Abstract
PFIC II is a subtype of progressive familial intrahepatic cholestasis (PFIC) that is associated with mutations in the ABCB11 gene encoding the bile salt export pump (BSEP). However it is not known how these mutations cause this disease. To evaluate these mechanisms, we introduced seven PFIC II–associated missense mutations into rat Bsep and assessed their effects on Bsep membrane localization and transport function in MDCK and Sf9 cells, respectively. Five mutations, G238V, E297G, G982R, R1153C, and R1268Q, prevented the protein from trafficking to the apical membrane, and E297G, G982R, R1153C, and R1268Q also abolished taurocholate transport activity, possibly by causing Bsep to misfold. Mutation C336S affected neither Bsep transport activity nor the apical trafficking of Bsep, suggesting that this mutation alone may not cause this disease. D482G did not affect the apical expression but partially decreased the transport activity of Bsep. Mutant G238V was rapidly degraded in both MDCK and Sf9 cells, and proteasome inhibitor resulted in intracellular accumulation of this and other mutants, suggesting proteasome-mediated degradation plays an important role in expression of these PFIC II mutants. Our studies highlight the heterogeneous nature of PFIC II mutations and illustrate the significance of these mutations in the function and expression of Bsep.
Introduction
Bile secretion is mediated by several ATP binding cassette (ABC) transporters located in the canalicular membrane of hepatocytes (for reviews, see refs. 1, 2). Among these ABC transporters, the bile salt export pump (BSEP, or ABCB11) represents the primary if not the sole transport system for the canalicular excretion of bile salts (3, 4). Bile secretory failure results in cholestasis. Progressive familial intrahepatic cholestasis (PFIC) of infancy represents a group of inherited cholestatic diseases that are classified into three subtypes. One of these subtypes, PFIC II, is associated with mutations in the BSEP gene (5, 6). PFIC I and PFIC III are caused by mutations in the genes encoding FIC I (ATP8B1) and MDR3 (ABCB4), respectively (7, 8).
PFIC patients with mutations in the BSEP gene have normal γ-glutamyltransferase activity, very low concentrations of bile salts in bile, and an absence of bile duct proliferation. Two human genetics studies have identified a number of premature termination, missense, and frame shift mutations in the BSEP gene in patients with PFIC II (5, 6). These data provided definitive evidence that BSEP plays a crucial role in bile salt–dependent bile secretion in humans.
In the present study we have examined the effects of a number of missense mutations in the human BSEP gene that have previously been associated with PFIC II. We chose to study missense mutations because these single-point mutations may reveal information about residues or regions in BSEP essential for its function or biogenesis. It remains unclear how these missense mutations identified from genetic studies affect BSEP expression. For example, some published missense mutations have been found on only one allele (i.e., heterozygous mutation). Yet in these patients BSEP was not detected by immunohistochemistry (6). Since in some cases the parents carrying the mutation were reported to be phenotypically normal, the identified missense mutations may not cause the disease. To clarify this issue, mutagenesis studies are also needed to directly determine whether these defined mutations are capable of impairing BSEP function. In addition, if a missense mutation results in PFIC II (i.e., a homozygous missense mutation or a heterozygous mutation that affects BSEP function), the mutation could affect BSEP expression in at least two ways. First, the mutation could affect the biogenesis of BSEP, in which case the BSEP mutant might be sequestered within the cell and unable to be targeted correctly to the canalicular membrane. Second, the mutation might not affect the trafficking of BSEP to the plasma membrane, but instead may impair the transport function of the protein, for example by preventing substrate binding.
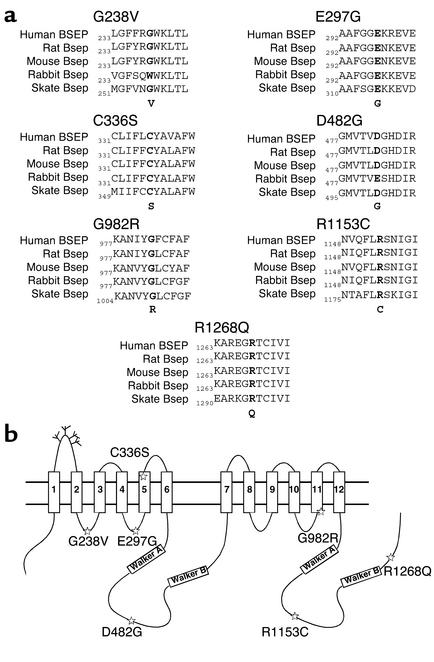
To examine these issues in greater detail, we have introduced seven published human BSEP missense mutations that are associated with PFIC II into rat Bsep. These mutations in human BSEP (Figure 1a) occur in regions that are highly conserved throughout the vertebrate phylum (9) and therefore are likely to be essential residues for Bsep function or membrane localization. We used rat Bsep as a model protein in our studies because the human gene had not been functionally expressed and rat Bsep is highly homologous to the human orthologue, with 82.3% identity and 88% similarity at the amino acid level (3, 5). Furthermore, the PFIC II mutations in human BSEP occur in regions highly conserved between human BSEP and rat Bsep (Figure 1a). We have expressed these Bsep PFIC II mutants in both Madin-Darby canine kidney (MDCK) cells and insect Sf9 cells. Cellular localization of these Bsep PFIC II mutants was analyzed in MDCK cells, and their ability to transport taurocholate was determined in the Sf9 cell expression system.
Figure 1.
PFIC II mutations reside in regions highly conserved throughout the vertebrate phylum. (a) Sequence alignment of mutation sites in BSEP/Bsep from human, rat, mouse, rabbit, and skate using the Clustal alignment software MegAlign (DNASTAR Inc., Madison, Wisconsin, USA). The PFIC II mutations in human BSEP and the corresponding residues in other Bseps are shown in bold. (b) The positions of G238V, E297G, C336S, D482G, G982R, R1153C, and R1268Q are indicated by stars in a predicted topology model of rat Bsep. The Walker A and Walker B regions are shown in boxes, and a number of potential glycosylation sites are indicated by the branch structures in the first extracellular loop of Bsep. The predicted 12 transmembrane-spanning segments in Bsep are shown in boxes labeled 1–12.
Methods
Chemicals, cells, and antibodies.
[3H]taurocholic acid (3 Ci/mmol) was obtained from NEN Life Science Products Inc. (Boston, Massachusetts, USA). Common chemicals and reagents were purchased from American Bioanalytical (Natick, Massachusetts, USA) or Mallinckrodt Baker Inc. (Phillipsburg, New Jersey, USA). Bile acids were from Sigma-Aldrich (St. Louis, Missouri, USA). The tissue culture reagents were from Invitrogen Corp. (Carlsbad, California, USA). MDCK cells and human embryonic kidney (HEK) 293 cells were generous gifts from Alan Fanning (Yale University). Antibodies raised against rat Bsep, protein gp-135 (an apical marker of MDCK cells) (10), and NaK-ATPase were kindly provided by Bruno Stieger (University Hospital, Zurich, Switzerland), George Ojakian (State University of New York, Brooklyn, New York, USA), and Michael Caplan (Yale University), respectively.
Plasmid construction.
For the construction of the pEGFP-Bsep plasmid incorporating green fluorescent protein (GFP), a full-length cDNA encoding rat Bsep (kindly provided by Peter Meier, University Hospital) was introduced into pEGFPC1 (Clontech Laboratories Inc., Palo Alto, California, USA) after the C-terminus of EGFP by digestion with restriction enzymes and PCR. The fusion construct (pEGFP-Bsep) encodes a chimera protein, Bsep-GFP, starting with EGFP and followed by a 17-amino-acid linker and the Bsep sequence minus the first amino acid, methionine. For construction of the PFIC II mutants of pEGFP-Bsep, site-directed mutagenesis was performed using the QuikChange PCR mutagenesis kit (Stratagene, La Jolla, California, USA). This generated the following missense mutants: G238V, E297G, C336S, D482G, G982R, R1153C, and R1268Q. The location of these mutations is shown in a topology model of rat Bsep (Figure 1b). Both EGFP-Bsep and PFIC II mutant constructs were confirmed by DNA sequencing at the W.M. Keck sequencing facility of Yale University.
Cell culture and transfection.
MDCK cells and HEK 293 cells were grown in DMEM supplemented with 10% FBS, containing 100 units of penicillin/ml and 100 μg of streptomycin sulfate/ml. For transient transfection, confluent MDCK cells and HEK 293 cells were transfected using LipofectAmine (Invitrogen Corp.) according to the manufacturer’s instructions. MDCK cells stably expressing Bsep-GFP were obtained by selection using antibiotic G-418 sulfate (Invitrogen Corp.). For proteasome inhibition, 5 μmol/ml MG-132 (Calbiochem-Novabiochem Corp., San Diego, California, USA) was added to the culture medium for various lengths of time as indicated in Figure 4.
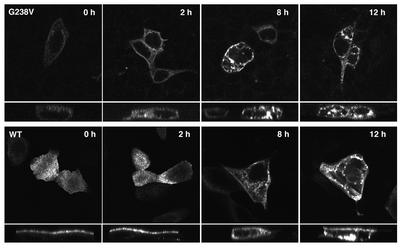
Figure 4.
G238V is degraded by proteasomes in MDCK cells. MDCK cells were transiently transfected with wild-type Bsep-GFP (bottom row) or G238V (top row) for 48 hours. The transfected cells were then either treated with 5 μmol/ml MG-132 for an additional 2, 8, or 12 hours or left untreated (controls, indicated as 0 h). All images were acquired under identical conditions, in which gain was optimized for the treated cells in order to avoid saturation of the signal. Under these conditions, G238V was barely detectable in MDCK cells not treated with MG-132 (G238V, 0 h). Protein aggregates were seen as a number of signals at perinuclear positions in the cytoplasm. In each panel, the top part shows the en face image and the bottom part shows the Z-sectioning image.
Membrane preparation.
Total membrane fractions were prepared from transfected MDCK and HEK 293 cells grown on 6-cm dishes (Corning-Costar Corp., Acton, Massachusetts, USA). The cells were scraped from the dish, homogenized in a hypotonic lysis buffer (10 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10 mM Tris/HEPES at pH 7.4, 5 μg/ml leupeptin, 1 μg/ml pepstatin A, and 2 mM PMSF) on ice and centrifuged at 400 g. The postnuclear supernatant was centrifuged for 20 minutes at 100,000 g to pellet the total membranes. The cytosol fraction (the supernatant after centrifugation) was removed and stored for separate analysis, and the membranes were resuspended in the lysis buffer supplemented with 0.25 M sucrose.
Expression of PFIC II mutants of rat Bsep in Sf9 cells.
The PFIC II mutant pEGFP-Bsep constructs described above were used to generate a series of pFastBac-Bsep constructs each containing a desired PFIC II mutation. The Bac-to-Bac baculovirus system (Invitrogen Corp.) was used to generate the respective recombinant baculovirus. The total membrane fractions were prepared from the infected Sf9 cells and resuspended in a taurocholate uptake buffer as described previously (9). Membrane aliquots were stored at –80°C. Protein concentration was determined using the Bradford assay with BSA as a standard (11).
Vesicle transport assays.
One hundred micrograms of Sf9 cell membrane vesicles were used in each assay. [3H]taurocholate uptake was measured by a rapid filtration assay as described previously (9). Briefly, vesicle uptake of [3H]taurocholate (2.5 μM) was determined in the presence and absence of ATP (5 mM) for 10 minutes at 28°C. The difference between taurocholate uptake measured in the presence and absence of ATP was defined as ATP-dependent taurocholate uptake. For inhibition studies, glycochenodeoxycholate, glycocholate, taurochenodeoxycholate, and taurocholate were each added to the uptake solution at a final concentration of 50 μM.
Electrophoresis and immunoblotting.
Total membrane and cytosol fractions from MDCK cells and total membranes from HEK 293 cells and Sf9 cells were analyzed by SDS-PAGE (12). The separated polypeptides were electrotransferred to PVDF membranes (Bio-Rad Laboratories Inc., Hercules, California, USA) and then subjected to Western blotting using a standard protocol. We used a rabbit polyclonal antibody (kindly provided by Bruno Stieger) raised against the C-terminal 14 amino acids of rat Bsep (3). The immunoreactive proteins were visualized using an enhanced chemiluminescence kit (Amersham Biosciences, Piscataway, New Jersey, USA).
Confocal microscopy.
After transfection, MDCK and HEK 293 cells grown on coverslips were fixed with ice-cold methanol for 10 minutes at –20°C. Coverslips were mounted on glass microscope slides using Vectashield containing propidium iodide (Vector Laboratories, Burlingame, California, USA). Cells were viewed on a Zeiss LSM 510 confocal microscope (Carl Zeiss Inc., Thornwood, New York, USA) using Multitracking with BP505-550 and LP585 filters. En face views were used to ascertain the expression level of Bsep-GFP, and then areas were selected to collect a Z section series to determine the cellular localization of the signal. Digital images were saved and analyzed with Adobe PhotoShop (Adobe Systems Inc., Mountain View, California, USA).
Results
Expression of Bsep-GFP in MDCK and HEK 293 cells.
To examine the cellular localization of the rat Bsep PFIC II mutants, we first created a fusion construct, Bsep-GFP, in which GFP was fused to the N-terminus of Bsep, and Bsep-GFP was expressed in MDCK cells. We chose MDCK cells, a well-characterized polarized kidney cell line, for our expression studies because many apical and basolateral proteins (e.g., P-glycoprotein and NaK-ATPase) have the same membrane localization in these cells as in hepatocytes.
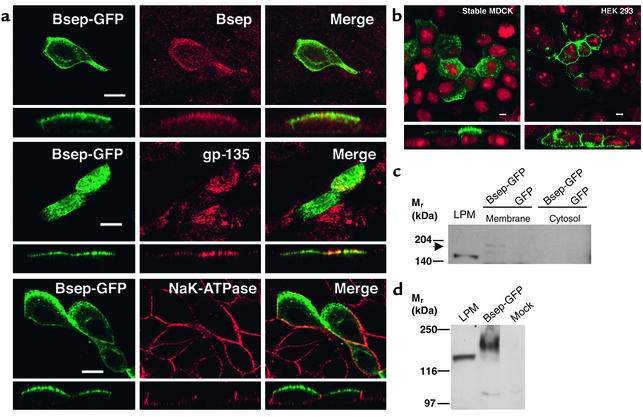
In transiently transfected MDCK cells, transfection efficiency was low (less than 5%). However, the green fluorescent signal colocalized well with the immunostaining signal detected using an antibody against rat Bsep (Figure 2a, top row). GFP also colocalized with gp-135, an apical marker of MDCK cells (Figure 2a, center row), but was distinct from the basolateral marker, NaK-ATPase (Figure 2a, bottom row), as shown by immunostaining. This demonstrates that Bsep-GFP was exclusively expressed on the apical membrane surface of MDCK cells. No specific apical fluorescent signal was detected in nontransfected MDCK cells, and the green fluorescent signal was observed throughout the cytoplasm when only GFP was transfected into MDCK cells (data not shown). Our data showing that Bsep-GFP was expressed at the apical membrane of MDCK cells is consistent with the normal canalicular/apical localization of Bsep in hepatocytes and suggests that the added GFP tag did not affect the membrane localization of Bsep as it trafficked through the secretory pathways in MDCK cells.
Figure 2.
Expression of Bsep-GFP in MDCK cells and HEK 293 cells. (a) After transient transfection with Bsep-GFP, MDCK cells were processed for immunofluorescence using antibodies against rat Bsep, gp-135, and NaK-ATPase, and imaged to locate the GFP fusion protein and apical and basolateral membranes of MDCK cells. In each panel, the top part shows the en face image and the bottom part shows the Z-sectioning image. Bsep-GFP colocalized with gp-135 but was distinct from NaK-ATPase. Bar, 5 μm. (b) Bsep-GFP was expressed in either a stable MDCK cell line or HEK 293 cells by transient expression. Both en face and Z-sectioning images of the GFP fusion protein are presented. Bar, 5 μm. Total membrane and cytosol fractions were prepared from (c) MDCK cell lines either stably expressing Bsep-GFP or expressing GFP alone, or (d) from transfected and mock-treated HEK 293 cells. These fractions (100 μg protein per lane) and a rat liver plasma membrane preparation (LPM) (100 μg protein per lane) were separated on a 6.5% Laemmli gel, and Bsep-GFP was detected by Western blotting using an antibody against rat Bsep. The position of Bsep-GFP in the total membrane fraction of MDCK cells is indicated by the arrow. Rat Bsep and Bsep-GFP have apparent molecular weights of approximately 160 kDa and 190 kDa, respectively.
Because the low transfection efficiency of transient transfection made detailed analyses of Bsep-GFP expression difficult, we next created an MDCK cell line that stably expressed Bsep-GFP. In the stable cell line, Bsep-GFP was exclusively expressed at the apical surface of MDCK cells (Figure 2b). When the total membranes from the stable cell lines were analyzed by SDS-PAGE followed by immunoblotting, the rat Bsep antibody recognized a band with an approximate molecular weight of 190 kDa, which is 30 kDa larger than the rat Bsep (∼160 kDa) detected from a rat liver plasma membrane fraction (Figure 2c). This difference in molecular weight is consistent with the size of the GFP tag. The Bsep-GFP band was not detected in MDCK cells stably expressing GFP alone, nor was it detected in the cytosolic fraction of MDCK cells.
We also compared the expression of Bsep-GFP in nonpolarized kidney epithelial cells (HEK 293 cells). After transient transfection, Bsep-GFP was expressed over the entire plasma membrane of HEK 293 cells and Bsep-GFP expression in plasma membrane was higher in HEK 293 cells than in MDCK cells, reflecting higher efficiency of transfection (Figure 2b). Bsep-GFP was detected by immunoblotting as a 190-kDa protein, consistent with the size of Bsep-GFP expressed in MDCK cells (Figure 2d). Taken together, expression studies in both MDCK and HEK 293 cells showed that Bsep-GFP was synthesized with the expected molecular weight and trafficked to the apical membrane surface only when expressed in the polarized kidney cells.
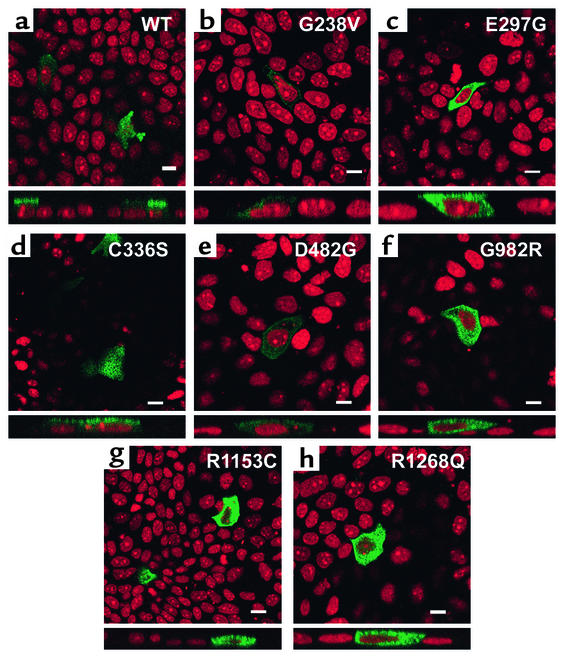
PFIC II mutations have a heterogeneous effect on Bsep-GFP localization in MDCK cells.
G238V, E297G, C336S, D482G, G982R, R1153C, and R1268Q are published mutations in human BSEP that are associated with PFIC II. To analyze whether these PFIC II mutations affect the membrane localization of Bsep, each mutation was introduced into Bsep-GFP and the mutant proteins were expressed in MDCK cells by transient transfection. Contrary to the apical expression of wild-type Bsep-GFP (Figure 3a), five of these PFIC II mutations, G238V, E297G, G982R, R1153C, and R1268Q, caused the protein to be sequestered within the cell (Figure 3, b, c, and f–h). The intracellular localization of these mutants was confirmed by immunostaining with antibody against gp-135, which did not colocalize with the green fluorescent signals (data not shown). In the cases of E297G, G982R, R1153C, and R1268Q, the green fluorescent signals were widely distributed throughout the cytoplasm. The level of expression was much lower in G238V, although this mutant was also detected in the cytoplasm rather than on the apical membrane. Surprisingly, mutation C336S did not alter the apical expression of Bsep-GFP (Figure 3d). In the cells expressing D482G, a significant green fluorescent signal was also detected at the apical surface as well as in the cytoplasm (Figure 3e). Other studies in polarized hepatic C2rev7 cells also showed that mutant C336S was expressed at the canalicular membrane of the C2rev7 cells in a manner similar to the wild-type Bsep-GFP (our unpublished observation).
Figure 3.
PFIC II mutations have a heterogeneous effect on the localization of Bsep-GFP in MDCK cells. MDCK cells were transiently transfected with wild-type (WT) Bsep-GFP (a) or Bsep PFIC II mutants (b–h) for 72 hours and imaged by confocal microscopy. In each panel, the top part shows the en face image and the bottom part shows the Z-sectioning image. Bar, 10 μm.
Ubiquitin/proteasome-mediated degradation plays a role in the expression of some PFIC II mutants.
Several studies with other ABC transporters, such as cystic fibrosis transmembrane conductance regulator (CFTR) and copper-transporting P-type adenosine triphosphatase (ATP7B), have shown that intracellularly sequestered membrane proteins that resulted from mutation-induced misfolding were subsequently degraded by the ubiquitin/proteasome system (13–15). This seemed a particularly interesting possibility for G238V because this mutant protein was expressed rather poorly in MDCK cells. Thus we examined the involvement of proteasomes in the expression of Bsep PFIC II mutants by adding MG-132, a specific proteasome inhibitor, for different lengths of time to the culture medium of the MDCK cells transfected with PFIC II mutants (Figure 4, top row). The effect of MG-132 on the expression of wild-type Bsep-GFP in MDCK cells was also examined (Figure 4, bottom row).
After treatment with MG-132 for 2 hours, we did not observe any increase in fluorescent signal intensity or any change in localization of wild-type Bsep-GFP. In contrast, a 2-hour treatment with MG-132 increased the fluorescent signal for G238V about 60% compared with untreated cells (after quantification by summation of the fluorescent signals from a series of Z-sectioning images); aggregate formation began to be seen in G238V-expressing cells after the 2-hour treatment. The presence of MG-132 significantly increased the fluorescence intensity of mutant G238V seen in the cells after an 8-hour incubation and persisted after 12 hours (Figure 4, top row). The G238V mutant accumulated at several perinuclear positions, reminiscent of the protein aggregates seen in a previous study of a mutant of ATP7B, which is associated with Wilson disease (15). These data together suggest that G238V in MDCK cells was very unstable and that active ubiquitin/proteasome-mediated degradation played a role in the expression of this mutant in MDCK cells. Mutants E297G, G982R, R1153C, and R1268Q (data not shown) also accumulated into aggregates after incubation with MG-132, suggesting the involvement of the ubiquitin/proteasome system in their expression, but the increase in fluorescence intensities was less significant with these mutants than with G238V. After a 12-hour treatment with proteasome inhibitor, some wild-type Bsep-GFP was detected in perinuclear aggregate structures, with some of the Bsep-GFP still localized on the apical surface (Figure 4, bottom row, far right). This is not surprising since several studies have shown that GFP chimeras containing wild-type proteins expressed in HEK 293 cells can form an aggresome or aggregates when the cells are treated with proteasome inhibitors for several hours (14, 16).
Taurocholate transport function of PFIC II mutants.
As described above, two mutations (C336S and D482G) did not affect the ability of Bsep to reach the apical membrane. Yet it is not clear whether C336S and D482G disturb the folded structure of Bsep in such a way that the bile salt transport function of Bsep is affected. Therefore we used a baculovirus expression system to characterize the transport function of these mutants. In contrast to C336S and D482G, G238V, E297G, G982R, R1153C, and R1268Q prevented the apical trafficking of Bsep-GFP in MDCK cells. We also examined whether these mutants were able to transport bile salt despite their inability to reach the cell surface when expressed in MDCK cells.
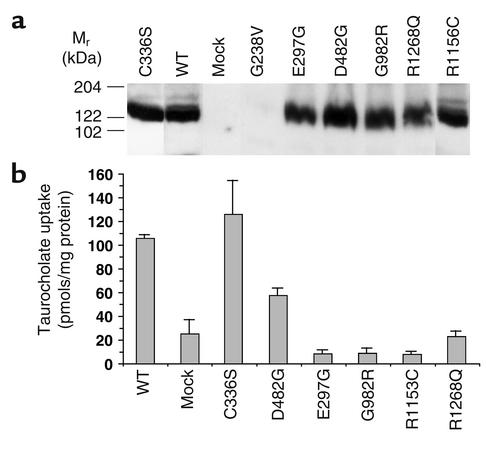
Bsep cDNAs carrying the seven PFIC II missense mutations were expressed in insect Sf9 cells using recombinant baculovirus. We manipulated the inocula of the baculovirus used in the experiment in order to achieve an expression level in all mutants that was comparable to that of wild-type protein. When total membranes from infected Sf9 cells were analyzed by SDS-PAGE followed by Western blotting, six of the PFIC II mutants were expressed at a level comparable to the wild-type rat Bsep as detected by the Bsep antibody. In contrast, G238V mutant protein was below the level of detection (Figure 5a).
Figure 5.
PFIC II mutations exhibit heterogeneous effects on Bsep taurocholate transport activity. (a) Sf9 cell vesicles (100 μg) expressing Bsep or Bsep PFIC II mutant were analyzed by Western blotting as indicated. (b) ATP-dependent [3H]taurocholate transport was measured using membrane vesicles isolated from Sf9 cells expressing Bsep or PFIC II mutants. Vesicle uptake of [3H]taurocholate (2.5 μM) was determined in the presence and absence of ATP (5 mM). Data represent the mean ± SD of three determinations. The difference between taurocholate uptake measured in the presence and absence of ATP was defined as ATP-dependent taurocholate uptake.
To examine whether these PFIC II missense mutations affected the bile salt transport function of Bsep, total membrane fractions were prepared from the infected Sf9 cells and analyzed for ATP-dependent taurocholate uptake. As can be seen in Figure 5b, significant [3H]taurocholate uptake (∼110 pmol/mg protein/10 min) was observed in membrane vesicles isolated from Sf9 cells infected with wild-type rat Bsep cDNA. Very little ATP-dependent taurocholate uptake was seen in vesicles from mock-treated Sf9 cells. The E297G, G982R, R1153C, and R1268Q mutations reduced taurocholate uptake of Bsep to the level of that in membrane vesicles from mock-treated Sf9 cells (Figure 5b). Because G238V decreased Bsep expression to an undetectable level (Figure 5a), we were unable to measure the taurocholate uptake by this mutant. The membrane vesicles expressing D482G showed a decrease in taurocholate uptake compared with wild-type Bsep, while the vesicles expressing C336S showed no significant difference from wild-type Bsep (Figure 5b).
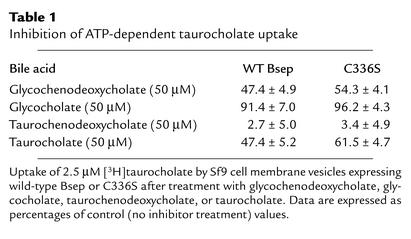
Since glycochenodeoxycholate and glycocholate rather than taurochenodeoxycholate and taurocholate are the major bile salt species in human bile, we also investigated whether glycochenodeoxycholate and glycocholate inhibited [3H]taurocholate transport in Sf9 vesicles expressing C336S mutant and wild-type Bsep (Table 1). Glycocholate had a small inhibitory effect on both the wild-type Bsep and C336S (inhibited less than 10% of taurocholate transport) in this assay, while taurochenodeoxycholate inhibited more than 90% of the transport. Glycochenodeoxycholate and taurocholate inhibited about 40–50% of the [3H]taurocholate transport by both wild-type Bsep and C336S. Thus the relative inhibitory effects of these bile salts on the C336S mutant and the wild-type rat Bsep in this study is similar to the relative affinities of these bile salts for the rat and human bile salt transporters (3, 17, 18, 19). The similarity of the inhibitory effects of these bile salts on the C336S mutant and wild-type rat Bsep is consistent with the finding that C336S in rat Bsep did not affect the transport of taurocholate by this protein (Figure 5b).
Table 1.
Inhibition of ATP-dependent taurocholate uptake
Discussion
In this study we characterized the effect of seven missense PFIC II mutations introduced into rat Bsep when expressed in MDCK cells and Sf9 cells. Taken together, the results from these two expression systems show that C336S affects neither the bile salt transport nor the apical localization of the protein. D482G also did not seem to prevent some of the Bsep from reaching the apical membranes in MDCK cells, but this mutation partially decreased the transport activity of the protein. In contrast, E297G, G982R, R1153C, and R1268Q abolished taurocholate transport by Bsep and also prevented the protein from trafficking to the apical membrane. G238V apparently led to rapid degradation of Bsep when expressed in MDCK and Sf9 cells, presumably by making the protein highly unstable. These mutations presumably have a similar effect on Bsep expression in hepatocytes.
Since C336S did not affect either apical localization or the transport function of Bsep, this mutation alone may not be the cause of the cholestatic condition seen in PFIC II. However, because we have not examined the effects of C336S on the transport function and trafficking behavior of human BSEP protein in a hepatic cell model, we cannot exclude the possibility that this mutation may have distinct effects on human BSEP expressed in vivo. We also noticed that in one earlier genetic study, this mutation was described as a heterozygous mutation in two patients (6). Because only single-strand conformation polymorphism analysis instead of full-length sequencing was used in the earlier study to screen for mutations in human BSEP, other yet-to-be-identified mutations in BSEP may cause the cholestatic condition in those patients.
Our expression studies in MDCK cells showed that G238V, E297G, G982R, R1153C, and R1268Q prevented the Bsep protein from trafficking to the apical membrane. In general, there are two types of mutations that can cause the intracellular retention of a protein. The first type impairs the specific sorting signals within the protein, leading to the intracellular retention of the mutant protein. The second type of mutation disrupts the global folding of the protein in such a way that misfolded proteins are detected by the quality control system of the secretory pathway and are subsequently sequestered in the cytoplasm. Several studies have shown that the second type of mutation often leads to loss of function as a result of misfolding, whereas the first type of mutation affects only specific regions of the protein and thus may not necessarily affect the protein’s function (20, 21). Therefore it was of interest to examine whether mutants G238V, E297G, G982R, R1153C, and R1268Q are capable of transporting bile salts despite their defect in apical trafficking. Our results showing that the Bsep mutants E297G, G982R, R1153C, and R1268Q also lost bile salt transport function (see Figure 5b) raise the possibility that the trafficking defect of these mutations could have resulted from misfolding of the protein.
In some of our experiments, we observed that mutant D482G was expressed in the cytoplasm of MDCK cells in addition to trafficking to the apical membranes. This could be due either to the protein overexpression that is often associated with transient transfection or to any trafficking signal in Bsep being adversely affected by this mutation. D482G partially decreased Bsep bile salt transport function. It remains to be investigated whether this mutation affects substrate binding to Bsep or the ATP hydrolysis of the protein since it lies between the Walker A and Walker B regions of the first ABC domain of the protein.
Several recent studies have shown that misfolded proteins such as the CFTR F508 deletion mutant are degraded by the ubiquitin/proteasome system (13, 14). Ubiquitin/proteasome-mediated degradation has also been implicated in the expression of a mutant of multidrug resistance protein 2 (ABCC2) that causes Dubin-Johnson syndrome (22) and a mutant of ATP7B, which is associated with Wilson disease (15). Our finding that the presence of proteasome inhibitor MG-132 induced the sequestered proteins to form protein aggregates suggests that ubiquitin/proteasome-mediated degradation was involved in the expression of G238V, E297G, G982R, R1153C, and R1268Q. The G238V Bsep mutant is particularly worth noting since the mutant protein was detectable only when the cells were treated with MG-132, suggesting that G238V probably made the protein highly unstable so that it was rapidly degraded in MDCK cells. This conclusion is also supported by the failure of G238V to be expressed in Sf9 cells, whereas the other six mutants and wild-type Bsep were expressed at relatively high levels. The reason only protein aggregates rather than a single aggresome were seen in the MDCK cells transfected with Bsep-GFP or the PFIC II mutants may be the relatively low expression of Bsep-GFP and PFIC II mutants in MDCK cells compared with the expression of Bsep-GFP in HEK 293 cells (see Figure 2). Several previous studies reporting aggresome formation have used HEK 293 cells, which normally have a higher expression level (14, 16). Similar protein aggregates (rather than aggresomes) were seen in human Huh7 cells expressing a mutant of ATP7B (15). An earlier study reported that human BSEP protein was not detected by immunohistochemistry of liver biopsies from patients who carried either homozygous mutation G238V, heterozygous mutation G982R, or compound heterozygous mutations E297G and R1057X (a premature termination mutation) (6). The possibility that rapid degradation contributes to the expression of these mutants in vivo may explain their absence in the liver biopsies of the PFIC II patients.
Two heterozygous mutations identified in some patients, G982R and E297G, have also been determined to be homozygous mutations in other PFIC II families (5). Our data showed that these mutations both impaired taurocholate transport by Bsep and prevented the protein from trafficking to the apical membranes. It would be of interest to determine whether the cholestatic phenotype was less severe in the patient who carries the heterozygous mutation G982R, since one would assume that some functional BSEP might still be expressed. Other mutations (G238V, D482G, R1153C, and R1268Q) have been identified as homozygous mutations in affected PFIC II patients.
In summary, our studies have characterized the effects of seven published PFIC II missense mutations that have previously been shown to be associated with this disease. The PFIC II missense mutations in Bsep fall into several categories: (a) mutations (such as C336S) that did not affect function and targeting and therefore may not cause the disease; (b) mutations such as D482G that potentially affected specific regions in the protein and led to impaired function (e.g., loss of substrate binding) or impaired trafficking (e.g., loss of sorting signals); and (c) mutations such as G238V, E297G, G982R, R1153C, and R1268Q that abolished both the protein’s function and apical expression, possibly by misfolding. Our results highlight the heterogeneous nature of the missense mutations identified from human genetics studies. Mutations G238V, E297G, G982R, R1153C, and R1268Q are located at various cytosolic and transmembrane domains of Bsep. Our studies suggest that these residues and regions are critical for the proper expression of Bsep. Further characterization of the functional effects of these mutations will help to determine the roles that these residues and regions play in the biogenesis and normal folding of Bsep.
Acknowledgments
This work was funded by a grant (DK-25636) and pilot grant (DK P30-34989)from the National Institute of Diabetes and Digestive and Kidney Diseases, and by the Morphology Core of the Yale Liver Center. We thank Alan Fanning, Peter Meier, Bruno Stieger, George Ojakian, and Michael Caplan for generously providing us critical reagents, and Kim Ng for helpful discussion and critical reading of the manuscript.
Footnotes
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: ATP binding cassette (ABC); bile salt export pump (BSEP); progressive familial intrahepatic cholestasis (PFIC); Madin-Darby canine kidney (MDCK); human embryonic kidney (HEK); green fluorescent protein (GFP), cystic fibrosis transmembrane conductance regulator (CFTR).
References
- 1.Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol. 2002;64:635–661. doi: 10.1146/annurev.physiol.64.082201.100300. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, J.L., and Nathanson, M.H. 1999. Bile formation. In Schiff’s diseases of the liver. 8th edition. E.R. Schiff, M.F. Sorrell, and W.C. Maddrey, editors. Lippincott-Raven Publishers. Philadelphia, Pennsylvania, USA. 119–146.
- 3.Gerloff T, et al. The sister P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- 4.Wang R, et al. Targeted inactivation of sister of P-glycoprotein gene (spgp) in mice results in non-progressive but persistent intrahepatic cholestasis. Proc Natl Acad Sci USA. 2001;98:2011–2016. doi: 10.1073/pnas.031465498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strautnieks SS, et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20:233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- 6.Jansen PL, et al. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology. 1999;117:1370–1379. doi: 10.1016/s0016-5085(99)70287-8. [DOI] [PubMed] [Google Scholar]
- 7.Bull LN, et al. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 8.deVree JML, et al. Mutations in the MDR3 gene cause progressive familial intrahepatic cholestasis. Proc Natl Acad Sci USA. 1998;95:282–287. doi: 10.1073/pnas.95.1.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai SY, Wang L, Ballatori N, Boyer JL. Bile salt export pump is highly conserved during vertebrate evolution and its expression is inhibited by PFIC type II mutations. Am J Physiol Gastrointest Liver Physiol. 2001;281:G316–G322. doi: 10.1152/ajpgi.2001.281.2.G316. [DOI] [PubMed] [Google Scholar]
- 10.Wang AZ, Wang JC, Ojakian GK, Nelson WJ. Determinants of apical membrane formation and distribution in multicellular epithelial MDCK cysts. Am J Physiol. 1994;267:C473–C481. doi: 10.1152/ajpcell.1994.267.2.C473. [DOI] [PubMed] [Google Scholar]
- 11.Smith PK, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 14.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada M, et al. Role of ATP7B in biliary copper excretion in a human hepatoma cell line and normal rat hepatocytes. Gastroenterology. 2000;118:921–928. doi: 10.1016/s0016-5085(00)70178-8. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stieger B, Fattinger K, Madon J, Kullak-Ublick GA, Meier PJ. Drug- and estrogen-induced cholestasis through inhibition of the hepatocellular bile salt export pump (Bsep) of rat liver. Gastroenterology. 2000;118:422–430. doi: 10.1016/s0016-5085(00)70224-1. [DOI] [PubMed] [Google Scholar]
- 18.Noé, J., Steiger, B., and Meier, P. 2002. Functional expression of the canalicular bile salt export pump (BSEP) of human liver. Gastroenterology. In press. [DOI] [PubMed]
- 19.Byrne, J.A., et al. 2002. The human bile salt export pump:characterization of substrate specificity and identification of inhibitors. Gastroenterology. In press. [DOI] [PubMed]
- 20.Ma D, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- 21.Trudeau MC, Zagotta WN. An intersubunit interaction regulates trafficking of rod cyclic nucleotide-gated channels and is disrupted in an inherited form of blindness. Neuron. 2002;34:197–207. doi: 10.1016/s0896-6273(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 22.Keitel V, et al. Impaired protein maturation of the conjugate export pump multidrug resistance protein 2 as a consequence of a deletion mutation in Dubin-Johnson syndrome. Hepatology. 2000;32:1317–1328. doi: 10.1053/jhep.2000.19791. [DOI] [PubMed] [Google Scholar]