Short abstract
Membrane-associated progesterone receptors are thought to mediate a number of rapid cellular effects not involving changes in gene expression. These receptors are here identified as distant homologs of cytochrome b5.
Abstract
Background
Membrane-associated progesterone receptors (MAPRs) are thought to mediate a number of rapid cellular effects not involving changes in gene expression. They do not show sequence similarity to any of the classical steroid receptors. We were interested in identifying distant homologs of MAPR better to understand their biological roles.
Results
We have identified MAPRs as distant homologs of cytochrome b5. We have also found regions homologous to cytochrome b5 in the mammalian HERC2 ubiquitin transferase proteins and a number of fungal chitin synthases.
Conclusions
In view of these findings, we propose that the heme-binding cytochrome b5 domain served as a template for the evolution of membrane-associated binding pockets for non-heme ligands.
Background
There are two main kinds of cellular effect mediated by steroids. One involves the alteration of gene expression, and is therefore characterized by a latency period between steroid signal reception and cellular effects. The second is associated with a much more rapid onset of cellular effects, and does not involve gene expression. A rapid anesthetic effect, induced by progesterone, was the first identified example of this group of effects [1]. Several rapid effects have now been described for all classes of steroids [2].
Several receptor types have been implicated in steroid action. The 'classical' receptors are members of the steroid/thyroid hormone receptor superfamily [3], and their ligand-binding domains have a characteristic helical sandwich structure around the steroid ligand [4]. They bind steroids in the nucleus or in the cytosol, dimerize, and migrate to the nuclear genome, where they act as transcription factors.
Such a mechanism cannot account for the rapid cellular effects of steroids, and a number of different receptors may be involved. In the case of progesterone, such rapid effects include: depolarization of rat hepatocytes by decreasing cell-membrane potassium conductance [5], calcium influx and chloride efflux in sperm during the acrosome reaction [6,7], calcium influx in Xenopus oocytes [8], as well as the anesthetic effect in the CNS. The latter has been shown to be mediated by an effect of progesterone on GABAA receptors [9]. More recently, progesterone was found to inhibit the action of oxytocin through direct binding to uterine oxytocin receptors [10].
A putative membrane-associated steroid receptor with high affinity for progesterone (MAPR) was first identified in porcine liver membranes [11] and later cloned and sequenced from porcine vascular smooth-muscle cells [12]. Subsequently, homologous human [13] and rat [14] sequences were cloned. These sequences were found to show no significant similarity to the classical intracellular receptors.
Results
We were interested in identifying distant homologs of these novel progesterone-binding proteins to help elucidate the function of this class of receptors. We used the porcine membrane progesterone receptor sequence (SWISS-PROT accession Q95250; ID PGC1_PIG [15]) to seed a PSI-BLAST search [16] at the National Center for Biotechnology Information (NCBI) website, and iterated with a default inclusion threshold of 0.005. In the second iteration we found a match to delta-5 fatty-acid desaturase from Mortierella alpina (GenBank accession AAC72755 [17]) with an E-value of 0.005. In the third iteration we found matches to cytochrome b5 from Borago officinalis (SWISS-PROT accession O04354; ID CYB5_BOROF) with an E-value of, 8 × 10-5 and chitin synthase from Blumeria graminis (GenBank accession AAF04279) with an E-value of 1 × 10-4.
We have also identified the reported human (SWISS-PROT accession O00264; ID PGC1_HUMAN) and rat (SWISS-PROT accession P70580; ID PGC1_RAT) MAPRs, as well as a number of other putative MAPRs from the following species: human, mouse (two sequences), cow, Caenorhabditis elegans, Arabidopsis thaliana (five sequences), and Oryza sativa (two sequences). Thus, at least in human, mouse, A. thaliana, and O. sativa, MAPR paralogs are present. The potential presence of MAPRs in plants suggests that they may also use steroids for rapid cell signaling.
Further study suggested that HERC2 proteins also have a region of sequence similarity to the cytochrome b5 domain. This was confirmed by seeding a reciprocal PSI-BLAST search with residues 1,209-1,286 of the human HERC2 protein (SWISS-PROT accession O95714), in which cytochrome b5 from A. thaliana (GenBank accession NP_173958) was matched in the first iteration with an E-value of 9 × 10-6. We have constructed a multiple sequence alignment of representative sequences whose similarity to cytochrome b5 we have identified (Figure 1), and will refer to the aligned region as the b5-domain.
Figure 1.
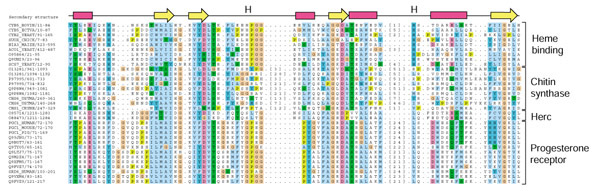
Multiple sequence alignment of b5-domains. This figure has been generated using the Jalview program written by Michele Clamp. Proteins are described by their SWISS-PROT identifier or accession number/start-end, and the class they belong to is indicated on the right. Alignments are colored using the ClustalX scheme in Jalview (orange, glycine (G); light green, proline (P); blue, small and hydrophobic amino acids (A, V, L, I, M, F, W in single-letter amino acid code); dark green, hydroxyl and amine amino acids (S, T, N, Q); magenta, negatively charged amino acids (D, E); red, positively charged amino acids (R, K); dark-blue, histidine (H) and tyrosine (Y). The top line indicates secondary structural elements obtained from bovine microsomal cytochrome b5 (PDB 1cyo): magenta rectangles represent α helices, and yellow arrows represent β strands. The two positions marked 'H' indicate the conserved pair of heme-coordinating histidines in the heme-binding b5-domains. The numbers in square brackets indicate the numbers of residues not shown in this illustration.
We have thus found conserved regions in MAPR, HERC2, and some fungal chitin synthases with significant similarity to the cytochrome b5 domain. Briefly, this domain has a mixed α + β structure with two pairs of α helices forming a binding pocket to one side of a β sheet (Figure 2).
Figure 2.
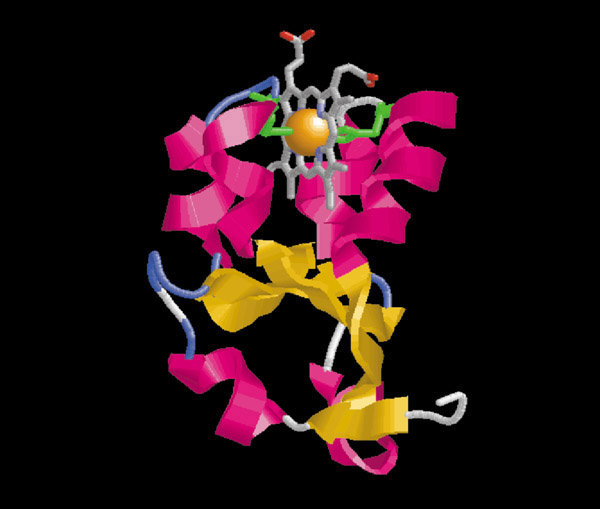
Structure of the heme-binding domain from bovine cytochrome b5 (PDB 1cyo). The illustration has been prepared using the Rasmol program [27]. The protein is colored by secondary structure. The heme group is shown in ball-and-stick representation and the axial heme ligand histidines are shown as green side chains.
We have also studied the domain organizations of the proteins aligned in Figure 1. Figure 3 shows schematically the domains as found in the Pfam database [18,19]. The b5-domain can be found in Pfam as the heme_1 family (Pfam accession PF00173). Essentially, we can divide the b5-domain proteins into two major classes: those with a heme-binding cytochrome b5 fold (including cytochrome b5, bacterial cytochrome b558, flavocytochrome b2, sulfite oxidase, nitrate reductase, and a number of other oxidoreducatases), and the probable non-heme-binding ones (chitin synthases, HERC2, and MAPR), as explained below.
Figure 3.
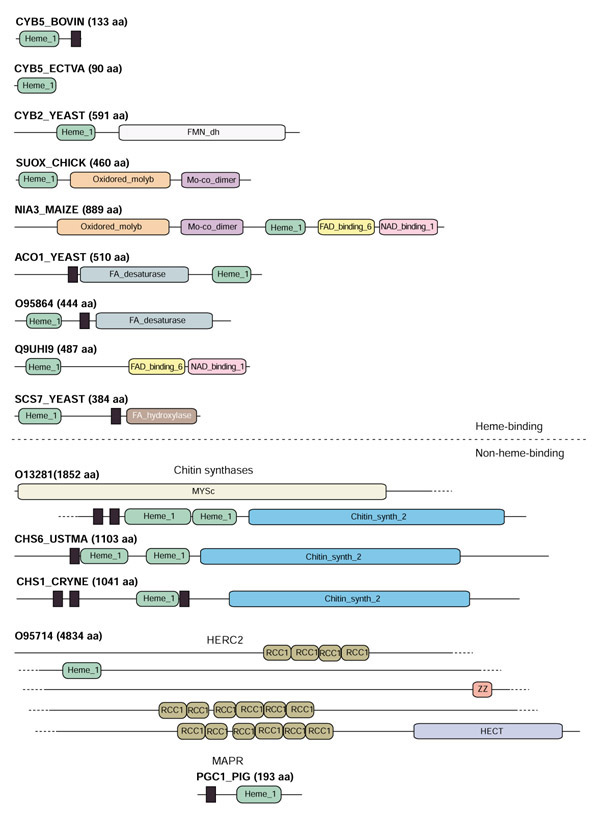
A schematic figure showing the architectures of proteins containing b5-domains. The order of the proteins in this illustration is the same as in Figure 1, except that, for proteins with a similar architecture, only one example is shown. All domain names correspond to homonymous Pfam-A families, except for the MYSc domain, which is a SMART database entry [28]. The black rectangles represent inter-domain transmembrane regions predicted by TMHMM [29]. The heme_1 domain corresponds closely to the b5-domain aligned in Figure 1. More information about each of the domains can be found in the Pfam database [18,19].
Discussion
Heme-binding b5-domain proteins are found in all major eukaryotic lineages. A bacterial cytochrome b5 homolog, cytochrome b558 from Ectothiorhodospira vacuolata, has also been identified and structurally characterized [20]. The heme-binding b5-domains are involved in electron transfer to and from other proteins, or other domains on the same protein (as in flavocytochrome b2 [21] and sulfite oxidase [22]). The heme ligand is buried in the binding pocket formed by the two pairs of α helices over the β sheet, and is coordinated by the NE atoms of two conserved histidine residues [21]. The positions of these two histidines are indicated in Figure 1.
Neither of the MAPR, HERC2, or chitin synthase b5-domains appear to bind heme or to be involved in redox reactions. Indeed, they all lack the pair of heme-coordinating histidine residues. Chitin synthases catalyze the polymerization of N-acetylglucosamine through β-1,4 linkage. Chitin is an important constituent of fungal cell walls. Chitin synthase genes are present in various numbers in different fungal species, and the relative importance of the individual isozymes in particular fungal species is only partially understood. We have identified several fungal chitin synthases with a b5-domain, which can be organized into three groups on the basis of domain organization (Figure 3).
The function of the giant HERC2 protein is not known. It has been predicted to act as a guanine-nucleotide-exchange factor and an E3 ubiquitin ligase, involved in intracellular protein trafficking and degradation [23].
As mentioned above, heme-binding b5-domains are found in all eukaryotic lineages. The MAPR proteins appear to be restricted to plants and metazoans, and the HERC2 proteins with a b5-domain are mammalian. The b5-domain chitin synthases are only found in fungi. Taken together, our findings suggest that the heme-binding cytochrome b5 domain may have served as a template for the more recent evolution of novel ligand-binding pockets, such as a steroid-binding site in the MAPR proteins. We predict that the b5-domains that we have identified in chitin synthases and HERC2 proteins might serve as binding sites for lipid ligands. The relatively narrow species distributions of b5-domain chitin synthases and HERC2 proteins suggests that these binding sites may have arisen as a result of relatively recent insertions of a b5-domain. We did not find a b5-domain in the HERC2-related proteins HERC1 or HERC3.
Most of the proteins with a b5-domain are linked to cell membranes, either directly or by forming part of membrane-associated complexes. We propose that the proximity of cell membranes to a heme-binding template pocket could have served to give rise to new ligand-binding pockets with specificity for membrane-soluble molecules such as steroids. Thus, MAPRs may represent an adaptation by which cells could start making use of steroids as triggers for rapid response mechanisms.
The MAPRs could be localized to both plasma and intracellular membranes [11,12,13,14], which would afford cells greater flexibility in utilizing progesterone and other ligands for rapid signaling. It seems likely that MAPRs bind to progesterone intracellularly: all the other b5-domains are intracellular, and there is no evidence to suggest that the MAPR b5-domain is extracellular. It is also probable that another class of membrane-associated progesterone receptors can bind to their ligand extracellularly: the rapid effects of progesterone on spermatozoa may be mediated by such a receptor, and are reproducible even with preparations of progesterone to which the plasma membrane is impermeable (reviewed in [24]). Antibodies raised against the carboxy-terminal ligand-binding domain of the nuclear progesterone receptor identified a surface protein in human spermatozoa, and were able to inhibit the rapid effects of progesterone [25]. Antibodies raised against the amino-terminal transactivation domain of the nuclear progesterone receptor did not identify any such protein in human spermatozoa [26]. These findings suggest that spermatozoa have a cell-surface progesterone receptor with a classical ligand-binding domain, but without a transactivation domain. Unfortunately, no sequence data for this protein are available, and it is not known whether a similar protein is expressed in other tissues.
The details of the physiological role of the MAPRs remain to be worked out. For example, mutagenesis studies should help better to define the importance of individual residues in the MAPR molecule. The determination of the molecular structure of MAPR would provide crucial information on the nature of the interaction of MAPR and its ligand, as well as its possible interaction with the intracellular signaling machinery. Sequencing and structural determination of the extracellular sperm progesterone receptor would allow comparison between it, MAPR, and the nuclear progesterone receptor. Supplementing these data with detailed studies of the tissue-expression patterns of these molecules, and their associated signal transduction mechanisms should help to provide a better understanding of rapid cellular signaling by steroids.
Conclusions
We have found significant sequence similarity between the cytochrome b5 domain and MAPRs. We have also identified regions of similarity to b5 in a number of fungal chitin synthases, and in mammalian HERC2 proteins, and propose that the cytochrome b5 domain served as an evolutionary template for the development of membrane-associated lipid ligand binding in these proteins.
Acknowledgments
Acknowledgements
We are grateful to Robert Finn for his valuable comments and help. A.B. is supported by the Wellcome Trust.
References
- Seyle H. Anaesthetic effects of steroid hormones. Proc Soc Biol Med. 1941;46:116–118. [Google Scholar]
- Falkenstein E, Norman AW, Wehling M. Mannheim classification of nongenomically initiated (rapid) steroid action(s). J Clin Endocr Metab. 2000;85:2072–2075. doi: 10.1210/jcem.85.5.6516. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- Waldegger S, Beisse F, Apfel H, Breit S, Kolb HA, Haussinger D, Lang F. Electrophysiological effects of progesterone on hepato-cytes. Biochim Biophys Acta. 1995;1266:186–190. doi: 10.1016/0167-4889(94)00236-8. [DOI] [PubMed] [Google Scholar]
- Blackmore PF, Beebe SJ, Danforth DR, Alexander N. Progesterone and 17 alpha-hydroxyprogesterone. Novel stimulators of calcium influx in human sperm. J Biol Chem. 1990;265:1376–1380. [PubMed] [Google Scholar]
- Turner KO, Meizel S. Progesterone-mediated efflux of cytosolic chloride during the human sperm acrosome reaction. Biochem Biophys Res Commun. 1995;213:774–780. doi: 10.1006/bbrc.1995.2197. [DOI] [PubMed] [Google Scholar]
- Wasserman WJ, Pinto LH, O'Connor CM, Smith LD. Progesterone induces a rapid increase in [Ca2+]in of Xenopus laevis oocytes. Proc Natl Acad Sci USA. 1980;77:1534–1536. doi: 10.1073/pnas.77.3.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan NC, Bolger MB, Gee KW. Identification and characterization of a pregnane steroid recognition site that is functionally coupled to an expressed GABAA receptor. Neurochem Res. 1991;16:347–356. doi: 10.1007/BF00966098. [DOI] [PubMed] [Google Scholar]
- Grazzini E, Guillon G, Mouillac B, Zingg HH. Inhibition of oxytocin receptor function by direct binding of progesterone. Nature. 1998;392:509–512. doi: 10.1038/33176. [DOI] [PubMed] [Google Scholar]
- Meyer C, Schmid R, Scriba PC, Wehling M. Purification and partial sequencing of high-affinity progesterone-binding site(s) from porcine liver membranes. Eur J Biochem. 1996;239:726–731. doi: 10.1111/j.1432-1033.1996.0726u.x. [DOI] [PubMed] [Google Scholar]
- Falkenstein E, Meyer C, Eisen C, Scriba PC, Wehling M. Full-length cDNA sequence of a progesterone membrane-binding protein from porcine vascular smooth muscle cells. Biochem Biophys Res Commun. 1996;229:86–89. doi: 10.1006/bbrc.1996.1761. [DOI] [PubMed] [Google Scholar]
- Gerdes D, Wehling M, Leube B, Falkenstein E. Cloning and tissue expression of two putative steroid membrane receptors. Biol Chem. 1998;379:907–911. doi: 10.1515/bchm.1998.379.7.907. [DOI] [PubMed] [Google Scholar]
- Nolte I, Jeckel D, Wieland FT, Sohn K. Localization and topology of ratp28, a member of a novel family of putative steroid-binding proteins. Biochim Biophys Acta. 2000;1543:123–130. doi: 10.1016/s0167-4838(00)00188-6. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2002;30:17–20. doi: 10.1093/nar/30.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfam protein family database http://www.sanger.ac.uk/Software/Pfam
- Kostanjevecki V, Leys D, Van Driessche G, Meyer TE, Cusanovich MA, Fischer U, Guisez Y, Van Beeumen J. Structure and characterization of Ectothiorhodospiravacuolata cytochrome b558, a prokaryotic homologue of cytochrome b5. J Biol Chem. 1999;274:35614–35620. doi: 10.1074/jbc.274.50.35614. [DOI] [PubMed] [Google Scholar]
- Xia ZX, Mathews FS. Molecular structure of flavocytochrome b2 at 2.4 A resolution. J Mol Biol. 1990;212:837–863. doi: 10.1016/0022-2836(90)90240-M. [DOI] [PubMed] [Google Scholar]
- Kisker C, Schindelin H, Pacheco A, Wehbi WA, Garrett RM, Rajagopalan KV, Enemark JH, Rees DC. Molecular basis of sulfite oxidase deficiency from the structure of sulfite oxidase. Cell. 1997;91:973–983. doi: 10.1016/s0092-8674(00)80488-2. [DOI] [PubMed] [Google Scholar]
- Ji Y, Walkowicz MJ, Buiting K, Johnson DK, Tarvin RE, Rinchik EM, Horsthemke B, Stubbs L, Nicholls RD. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- Blackmore PF. Extragenomic actions of progesterone in human sperm and progesterone metabolites in human platelets. Steroids. 1999;64:149–156. doi: 10.1016/S0039-128X(98)00109-3. [DOI] [PubMed] [Google Scholar]
- Sabeur K, Edwards DP, Meizel S. Human sperm plasma membrane progesterone receptor(s) and the acrosome reaction. Biol Reprod. 1996;54:993–1001. doi: 10.1095/biolreprod54.5.993. [DOI] [PubMed] [Google Scholar]
- Castilla JA, Gil T, Molina J, Hortas ML, Rodriguez F, Torres-Munoz J, Vergara F, Herruzo AJ. Undetectable expression of genomic progesterone receptor in human spermatozoa. Hum Reprod. 1995;10:1757–1760. doi: 10.1093/oxfordjournals.humrep.a136169. [DOI] [PubMed] [Google Scholar]
- Sayle R, Milner-White E. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;1995:374. doi: 10.1016/S0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]


