus of much current attention, centered largely on the possibility that the nucleosome, with its modified tail domains, is not just a humble packer of DNA, but a carrier of epigenetic information that determines both how genes are expressed and how their expression patterns are maintained from one cell generation to the next.
Figure 1.
A nucleosome core particle, showing the core histone N-terminal tail domains and sites of posttranslational modification. Residue numbers for modified residues are shown. Note that H3 lysine 9 (star) can be either acetylated or methylated. The C-terminal tail domain of one H2A molecule is shown (dashed line) with the site of ubiquitination at lysine 119 (yellow square). Modifications are shown on only one of the two copies of histones H3 and H4, and only one tail is shown for H2A and H2B. Numbers along the DNA indicate each complete helical turn on either side of the dyad axis. Sites marked by green arrows are susceptible to cutting by trypsin in intact nucleosomes. Red circles, acetyl lysine (acK); blue circles, methyl lysine (meK); white circles, methyl arginine (meR); green squares, phosphoryl serine (PS); yellow square, ubiquitinated lysine (uK).
An epigenetic code
The first association between a histone tail modification and a particular functional state of chromatin came with the demonstration that transcriptionally active chromatin fractions are enriched in acetylated histones (4, 5). Subsequently, regions of transcriptionally silent constitutive and facultative heterochromatin were found to be underacetylated (6), consistent with the idea that acetylation of the histone tails, with the associated loss of positive charge and weaker DNA binding, somehow caused chromatin to become more “open” (or less “condensed”) and thereby more conducive to transcription. While such charge-mediated structural changes are likely to contribute to altered patterns of gene expression, it has now become clear that the story is much more complex and that the functional effects of tail modifications depend on the specific amino acids that are modified.
An early indication that modification of specific tail residues was linked to chromatin functional states came from immunostaining of Drosophila polytene chromosomes. Unlike in mammals, where female cells inactivate one of their two X chromosomes to avoid unbalanced expression of X-linked genes, Drosophila equalizes the levels of X-linked gene products between XY males and XX females by transcribing X-linked genes in males at twice the rate seen in females. Analyses using residue-specific antibodies show that H4 acetylated at lysine 16 (H4Ac16) occurs almost exclusively on the transcriptionally hyperactive male X chromosome. In addition, the Drosophila H4 lysine 12 remains acetylated in centric heterochromatin, whereas lysines 5, 8, and 16 are all underacetylated (7). These observations led to the suggestion that the histone N-terminal tails constitute nucleosome surface markers that can be recognized by nonhistone proteins in a modification-dependent manner to alter the functional state of chromatin (7). More importantly, these observations also raised the possibility that tail modifications not only contribute to the ongoing up- and downregulation of transcription but also carry information regarding the potential for transcription (or lack of it) in response to specific factors. In other words, the nucleosome may have a role not only in DNA packaging, but also in the transmission from one cell generation to the next of epigenetic information regarding transcriptional potential (8, 9).
The amount of epigenetic information that can be carried in the histone tails is remarkable. For example, there are 50 different acetylated isoforms of the four core histones (H2B, H3, and H4 have 16 each and H2A has two). These isoforms can be modified further by methylation of selected lysines and arginines (H3 and H4) and phosphorylation of serine (H3, H4, H2B). Further, methylation can involve attachment of one, two, or three methyl groups (see below), and there are other modifications, such as ubiquitination and ADP-ribosylation (10). The total number of possible histone isoforms, carrying different combinations of tail modifications, that can mark the nucleosome surface, runs into many thousands. This has given rise to the idea that the tail modifications constitute a histone code (11, 12) or epigenetic code (13), which is set and maintained by tail-modifying and -demodifying enzymes and read by nonhistone proteins.
The histone tail modifications are likely to act in concert with a rather more widely known mediator of chromatin structure and gene expression, namely methylation of cytosine residues in CpG dimers through the action of DNA methyltransferases. Long-term silencing, as found in imprinted genes or the female inactive X chromosome, is generally associated with relatively high levels of CpG methylation. The mechanism(s) by which CpG methylation leads to gene silencing remains to be defined, but there is evidence that in at least some situations, histone modifications are involved. The methyl DNA-binding protein MeCP2 can bind histone deacetylases, thereby targeting them to methylated DNA and resulting in local histone deacetylation and suppression of transcription (14). Conversely, experiments in the filamentous fungus Neurospora crassa have shown that DNA methylation is dependent upon methylation of histone H3 lysine 9 (15).
Dynamics of histone modification
The modifications to the N-terminal histone tails in chromatin are put in place and maintained by the action of families of modifying and demodifying enzymes. For example, most histone acetates are undergoing a continuous cycle of acetylation and deacetylation and turn over with half-lives ranging from just a few minutes to several hours (16). Thus, the level of acetylation of a particular lysine in a particular histone is likely to be a steady state maintained by the ongoing activities of histone acetyltransferases (HATs) and deacetylases (HDACs). While this makes continuous demands on the cell’s energy resources, it also offers an advantage to those genes whose expression may need to be up- or downregulated relatively quickly, such as those that respond to hormonal cues or growth factor stimulation. Removal or inhibition of either HATs or HDACs will precipitate a rapid change in acetylation, and hence transcription or transcriptional potential. Conversely, where levels of gene expression are to be stably maintained over long periods, patterns of histone modification are stabilized by removing (or inhibiting) the HATs and HDACs targeted to that genomic region. In the case of the imprinted gene U2af1-rs1 in mice, differential acetylation levels on the maternal and paternal alleles are maintained dynamically early in development but become static (operationally defined by resistance to HDAC inhibitors) as development proceeds (17).
Lysine methylation seems to be an exceptionally stable modification. Early studies showed that turnover of histone methyl groups was even slower than turnover of the histones themselves (see, for example, ref. 18). No conclusive evidence has yet been found for histone-demethylating enzymes, which may indeed not exist. Removal of methylated histones may simply occur passively through postreplication chromatin assembly and replacement of old, methylated histones with new, unmethylated ones. However, the possibility remains that local methylation patterns may be more dynamic and may involve novel mechanisms for removal of methylated tails, such as proteolysis (19) or histone replacement uncoupled from DNA replication (20).
Setting the code
Among the various tail modifications, setting patterns of histone phosphorylation might seem relatively straightforward in that only a few residues are typically involved (Figure 1). However, the same modification has been implicated in two different functions. Phosphorylation of H3S10, precisely targeted to control regions, accompanies activation of immediate-early genes, while a genome-wide phosphorylation of the same residue occurs just prior to entry into mitosis (21, 22). Different kinases are involved in each case (22). At first sight, it seems odd that the same modification is associated with both progression through mitosis (a time of chromatin condensation and transcriptional silence) and gene induction. Perhaps H3S10 phosphorylation is involved in the chromatin decondensation and reinitiation of transcription that accompany progression from mitosis into G1.
In theory, the setting of specific patterns of acetylation (i.e., residue-specific modifications) could reflect the specificities of HATs or HDACs. Both HATs and HDACs constitute extensive enzyme families and are often found as a catalytic subunit in multiprotein complexes whose other components confer genomic targeting and other capabilities (23, 24). In vitro assays have shown specificity, or at least substrate preferences, for most HATs. The most extreme example is provided by the Drosophila enzyme MOF, which acetylates H4 specifically at lysine 16 and is responsible for the acetylation of this residue exclusively on the male X chromosome (ref. 25 and references therein). The HDACs so far tested show less specificity for individual histones or residues, though there are certainly preferences (ref. 26 and references therein). It is, of course, difficult to be sure that in vitro assays provide a true reflection of specificity in vivo. Both HATs and HDACs can show quite different specificities, or levels of catalytic activity, depending on whether they are assayed against peptide, whole histone, or chromatin substrates. Linking in vitro specificity to in vivo effects, as in the case of MOF, is reassuring.
Histone methyltransferases (HMTs) are specific for either lysine or arginine and are the subject of two excellent recent reviews (27, 28). There are five arginine HMTs (four of which are confirmed as enzymes). All have the same, highly conserved catalytic domain and S-adenosyl methionine–binding region, but little else in the way of known protein domains. The enzymes studied so far show varying degrees of specificity for particular histone arginines. All can attach one or two methyl groups to a single arginine, but they differ in whether the two methyl groups are distributed symmetrically (i.e., one on each of the two terminal nitrogens) or asymmetrically (i.e., both on the same nitrogen) (27).
Lysine HMTs all contain a SET domain, although not all SET domain proteins have HMT activity (29). Database searches reveal 73 SET domain proteins in humans, of which about one-third have been characterized and grouped into four families on the basis of sequence homology and relationship to the SET domain proteins in yeast (28). Several have confirmed HMT activity, and some are highly specific. For example, four members of the SUV39 family methylate only H3 lysine 9 (30–32). Importantly, this specificity has been linked to a particular chromatin state, namely, formation of heterochromatin.
The mammalian enzymes SUV39H1 and SUV39H2 have been shown to locate to constitutive heterochromatin, which also, by immunofluorescent labeling, appears to be relatively rich in H3 methylated at lysine 9 (33, 34). The protein HP-1 is also enriched in heterochromatin, where it plays a crucial, although as-yet unspecified, role in heterochromatin assembly. HP-1 has recently been shown to bind, through its chromodomain, specifically to H3 peptides methylated at lysine 9 (35, 36). This is the first example in which the enzymes that put a specific modification in place, the protein that reads it, and the functional effects that result are all known, at least in outline. As always, there are complications, as discussed below, and much remains to be worked out. In particular, we need to understand how the appropriate HMTs are targeted to defined genomic regions. Lysine HMTs are rich in known protein domains (28), some of which may have a targeting function. Intriguingly, some SET domain proteins also have a methyl DNA–binding domain, which would be predicted to target them to methylated CpG, and which may play a role in establishing the transcriptionally silent state usually associated with DNA methylation.
Modifications can interact, both in determining whether or not a particular residue can be modified and in providing a meaningful code. For example, H3 tail peptides phosphorylated at S10 are not methylated at lysine 9 by SUV39H1 in vitro (29). Conversely, peptides methylated at lysine 9 are poor substrates for phosphorylation of serine 10 by the IpI1/aurora kinase. Unsurprisingly, acetylation of lysine 9 prevents methylation of the same residue by SUV39H1 (29), but it is interesting that the homologous enzyme in Drosophila has been shown to exist in a protein complex containing dHDAC1 (37). The deacetylase activity in the complex may serve both to clear away any potentially inhibitory acetates and to create a globally underacetylated chromatin context in which the H3 methylation signal can be most appropriately read.
Attention to detail and creating a context
The lysine ε-amino group can be mono-, di-, or trimethylated, and all three methylation levels occur at high frequency in vivo. In mammalian cultured cells, the ratio of mono-, di-, and trimethylated lysines in H3 has been estimated as 1:2:1, with the overall methylation of H3 lysines 4 and 9 being close to 100% (18). Whether different enzymes are responsible for putting the different levels of methylation in place remains to be determined, but it is clear from recent results that these extremely detailed differences are significant in vivo. Studies in yeast using antisera that distinguish between H3 dimethylated and H3 trimethylated at lysine 4 show that only the trimethylated isoform, not its dimethylated counterpart, is consistently associated with transcriptionally active chromatin (38).
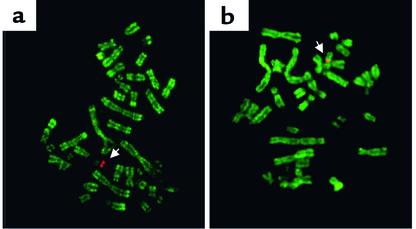
One of the most consistent and widespread associations between histone modification and chromatin structure is the general underacetylation of all four core histones in constitutive and facultative heterochromatin (ref. 6; ref. 39 and references therein). This underacetylation is not reversible by HDAC inhibitors such as butyrate or trichostatin A suggesting that the underacetylated state is static and that HATs are absent from heterochromatin. The inactive X chromosome (Xi) in female human cells is also deficient in H3 dimethylated at lysine 4 (40) but retains H3 dimethylated at lysine 9 at levels at or above those found elsewhere in the genome (41). Thus, Xi exemplifies both extremes of the histone code, namely the general deacetylation of all four core histones and the specific retention of H3 dimethylated at lysine 9. It also illustrates the great stability of epigenetic signals carried by the histone tails. The general underacetylation of Xi, the demethylation of H3 lysine 4, and the selective retention of H3 dimethylated at lysine 9 are all maintained in human x hamster somatic cell hybrids, in which Xi is maintained in an almost completely foreign cellular environment (Figure 2).
Figure 2.
Metaphase chromosome spreads from human X hamster somatic cell hybrids immunostained with rabbit antibodies (green) to H3 dimethylated at lysine 4 (a) and H3 dimethylated at lysine 9 (b). The inactive X (arrow) is the only human chromosome retained by these cells and is detected with a DNA probe specific for the human X centromere (red). The inactive X retains H3 dimethylated at lysine 9 but lacks H3 dimethylated at lysine 4.
It may be that the general deacetylation of all four histones provides a molecular or structural context in which the specific methylation signal can be most effectively read. Perhaps deacetylated tails encourage binding of a protein that recognizes H3 dimethylated at lysine 9, just as selective deacetylation of some H4 tail lysines encourages binding of the proteins Sir2 and Sir3 that assemble heterochromatin-like structures in yeast (42). This hypothetical protein remains to be identified. It seems not to be HP-1, which has not been detected on Xi (43). The fact that HP-1 binds to constitutive centric heterochromatin, but not to Xi, even though both are underacetylated and retain H3 dimethylated at K9, suggests that there is an additional determinant of HP-1 binding.
Reading the code
Antibodies can distinguish modified tails, combinations of modifications, and even lysines carrying two or three methyl groups. Other proteins appear to make similar discriminations. The clearest example may be HP-1’s ability to bind specifically, but with different affinities, to H3 peptides di- or trimethylated at lysine 9 (35, 36). Also, certain bromodomains (sequence elements found in proteins associated with maintenance of transcriptionally active states) bind preferentially to acetylated tail peptides in vitro (44, 45). Recent results have added a new twist to the question of how tail modifications might be read by implicating the histone tails in targeting of chromatin-remodeling enzymes. Firstly, immobilized H3 tail peptides lacking lysine methylation can remove components of the chromatin remodeling complex NuRD from crude cell extracts in pull-down assays. Remarkably, peptides methylated at lysine 4 lose this NuRD-binding ability, while peptides methylated at lysine 9 do not (46). It seems that binding of the NuRD complex to the nucleosome requires a specific region of the H3 tail and is methylation-sensitive.
Secondly, a genetic approach has provided evidence that the selective accumulation of H4 acetylated at lysine 16 on the Drosophila male X chromosome counteracts the condensing effect of the chromatin-remodeling ATPase ISWI (47). Related work shows that the site on chromatin recognized by ISWI consists of the H4 tail bound to DNA, and that acetylation of H4 at lysine 16 reduces the ability of ISWI to interact productively with its substrate (47, 48). Together, these results provide a plausible mechanism by which H4 acetylated at lysine 16 can lead to decondensation and transcriptional upregulation of the Drosophila male X chromosome. The results also provide an important example of how signals on the nucleosome surface, read by nonhistone proteins, may require a combination of both DNA and the appropriately modified histone tail.
To take full advantage of the information potential of histone tail modifications, the histone code must encompass combinations of modifications. Ideally, combinations will involve not only modifications on the same tail, but those on different tails as well. Recent studies in yeast have provided a dramatic example of how modification of one histone tail can influence that of another and have brought an additional player, the 76–amino acid peptide ubiquitin, into the limelight. Ubiquitin can be attached to specific lysines in the C-terminal tails of H2A (lysine 119) and H2B (lysine 120) in higher eukaryotes, and to H2B lysine 123 alone in yeast (49). By a combination of genetic and biochemical assays, Sun and Allis (50) have shown that yeast mutants that cannot ubiquitination H2B show no detectable methylation of lysine 4 in the N-terminal tail of H3. This is true whether the ubiquitination defect is due to mutation of the enzyme responsible (RAD6) or to substitution of lysine 123 of H2B with arginine. RAD6 is involved in many cellular processes, including DNA repair and gene silencing. The mechanism that links H2B ubiquitination and H3 methylation remains uncertain, though one can hazard a guess that ubiquitination somehow enhances the ability of the SET1 histone methylase to access its chromation substrate. It also remains to be determined whether this, or similar, interactions occur in higher eukaryotes.
Manipulating the code: new routes to therapy
Like all enzymes, the enzyme families that modify core histones or reorganize chromatin structure are susceptible to inhibition by appropriately designed reagents. Since these enzymes regulate patterns of gene expression by targeting selected genes or chromosome regions, the therapeutic potential of such inhibitors is enormous, particularly for treating cancers or other diseases showing aberrant patterns of gene expression. The enzymes most closely studied to date are the HDACs, for which a wide range of inhibitors is now available. Some of these agents have proved to be remarkably effective at selectively inhibiting the growth of human tumor cells, and several are now in clinical trials against a variety of tumor types (51). Unfortunately, we know rather little about the in vivo roles of the various deacetylases and their inhibitor sensitivities. The different members of the deacetylase family are likely to have very different functions in vivo, which provides a strong incentive to identify more selective inhibitors. Inhibitors specific for the NAD-dependent deacetylase Sir2 have recently been identified from screens of relatively small numbers of compounds using an in vivo assay (52). Initial experiments showed no increase in histone acetylation in cultured cells treated with these inhibitors, suggesting that acetylated histones are not a major substrate of the Sir2 enzyme family in vivo (52).
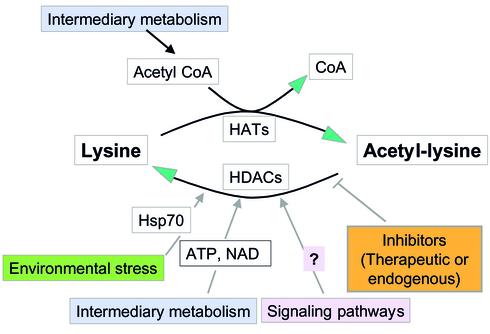
When considering therapeutic approaches, it is worth bearing in mind that factors other than enzyme inhibitors (natural or synthetic) can influence patterns of histone modification in vivo. The situation for histone acetylation is summarized in Figure 3. Metabolic factors include intranuclear levels of acetyl-CoA for HAT activity, NAD for Sir2 deacetylases (ref. 52 and references therein), and ATP for deacetylation of chromatin substrates by at least some HDACs (26). The HDAC inhibitor butyric acid occurs in the large intestine in millimolar concentrations that may be influenced by diet (53). HDAC activity can also be influenced indirectly by targeting of proteins that modify or interact with the HDACs themselves. For example, HDACs are phosphorylated in vivo by protein kinases such as CK2 in ways that influence both their catalytic activity and their ability to interact with other proteins in functional complexes (54, 55). This links HDAC function to intracellular signaling pathways. Also, the stress response (heat shock) protein HSP70 has recently been shown to be present in HDAC complexes, where it may play a role in the ATP-dependent deacetylation of chromatin substrates (22). Levels of HSP70 and other stress response proteins rise in response to a variety of toxins and other adverse environmental conditions, and it will be of interest to determine to what extent this influences HDAC activity in vivo. Finally, patterns of acetylation are inevitably influenced by, and exert an effect on, other histone tail modifications. These interactions, some of which are discussed above, will make it difficult to predict the functional effects of even highly specific enzyme inhibitors.
Figure 3.
Environmental and metabolic factors that can influence patterns of histone acetylation. Levels of this modification reflect the competing action of acetylating (HAT) and deacetylating (HDAC) enzymes in a given chromosomal region. Both of these classes of enzyme are under complex control.
Pressing questions
With new results appearing at such a pace, prediction is a risky business. However, there remain some fairly obvious gaps in our knowledge. Firstly, how is histone methylation removed? Is there a histone demethylase, or are methylation levels regulated by histone replacement or even removal of N-terminal tails by proteolysis? What is the significance of H3 isoforms mono-methylated at lysines 4 and 9, and how do we reconcile the frequency and stability of histone methylation with a regulatory role in gene expression? We also need more information on the relatively neglected histones H2A and H2B: The importance of H2B ubiquitination has been noted, and H2B phosphorylation has been associated with the chromatin reorganization that accompanies apoptosis (56), but studies on these histones still lag behind those on H3 and H4. Finally, we need to refine our ideas about the histone code and what it does, and to continue to search for specific examples of how it operates. The “epigenome” provides an attractive target for those seeking new work for soon-to-be-redundant DNA sequencing factories, but revealing the secrets of such a complex, dynamic, and multifaceted entity presents a challenge even more daunting than that of the genome itself.
Acknowledgments
The authors thank Darren White for advice on the preparation of illustrations. H.T. Spotswoodis supported by a Prize Studentship from the Wellcome Trust.
Footnotes
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: histone acetyltransferase (HAT); histone deacetylase (HDAC); histone methyltransferase (HMT).
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. Histones as regulators of genes. Sci Am. 1992;267:68B–74B. doi: 10.1038/scientificamerican1092-68. [DOI] [PubMed] [Google Scholar]
- 3.Langst G, Becker PB. Nucleosome mobilization and positioning by ISWI-containing chromatin remodeling factors. J Cell Sci. 2001;114:2561–2568. doi: 10.1242/jcs.114.14.2561. [DOI] [PubMed] [Google Scholar]
- 4.Pogo BG, Allfrey VG, Mirsky AE. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci USA. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebbes T, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1403. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by lack of histone H4 acetylation, a marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 7.Turner BM, Birley AJ, Lavender JS. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophilapolytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 8.Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 9.Tordera V, Sendra R, Pérez-Ortin JE. The role of histones and their modifications in the informative content of chromatin. Experientia. 1993;49:780–788. doi: 10.1007/BF01923548. [DOI] [PubMed] [Google Scholar]
- 10.Hansen JC, Tse C, Wolffe AP. Structure and function of the core histone N-termini. More than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 11.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 13.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 14.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 16.Covault J, Chalkley R. The identification of distinct populations of acetylated histones. J Biol Chem. 1980;255:9110–9116. [PubMed] [Google Scholar]
- 17.Gregory RI, et al. Inhibition of histone deacetylases alters allelic chromatin conformation at the imprinted U2af1-rs1locus in mouse embryonic stem cells. J Biol Chem. 2002;277:11728–11734. doi: 10.1074/jbc.M105775200. [DOI] [PubMed] [Google Scholar]
- 18.Thomas G, Lange HW, Hempel K. Kinetics of histone methylation in vivo and its relation to the cell cycle in Ehrlich ascites tumor cells. Eur J Biochem. 1975;51:609–615. doi: 10.1111/j.1432-1033.1975.tb03963.x. [DOI] [PubMed] [Google Scholar]
- 19.Lin R, Cook RG, Allis CD. Proteolytic removal of core histone amino termini and dephosphorylation of histone H1 correlate with the formation of condensed chromatin and transcriptional silencing during Tetrahymenamacronuclear development. Genes Dev. 1991;9:1601–1610. doi: 10.1101/gad.5.9.1601. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad K, Henikoff S. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 21.Thomson S, Mahadevan LC, Clayton AL. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin Cell Dev Biol. 1999;10:205–214. doi: 10.1006/scdb.1999.0302. [DOI] [PubMed] [Google Scholar]
- 22.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 23.Ng H-H, Bird A. Histone deacetylases: silencers for hire. TIBS. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 24.Grant PA, Berger SL. Histone acetyltransferase complexes. Semin Cell Dev Biol. 1999;10:169–178. doi: 10.1006/scdb.1999.0298. [DOI] [PubMed] [Google Scholar]
- 25.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 26.Johnson CA, White DA, Lavender JS, Turner BM. Human class I histone deacetytlase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem. 2002;277:9590–9597. doi: 10.1074/jbc.M107942200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 29.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 30.O’Carroll D, et al. Isolation and characterization of Suv39H2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, et al. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. J Biol Chem. 2002;277:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- 32.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 33.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromodomain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 34.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 37.Czermin B, et al. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2001;2:915–919. doi: 10.1093/embo-reports/kve210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos-Rosa, H., et al. 2002. Lysine tri-methylation in active genes extends histone code. Nature. In press.
- 39.Maison C, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 40.Boggs BA, et al. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 41.Heard E, et al. Methylation of histone H3 at lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 42.Hecht A, Laroche T, Strahl-Bosinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 43.Cowell IG, et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- 44.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 45.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 46.Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]
- 47.Corona DF, Clapier CR, Becker PB, Tamkun JW. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 2001;3:242–247. doi: 10.1093/embo-reports/kvf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jason LJM, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquintination: a tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- 50.Sun Z-W, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 51.Marks P, et al. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 52.Grozinger CM, Chao ED, Blackwell HE, Moazad D, Schreiber SL. Identification of a class of small molecule inhibitors of the Sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 53.Boffa LC, et al. Modulation of colonic epithelial cell proliferation, histone acetylation and luminal short chain fatty acids by variation of dietary fiber (wheat bran) in rats. Cancer Res. 1992;52:5906–5912. [PubMed] [Google Scholar]
- 54.Tsai, S.C., and Seto, E. 2002. Regulation of histone deacetylase-2 by protein kinase CK2. J. Biol. Chem. In press. [DOI] [PubMed]
- 55.Galasinski SC, Resing KA, Goodrich JA, Ahn NG. Phosphatase inhibition leads to histone deacetylases 1 and 2 phosphorylation and disruption of corepressor interactions. J Biol Chem. 2002;277:19618–19626. doi: 10.1074/jbc.M201174200. [DOI] [PubMed] [Google Scholar]
- 56.Ajiro K. Histone H2B phosphorylation in mammalian apoptotic cells. An association with DNA fragmentation. J Biol Chem. 2000;275:439–443. doi: 10.1074/jbc.275.1.439. [DOI] [PubMed] [Google Scholar]