Abstract
Steroid receptor coactivator-1 (SRC-1) family members interact with steroid receptors, including estrogen receptor α (ERα) and progesterone receptor (PR), to enhance ligand-dependent transcription. However, the expression of ERα and SRC-1 was found to be segregated in distinct subsets of cells within the epithelium of the estrogen-responsive rat mammary gland. This finding was in contrast to the finding for the stroma, where significant numbers of cells coexpressed ERα and SRC-1. Treatment of animals with estrogen induced PR expression in the ERα-expressing mammary epithelial cells in the absence of detectable SRC-1 and did not affect the segregated pattern of SRC-1 and ERα expression. PR was neither expressed nor induced by estrogen treatment in stroma, despite the coexpression of ERα and SRC-1. These results suggest that SRC-1 is not necessary for ERα-mediated induction of PR in mammary epithelial cells and is also not sufficient for PR induction in stromal cells expressing both ERα and SRC-1. Furthermore, the expression of SRC-1 in a subpopulation of mammary epithelial cells distinct from those expressing ERα or PR raises the possibility that SRC-1 has cell type-specific functions other than simply to act as coactivator for ERα or PR in the mammary epithelium.
Ovarian steroids play a critical role in mammary gland development, acting through specific receptors expressed in target cells. When steroid receptors become bound to hormones, the receptors undergo a conformational change, bind to their cognate DNA response elements in target genes, recruit coactivators and general transcription factors, and subsequently activate target gene expression. Several coactivators have been cloned, and many of these coactivator proteins were initially identified biochemically as nuclear receptor-interacting proteins of approximately 160 kDa based on their ability to interact with agonist-bound estrogen receptor α (ERα) (1, 2). To date, three distinct but related p160 family members have been identified, with each family member having a number of splice variants. This family includes steroid receptor coactivator-1 (SRC-1) [also designated nuclear receptor coactivator 1 (NCoA-1)], glucocorticoid receptor-interacting protein 1 (GRIP1) [also designated nuclear receptor coactivator 2 (NCoA-2), transcriptional intermediary factor 2 (TIF2), or steroid receptor coactivator-2 (SRC-2)], and p300/cAMP response element-binding protein (CREB)-binding protein cointegrator associate protein (p/CIP) [also designated amplified in breast cancer-1 (AIB1), receptor-associated coactivator 3 (RAC3), activator of thyroid and retinoic acid receptor (ACTR), thyroid hormone receptor activator molecule 1 (TRAM-1), or steroid receptor coactivator-3 (SRC-3)] (3–11). Members of this SRC-1 family have been shown to act as coactivators for steroid, retinoid, and thyroid hormone receptors, perhaps by modifying transcriptionally repressed chromatin or by enhancing stabilization of transcriptional preinitiation complexes (12–14). This coactivation function is mediated through direct ligand-dependent interaction with these receptors, which enhances the ligand-dependent transcription of target genes (15). Factors such as p300 and CREB-binding protein have been identified as part of the coactivator complex, functioning as a point of integration between ERα and other signaling pathways (16–18). The enhancement of the transcriptional activation of the steroid receptor superfamily by SRC-1 family members (19–21), as demonstrated in transfection experiments, suggests a potential role of SRC-1 family members in the development of normal estrogen target tissues and potentially in breast cancer formation. This is supported by the phenotype of SRC-1 null mice (22), which exhibit decreased growth and development of target organs (such as the uterus, prostate, testis, and mammary gland) in response to steroid hormones and by the finding of amplification of the SRC-1 family member AIB1 in some breast cancers.
Given the ability of SRC-1 to interact directly with steroid receptors and to enhance steroid receptor-dependent signaling, we expected that SRC-1 would be expressed in the same cells as the activator proteins such as ERα or progesterone receptor (PR) in normal hormone-responsive tissues. Although SRC-1 mRNA was detected in many tissues and cell lines (8, 23), the expression of SRC-1 protein at the cellular level has not previously been addressed. Experiments were designed to test the hypothesis that SRC-1 was expressed in the same cells as the ERα or PR. We chose the rat mammary gland as the model system because the development of the mammary gland is influenced by hormonal and growth factor signals, and this estrogen-responsive tissue has been a unique organ for the study of hormonal action, development, and tumorigenesis (24–26). The essential role of ERα and PR in mammary gland development has been confirmed by knockout mice lacking functional receptors. ERα knockout mice display grossly impaired ductal epithelial proliferation and branching (27, 28), and PR knockout mice display significant ductal development but decreased arborization and an absence of alveolar differentiation (29). To determine the precise localizations of ERα and SRC-1 within the mammary gland, we examined the expression of ERα and SRC-1 in the rat mammary gland by immunohistochemistry.
MATERIALS AND METHODS
Animals.
Wistar–Furth and Sprague–Dawley female rats were purchased from Harlan Sprague–Dawley (Indianapolis) and treated according to National Institutes of Health and University of Virginia guidelines for the care and use of animals. An s.c. injection of 1 μg of estrogen benzoate or vehicle (sesame oil) was given to the rats. Later (24 h), the mammary gland and uterus were removed and processed for antibody staining or Western blot analysis.
Generation of Antibody.
Human SRC-1 from amino acid 363 to the carboxyl terminus was fused to glutathione S-transferase (GST), and Escherichia coli-expressed GST–SRC-1 fusion protein was used to generate the monoclonal SRC-1 antibodies (GT12 and GT111). Tissue culture supernatants of GT12 and GT111 were used for both Western blot analysis and immunohistochemical staining.
Western Blot Analysis.
Whole uterine tissue lysate was prepared by homogenization in RIPA buffer (50 mM Tris⋅HCl, pH 7.5/1 mM EDTA/150 mM NaCl/1% Triton X-100/1% deoxycholic acid/1 mM DTT/1 μg/ml leupeptin/1 μg/ml aprotinin/100 μg/ml PMSF). Protein of whole cell lysate (300 μg) was separated by electrophoresis on 7.5% polyacrylamide gels containing 1% SDS. Western blot analysis was carried out according to procedures previously published (30) with GT12 at a 1:10 dilution.
Immunohistochemistry.
An indirect immunoperoxidase method was used to identify ERα-, SRC-1-, or PR-positive cells. Tissues were fixed in cold 2% paraformaldehyde in PBS for 2 h, and 5-μm paraffin sections were heated in a microwave oven (900 watt, high power) in 10 mM citric buffer, pH 6.0, for antigen retrieval, treated with 0.3% H2O2 in methanol, and blocked with avidin D/biotin blocking solutions. Sections were then incubated with appropriate 10% normal serum, primary antibodies, appropriate secondary biotinylated antibodies, avidin-biotin complex, and diaminobenzidine substrates. ERα was detected with a rabbit anti-ERα IgG (MC-20; 1:400; Santa Cruz Biotechnology) or a mouse anti-ERα IgG (6F11; 1:50; NovoCastra, Newcastle, U.K.). SRC-1 was detected with mouse anti-SRC-1 antibodies (GT12; 1:2 or GT111; 1:1) or a goat anti-SRC-1 IgG (M-20; 1:50; Santa Cruz Biotechnology). PR was detected with a mouse anti-PR IgG (MA1–410; 1:25; Affinity BioReagents, Neshanic Station, NJ).
For SRC-1/ERα dual immunofluorescent labeling, sections were incubated sequentially with GT12 (1:2), biotinylated horse anti-mouse IgG (1:1000), Texas Red-conjugated streptavidin (1:100; Vector Laboratories), MC-20 (1:100), and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch). Alternatively, sections were incubated sequentially with 6F11 (1:50), biotinylated horse anti-mouse IgG (1:500), FITC-conjugated streptavidin (1:200; Vector Laboratories), M-20 (1:25), and Texas Red-conjugated donkey anti-goat IgG (1:100; Jackson ImmunoResearch). For PR/ERα dual labeling, sections were incubated sequentially with MA1–410 (1:25), biotinylated horse anti-mouse IgG (1:500), FITC-conjugated streptavidin (1:100), MC-20 (1:100), and Texas Red-conjugated goat anti-rabbit IgG (1:200; Jackson ImmunoResearch). For PR/SRC-1 dual labeling, sections were incubated sequentially with MA1–410, biotinylated horse anti-mouse IgG, FITC-conjugated streptavidin, M-20, and Texas Red-conjugated donkey anti-goat IgG. All procedures were done at room temperature. Slides were examined with a Zeiss Axioskop microscope equipped with appropriate fluorescence filter sets. Images were taken with a SenSys charge-coupled device camera (Photometrics, Tuscon, AZ) and iplab spectrum software (Signal Analytics, Vienna, VA).
RESULTS
Expression of SRC-1 and ERα in Mammary Gland.
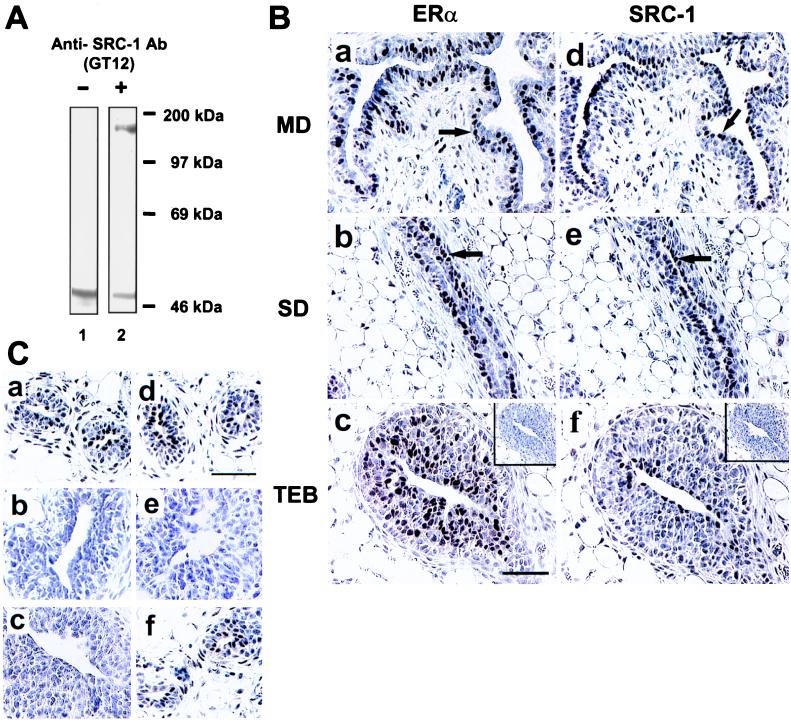
Human SRC-1 from amino acid 363 to the carboxyl terminus was fused to GST, and the GST–SRC-1 fusion protein was used to generate the mouse monoclonal antibodies GT12 and GT111. The ability of the mouse monoclonal antibody GT12 to specifically recognize rat SRC-1 was confirmed by Western blot analysis in which a single 160-kDa band was detected (Fig. 1A). Immunohistochemical staining performed with GT12 and rabbit anti-ERα (MC-20) antibodies on adjacent sections of the rat mammary gland demonstrated that both SRC-1 and ERα antibodies stained subpopulations of mammary epithelial cells in various regions of the gland (Fig. 1B, arrows). The ERα-positive cells were found in a layer closer to the basement membrane, and the SRC-1-positive cells existed in a more luminal layer.
Figure 1.
Expression of SRC-1 and ERα in the rat mammary gland. (A) Mouse anti-SRC-1 (GT12) antibody recognized rat SRC-1 (lane 2) as demonstrated by Western blot analysis of GT12 using rat uterine tissue lysate. The endogenous rat IgG from tissues was detected by the secondary antibody, and this signal was also detected without primary antibody (lane 1). (B) Immunohistochemical staining of ERα (a–c) and SRC-1 (d–f) on adjacent sections from main ducts (MD), small ducts (SD), and terminal end buds (TEB) of the mammary gland from 3-week-old virgin female rats. Control specimens stained without primary antibodies are shown in c and f (Insets). The arrows are pointing to ERα-positive cells that are found in a layer closer to the basement membrane and SRC-1-positive cells that exist in a more luminal layer. (C) Immunoreactive SRC-1 was detected by both anti-SRC-1 antibodies, GT12 (a) and M-20 (d), in rat mammary gland. A control specimen (c) stained without primary antibody showed no staining signal. Preabsorption of GT12 or M-20 antibodies with SRC-1 fusion protein (b) or M-20-specific peptide (e), respectively, out-competed the staining signal. M-20 peptide did not diminish the staining signal detected by GT12 (f). Sections were counterstained with Harris hematoxylin. (Bars = 50 μm in B and C.)
Control specimens stained with antibodies previously preabsorbed by the GST–SRC-1 fusion protein (Fig. 1Cb) or the ERα-specific MC-20 antigen (data not shown) yielded only background staining. Preabsorption of the GT12 antibody with GST protein alone did not diminish the specific immunostaining signal (data not shown). To ensure that the immunostaining profile was valid, we tested a second SRC-1 monoclonal antibody, GT111, and observed the same staining pattern (data not shown). In addition, we tested a goat anti-SRC-1 IgG (M-20; epitope corresponding to mouse amino acids 1386–1405) that recognizes a different portion of SRC-1 than GT12 or GT111 and again saw the same pattern of staining (Fig. 1Cd). As anticipated, the immunoreactivity could be eliminated by preabsorption of this antibody with the specific SRC-1 peptide antigen (Fig. 1Ce). The SRC-1 M-20-specific peptide did not compete with the stained signal detected by GT12 (Fig. 1Cf), confirming that M-20 and GT12 indeed recognize different epitopes of the rat SRC-1 protein.
We and others previously found that ERα was expressed in only a subset of cells in mammary epithelium (Fig. 1 Ba–Bc, arrows) (31, 32). In this study, we found that SRC-1 was also expressed in only a subset of mammary epithelial cells, with distinct patterns of expression when compared with ERα expression (Fig. 1 Bd–Bf, arrows). The percentage of mammary epithelial cells from 3-week-old female rats expressing ERα or SRC-1 in the main ducts, small ducts, and end buds is quantitated in Table 1. We found that ERα was most highly expressed in the small ducts and end buds, and SRC-1 was expressed in a complementary fashion, with the main duct epithelium having the highest percentage of SRC-1 positive cells. These initial results suggested that ERα and SRC-1 might have distinct patterns of expression within the mammary epithelium.
Table 1.
Distribution of ERα or SRC-1 immunoreactive cells in the epithelium of mammary gland in 3-week-old female rats
| Region | ERα | SRC-1 |
|---|---|---|
| Main duct | 29.46 ± 3.46 | 41.98 ± 3.10 |
| Small duct | 42.37 ± 1.77 | 32.43 ± 2.09 |
| End bud | 40.29 ± 3.11 | 18.21 ± 1.26 |
Cells expressing immunoreactive ERα or SRC-1 were counted from various regions (end buds, small ducts, and main ducts) of the mammary glands from adjacent sections as indicated in Fig. 1B. Values are mean percentages of positive nuclei (±SD), with four animals in each group. For ERα immunoreactive cells, a total of 2,831 cells from 6 main ducts, 5,690 cells from 33 small ducts, and 7,448 cells from 31 end buds was counted. For SRC-1 immunoreactive cells, a total of 2,657 cells from 6 main ducts, 5,437 cells from 33 small ducts, and 6,954 cells from 31 end buds was counted.
Discrete Pattern of SRC-1 and ERα Distribution in Mammary Gland.
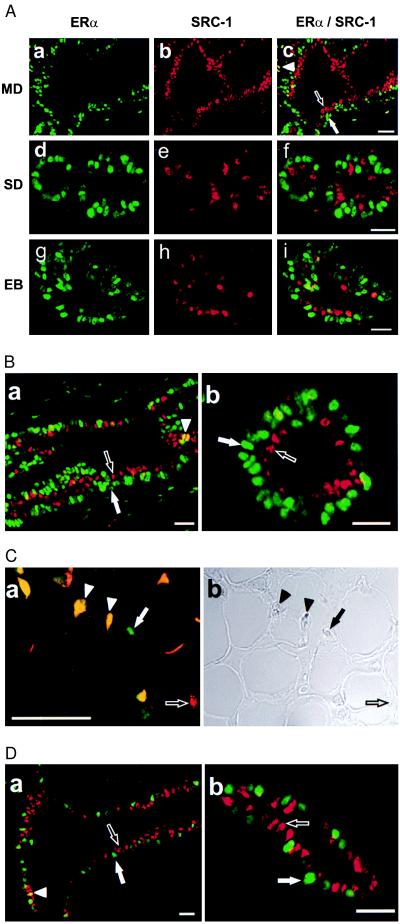
To test whether ERα and SRC-1 are expressed in the same or different mammary epithelial cells, we undertook dual immunofluorescent labeling studies with a combination of rabbit anti-ERα (MC-20) and mouse anti-SRC-1 (GT12) antibodies. The immunostaining signals were detected by FITC-conjugated anti-rabbit IgG in green for ERα (Fig. 2 Aa, Ad, and Ag) and biotinylated anti-mouse IgG and Texas Red-conjugated streptavidin in red for SRC-1 (Fig. 2 Ab, Ae, and Ah). Surprisingly, we found that cells expressing SRC-1 are nearly completely segregated in the mammary epithelium from cells expressing ERα in all regions of the mammary gland examined, including the main ducts (Fig. 2 Aa–Ac), small ducts (Fig. 2 Ad–Af), and end buds (Fig. 2 Ag–Ai). Merging the two images (Fig. 2 Ac, Af, and Ai) revealed only very rare cells coexpressing ERα and SRC-1 as would be indicated by a yellow signal. When these results were quantitated, less than 2% of the mammary epithelial cells in ducts or end buds expressed both ERα and SRC-1 (Table 2). These immunostaining results were confirmed with a second set of antibodies that recognize different epitopes of SRC-1 and ERα (Fig. 2B). In these fluorescent images from immature rats, it appears as though ERα and SRC-1 define two distinct layers in the ductal epithelium. This is consistent with our initial observation (Fig. 1B). The ERα-positive cells are found in a layer closer to the basement membrane, whereas the SRC-1-positive cells exist in a more luminal layer, suggesting that SRC-1 and ERα are markers of distinct subpopulations of the mammary epithelium.
Figure 2.
Segregation of SRC-1 expression from ERα-positive cells as illustrated by the dual immunofluorescent labeling of SRC-1 and ERα. (A) Mammary glands from 3-week-old virgin female rats were stained simultaneously for SRC-1 (b, e, and h; red) with GT12 and for ERα (a, d, and g; green) with MC-20. Green and red images were superimposed (c, f, and i). Main duct (MD), small duct (SD), and end bud (EB) are shown. (B) The discrete distribution pattern of ERα (green) and SRC-1 (red) was confirmed with the combination of 6F11 and M-20 antibodies (a and b). (C) Stroma expressing ERα alone (green), SRC-1 alone (red), or both (yellow) were detected with GT12 and MC-20 antibodies (a) as in A. The phase contrast image from a is shown in b. (D) Staining from 10-week-old virgin female rat mammary gland also demonstrated the segregation of SRC-1 from ERα in epithelial cells. MC-20 and GT12 were used as the primary antibodies. Solid arrow, Cells expressing only ERα; open arrow, cells expressing only SRC-1; solid arrowhead, cells expressing both ERα and SRC-1. (Bar = 100 μm.)
Table 2.
ERα and SRC-1 immunoreactive cells on dual fluorescent labeling in the epithelium of mammary gland in 3-week-old female rats
| Region | ERα-positive/SRC-1-negative | ERα-negative/SRC-1-positive | ERα-positive/SRC-1-positive | ERα-negative/SRC-1-negative |
|---|---|---|---|---|
| Main duct | 26.62 ± 5.85 | 37.12 ± 4.05 | 1.44 ± 0.67 | 34.82 ± 9.22 |
| Small duct | 34.82 ± 2.81 | 33.17 ± 4.04 | 1.64 ± 0.59 | 28.13 ± 4.28 |
| End bud | 34.60 ± 2.49 | 17.31 ± 2.44 | 1.31 ± 0.25 | 45.84 ± 2.54 |
Cells expressing immunoreactive ERα, SRC-1, or both were counted from various regions (end buds, small ducts, and main ducts) of the mammary glands as indicated in Fig. 2A. Values are mean percentages of positive nuclei (±SD), with four animals in each group. A total of 2,183 cells from 4 main ducts, 3,911 cells from 19 small ducts, and 4,274 cells from 18 end buds was counted.
We next examined whether the expression of ERα and SRC-1 was mutually exclusive in all cell types within the mammary gland. In contrast to the mammary epithelium, a substantial number of cells within the mammary stroma expressed both SRC-1 and ERα (Fig. 2C), implying a potential different function for SRC-1 in the mammary epithelium compared with mammary stroma. In addition, we examined whether the segregation of ERα and SRC-1 expression to distinct cells in the mammary epithelium was unique to early postnatal mammary development in the rat or was preserved in adult rats. We found that, as was the case in immature rats, ERα and SRC-1 were not coexpressed in the same cells in the mammary epithelium of mature virgin female rats (Fig. 2D). This finding indicates that the segregation of expression of ERα and SRC-1 was not altered during the maturation of the mammary gland under the control of ovarian steroids.
Estrogen-Stimulated PR Induction Does Not Require SRC-1.
The discovery of a subpopulation of mammary epithelial cells expressing ERα but lacking SRC-1 allowed us to ask whether SRC-1 expression was required for one of the critical ERα functions in the epithelium of the mammary gland, namely, the induction of PR. The PR gene has been shown to be a direct target of ERα regulation, is detected only in a subset of epithelial cells of the mouse mammary gland, and has been colocalized to ERα-expressing cells in normal human breast epithelium (33–35). Immature female rats were treated with vehicle alone (Fig. 3 Aa–Ac) or with estrogen benzoate (Fig. 3 Ad–Af) and the expression of SRC-1, ERα, and PR was examined in the mammary gland. We found that, as with mature rats, exposure of the mammary epithelium to estrogen did not alter the segregated pattern of expression of SRC-1 and ERα (Fig. 3 Aa and Ad). In vehicle-treated animals only a minority of mammary epithelial cells expressed PR, and these were also uniformly ERα-expressing cells (Fig. 3Ab). Significantly, treatment of animals with estrogen led to a striking increase in the proportion of cells in which PR was detectable, and again these were exclusively the ERα-positive cells (Fig. 3Ae). Consistent with the coexpression of ERα and PR and the segregated pattern of expression of ERα and SRC-1, PR and SRC-1 expression was also mutually exclusive (Fig. 3 Ac and Af) in the mammary epithelium. No PR was detected in the stromal compartment of the rat mammary gland and estrogen treatment did not induce PR expression in these stromal cells expressing both ERα and SRC-1 (Fig. 3 B and C; data not shown), suggesting that coexpression of ERα and SRC-1 was neither necessary nor sufficient to induce PR expression.
Figure 3.
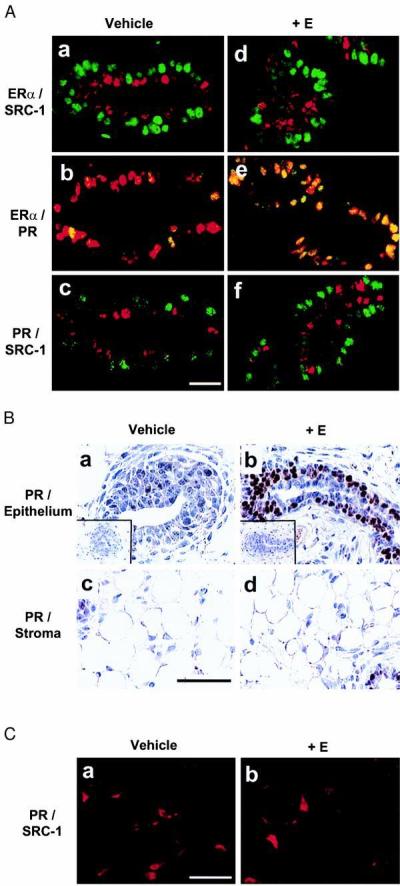
Segregation of SRC-1 from both ERα and PR in rat mammary epithelial cells when treated with estrogen. Mammary gland from 3-week-old virgin female rats treated with vehicle (sesame oil) or 1 μg of estrogen benzoate (+E) for 24 h were used. (A) Dual immunofluorescent labeling of ERα/SRC-1, ERα/PR, and PR/SRC-1. (a and d) ERα (green) and SRC-1 (red) were stained simultaneously with MC-20 and GT12. (b and e) ERα (red) and PR (green) were stained simultaneously with MC-20 and anti-PR IgG (MA1–410). (c and f) PR (green) and SRC-1 (red) were stained simultaneously with MA1–410 and M-20. (Bar = 100 μm.) (B) Expression of PR in both epithelium and stroma of the mammary glands. Control specimens stained without primary antibody are shown in insets. (Bar = 50 μm.) (C) Dual immunofluorescent labeling of PR and SRC-1 in stroma of the mammary glands. The staining was performed as in A. (Bar = 100 μm.)
DISCUSSION
SRC-1 has been identified as a coactivator for ERα based on its ability to interact directly with agonist-bound ERα and to potentiate ERα-dependent signaling in transient transfection experiments. The coactivation function of SRC-1 has also been documented for PR. Hence, we expected that SRC-1 would be expressed in the same cells in which ERα or PR was present in normal hormone-responsive tissues. In this study, we found unexpectedly that expression of SRC-1 and ERα is segregated in the rat mammary epithelium, an estrogen-responsive tissue. In addition, we have demonstrated that induction of PR expression in mammary epithelium does not require coexpression of SRC-1 and that the presence of SRC-1 in stromal cells coexpressing ERα was not sufficient to facilitate PR induction by estrogen. Finally, the pattern of expression of ERα in cells closer to the basement membrane and the contrasting expression of SRC-1 in cells that are more luminal implies the existence of at least two distinct cell subpopulations in the rat mammary ductal epithelium.
The recent description of SRC-1 null mice suggests that SRC-1 is required for efficient proliferation and differentiation of the mammary gland in response to estrogen and progesterone, because the mammary glands of these mice showed less alveolar development during pregnancy and responded to estrogen and progesterone treatment with only partial ductal growth (22). Our results here, showing that SRC-1 and ERα (or PR) are expressed in distinct subpopulations of mammary epithelial cells and that the coexpression of SRC-1 and ERα does not facilitate the PR gene expression in mammary stromal cells, are in concert with the mammary gland phenotype observed in the SRC-1 null mice. Because SRC-1 and ERα/PR are expressed in distinct cells within the mammary epithelium, SRC-1 is apparently not directly involved in estrogen- or progesterone-initiated signaling in the mammary epithelium. This strongly indicates that, conversely, the role of SRC-1 in morphogenesis of the epithelium is likely to interact with other signaling molecules whose identities remain to be defined. To the extent that morphogenesis is affected in the SRC-1 null mice, it remains unclear whether this is caused by a malfunctioning of the SRC-1-positive epithelial cells, the SRC-1-positive stromal cells, or both. Paracrine effects including epithelial–stromal and epithelial–epithelial interactions have been implicated in the action of ovarian steroids in the morphogenesis of the normal breast (33–39) and could play a role in the mammary gland phenotype observed in SRC-1 null mice.
The demonstration that the segregation of expression of ERα and SRC-1 was not altered during the maturation of the mammary gland under the control of ovarian steroids indicates that ovarian steroids are not able to increase the expression of SRC-1 in ERα-positive cells to potentiate the estrogen signaling. In the immature gland, two distinct layers of epithelial cells were clearly visualized in small ductal areas (Fig. 2B). Maturation of the mammary gland after puberty results in a less clear distinction of the layers of epithelial cells in small ductal areas (Fig. 2D). Interestingly, SRC-1-expressing cells are still adjacent to the ERα-expressing cells in adult rats. This suggests that the organization of the mammary epithelium may be important in mediating its complex paracrine response to estrogen. In addition, SRC-1 and ERα can serve as cellular markers of distinct subpopulations of the mammary epithelium, with unknown function.
Our observations that PR was not induced in ERα- and SRC-1-coexpressing stromal cells or in SRC-1-expressing epithelial cells during estrogen treatment support the hypothesis that SRC-1-coactivating function for ERα is neither necessary nor sufficient for PR expression in normal mammary gland. It is possible that other SRC-1 family members such as glucocorticoid receptor-interacting protein 1/transcriptional intermediary factor 2 or p300/CREB-binding protein cointegrator associate protein/AIB1 are involved in ERα coactivation in cells that do not express SRC-1, as suggested by other investigators (22). Examination of the expression of other SRC-1 family members in ERα-positive cells will be necessary to test this hypothesis. In vitro studies suggest that SRC-1 could partner with various nuclear receptors for its coactivation function. Based on our observation that SRC-1 is segregated from ERα and PR in mammary epithelium, it is interesting to speculate that SRC-1 may have cell- or tissue-specific partners for its coactivation function. The second form of ER, ERβ (40), or other nuclear receptors could be the partner for SRC-1 coactivation function in the mammary gland. On the other hand, we have found that the segregation of SRC-1 from ERα is tissue-specific and may serve to expand the spectrum of hormonal response in various estrogen target organs. This view is supported by our observation that in uterine epithelium SRC-1 and ERα are colocalized in the vast majority of uterine epithelial cells (M.-H.J., unpublished data).
One possible limitation of our findings, which are based on immunohistochemical staining, is that cells expressing extremely low levels of SRC-1 or ERα might not be detectable by this technique. We cannot rule out the possibility that levels of SRC-1 undetectable by immunohistochemical means may be sufficient for coactivation in cells expressing a high level of ERα. However, estrogen-stimulated PR induction occurred only in mammary epithelial cells expressing ERα but not in adjacent cells where the SRC-1 was easily detectable. Rather than playing the role of an ERα or PR coactivator in cells in which it cannot be detected, it is more likely that SRC-1 is in fact playing some cell type-specific role in the mammary epithelial cells, which do express SRC-1 to high levels. This view is further supported by the observation that PR was not induced in stromal cells that coexpressed both ERα and SRC-1. Our data strongly suggest a potential cell type-specific role of SRC-1 that does not involve direct interaction of SRC-1 with ERα or PR.
Finally, although ERα and SRC-1 are not coexpressed in the normal mammary epithelium, it is interesting to speculate, given the finding of amplification of the SRC-1- related gene AIB1 in some breast cancers (7), that the ectopic coexpression of ERα and an SRC-1 family coactivator may play a role in the growth stimulatory properties of estrogen in breast cancer.
Acknowledgments
We thank Dr. L. W. K. Chung for critical reading of the manuscript, Dr. C. Kao for advice during the course of these studies, W. K. Hong and A. C. Eischeid for skillful technical assistance in tissue section preparation, and J. M. Sanders and C. F. Murphy for assistance with the dual labeling studies. This work was supported by Department of Defense Breast Cancer Research Program Grants DAMD17-96-1-6233 and DAMD17-97-1-7066 (to M.-H.J.) and Grant DAMD 17-97-1-7096 (to J.D.), by National Institutes of Health Grant CA57374 (to M.B.), and by the Susan G. Komen Breast Cancer Foundation (to M.B).
ABBREVIATIONS
- SRC-1
steroid receptor coactivator 1
- ERα
estrogen receptor α
- PR
progesterone receptor
- AIB1
amplified in breast cancer-1
- GST
glutathione S-transferase
- FITC
fluorescein isothiocyanate
References
- 1.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 2.Cavailles V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 4.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 5.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 6.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 8.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Gomes P J, Chen J D. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 12.Beato M, Herrlich P, Schütz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 13.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 14.Jenster G, Spencer T E, Burcin M M, Tsai S Y, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M-J, O’Malley B W. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 16.Smith C L, Oñate S A, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oñate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M-J, Edwards D P, O’Malley B W. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 20.Henttu P M, Kalkhoven E, Parker M G. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McInerney E M, Tsai M-J, O’Malley B W, Katzenellenbogen B S. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 23.Misiti S, Schomburg L, Yen P M, Chin W W. Endocrinology. 1998;139:2493–2500. doi: 10.1210/endo.139.5.5971. [DOI] [PubMed] [Google Scholar]
- 24.Medina D. J Mammary Gland Biol Neoplasia. 1996;1:5–19. doi: 10.1007/BF02096299. [DOI] [PubMed] [Google Scholar]
- 25.Hennighausen L. Mol Biol Rep. 1997;24:169–174. doi: 10.1023/a:1006851531360. [DOI] [PubMed] [Google Scholar]
- 26.Hennighausen L, Robinson G W. Genes Dev. 1998;12:449–455. doi: 10.1101/gad.12.4.449. [DOI] [PubMed] [Google Scholar]
- 27.Lubahn D B, Moyer J S, Golding T S, Couse J F, Korach K S, Smithies O. Proc Natl Acad Sci USA. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bocchinfuso W P, Korach K S. J Mammary Gland Biol Neoplasia. 1997;2:323–334. doi: 10.1023/a:1026339111278. [DOI] [PubMed] [Google Scholar]
- 29.Lydon J P, DeMayo F J, Funk C R, Mani S K, Hughes A R, Montgomery C A J, Shyamala G, Conneely O M, O’Malley B W. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 30.Jeng M-H, Shupnik M A, Bender T P, Westin E H, Bandyopadhyay D, Kumar R, Masamura S, Santen R J. Endocrinology. 1998;139:4164–4174. doi: 10.1210/endo.139.10.6229. [DOI] [PubMed] [Google Scholar]
- 31.Jeng M-H, Kao C, Sivaraman L, Krnacik S, Chung L W K, Medina D, Conneely O M, O’Malley B W. Endocrinology. 1998;139:2916–2925. doi: 10.1210/endo.139.6.6073. [DOI] [PubMed] [Google Scholar]
- 32.Haslam S Z. Endocrinology. 1989;125:2766–2772. doi: 10.1210/endo-125-5-2766. [DOI] [PubMed] [Google Scholar]
- 33.Silberstein G B, Van Horn K, Shyamala G, Daniel C W. Cell Growth Differ. 1996;7:945–952. [PubMed] [Google Scholar]
- 34.Shyamala G, Barcellos-Hoff M H, Toft D, Yang X. J Steroid Biochem Mol Biol. 1997;63:251–259. doi: 10.1016/s0960-0760(97)00128-3. [DOI] [PubMed] [Google Scholar]
- 35.Clarke R B, Howell A, Potten C S, Anderson E. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- 36.Cunha G R, Hom Y K. J Mammary Gland Biol Neoplasia. 1996;1:21–35. doi: 10.1007/BF02096300. [DOI] [PubMed] [Google Scholar]
- 37.Cunha G R, Young P, Hom Y K, Cooke P S, Taylor J A, Lubahn D B. J Mammary Gland Biol Neoplasia. 1997;2:393–402. doi: 10.1023/a:1026303630843. [DOI] [PubMed] [Google Scholar]
- 38.Brisken C, Park S, Vass T, Lydon J P, O’Malley B W, Weinberg R A. Proc Natl Acad Sci USA. 1998;95:5076–5081. doi: 10.1073/pnas.95.9.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo I H, Russo J. J Mammary Gland Biol Neoplasia. 1998;3:49–61. doi: 10.1023/a:1018770218022. [DOI] [PubMed] [Google Scholar]
- 40.Kuiper G G J, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J-Å. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]