Abstract
Using differential display PCR, we have identified a gene [NOEY2, ARHI (designation by the Human Gene Nomenclature Committee)] with high homology to ras and rap that is expressed consistently in normal ovarian and breast epithelial cells but not in ovarian and breast cancers. Reexpression of NOEY2 through transfection suppresses clonogenic growth of breast and ovarian cancer cells. Growth suppression was associated with down-regulation of the cyclin D1 promoter activity and induction of p21WAF1/CIP1. In an effort to identify mechanisms leading to NOEY2 silencing in cancer, we found that the gene is expressed monoallelically and is imprinted maternally. Loss of heterozygosity of the gene was detected in 41% of ovarian and breast cancers. In most of cancer samples with loss of heterozygosity, the nonimprinted functional allele was deleted. Thus, NOEY2 appears to be a putative imprinted tumor suppressor gene whose function is abrogated in ovarian and breast cancers.
Germline abnormalities of several tumor suppressor genes have been implicated in hereditary breast and ovarian cancer syndromes, including BRCA1, BRCA2 (1, 2), and p53 (3). However, previously identified genes are silenced or mutated in only a fraction of ovarian and breast cancers; a number of relevant tumor suppressor genes may not yet have been identified. Deletions of regions on chromosome 1p have been observed in a variety of human malignancies. 1p31 has been found to have allelic deletion in 28–50% of breast cancers (4–6) and, as described below, in ≈40% of ovarian cancers. To date, the putative tumor suppressor gene(s) located within this region have not been reported.
Gene imprinting, defined as gene expression based on the gamete of origin, has been implicated in oncogenesis through loss of tumor suppressor gene regulation. The WT1 gene, in particular, is part of an imprinted cluster with INS, IGF2, H19, and p57KIP2 on chromosome 11p (7). Recent studies have suggested that portions of chromosome 1p might also be imprinted. In a subset of neuroblastoma tumors without N-myc amplification, allelic loss of 1p36 is preferentially of maternal origin. In contrast, the paternal allele of N-myc was amplified preferentially in those tumors with N-myc amplification (8).
The ras family of protooncogenes is among the most commonly activated in human cancers. Functional activation of the ras pathway in the absence of genetic mutations has been reported in both breast and ovarian cancer cells (9). Members of the rap family share high homology with ras proteins and have been shown to antagonize ras-mediated activation of mitogen-activated protein kinase (10) and cellular transformation in NIH 3T3 fibroblasts (11). Rap proteins also may stimulate mitogen-activated protein kinase activation and cell proliferation, depending on cell types. For instance, Rap1 positively regulates sustained mitogen-activated protein kinase activation through B-Raf in PC12 cells (12) and induces cell proliferation and transformation in Swiss 3T3 cells (13). Thus, the ras superfamily proteins play an important role in the control of cell growth and differentiation. Here, we report a ras-related, maternally imprinted gene mapped to 1p31, which acts as a negative growth regulator in both ovarian and breast cancers.
MATERIALS AND METHODS
Northern Blot Analysis.
NOEY2 cDNA probe was labeled with 32P-dCTP by using a random primer. Total cellular RNA (15 μg) was separated in 1.2% formaldehyde-agarose gels and was immobilized on a Hybond-N+ membrane by standard capillary transfer and UV crosslinking, and then was hybridized to NOEY2 probe by using standard protocol. The membrane was rehybridized to the DNA probe of 18S RNA to confirm equal loading among samples.
Transfection and Clonogenic Assays.
NOEY2 cDNA [1 kilobase (kb)] was released from the EcoRI cloning site in the Bluescript/Lambda ZapII vector and was inserted into the pcDNA3-neo eukaryotic expression vector (Invitrogen) in sense and antisense orientations. The constructs were transfected into different cell lines by using lipofectamine. After incubation for 48 hours at 37°C, transfected cells were trypsinized and seeded into 100-mm dishes. Selection medium with G418 (400 μg/ml to 1000 μg/ml) was added. Two weeks later, colonies were stained by 0.1% Coomassie blue in 30% methanol and 10% acetic acid.
Luciferase Assay.
The vector containing the luciferase reporter gene under the control of the human cyclin D1 promoter (-1745CD1LUC) was provided by R. G. Pestell (Albert Einstein College of Medicine, New York). NOEY2 sense and antisense constructs were cotransfected with the luciferase reporter, and luciferase activity was measured 48 hr post-transfection as described (14). In control experiments, a vector expressing β-galactosidase driven by the CMV promoter was cotransfected to ensure comparable transfection efficiency between the NOEY2 sense and antisense construct-transfected cells (through staining).
Western Blot Analysis.
The confluent cells in 60-mm culture dishes were lysed by adding boiling SDS sample buffer. The cell lysates were boiled for an additional 5 min, were passed several times through a 26-gauge needle, and were centrifuged for 5 min. Equal amounts of total cellular protein (15 μg) were electrophoresed in 15% SDS/PAGE and were transferred to polyvinylidene fluoride membrane (Millipore). The membrane was immunoblotted with anti-NOEY2 mAb (5 μg/ml) and was developed by using an ECL system (Amersham).
Detection of Loss of Heterozygosity (LOH).
One intragenic TA repeat marker was fluorescein isothiocyanate-labeled and was used for PCR-LOH analysis in 3 ovarian cancer cell lines, 38 primary ovarian cancers, and 8 primary breast cancers tumor DNA samples paired with normal DNA. DNA fragment analysis was done by Applied Biosystems 377 Automated sequencer. The data were collected automatically and were analyzed by genescan (Applied Biosystems). genotyper ii software (Applied Biosystems) was used for allele scoring and assessment of LOH quantitatively according to Canzian’s formula (15).
PCR–Single-Strand Conformation Polymorphism Sequencing Analysis.
The single-strand conformation polymorphism analysis was carried out essentially as described (16). After single-strand conformation polymorphism analysis of the coding region, PCR fragments with abnormal mobility were cloned into the PCR-Script Amp SK(+) cloning vector (Stratagene). More than 10 clones of each sample were sequenced by using an Applied Biosystems PRISM 377 DNA automatic sequencer and a Big Dye terminator cycle sequencing kit. Upstream promoter region sequences (1.9 kb) were amplified by four pairs of primers with a −21M13 tail. Purified PCR products were directly sequenced by using a 21M13 Dye primer cycle sequencing kit or a Big Dye terminator cycle sequencing kit (Applied Biosystems).
Methylation Analysis.
Genomic DNA (5 μg) was digested by using restriction enzymes XbaI/SacII in CpG islands surrounding the TATA box by Southern blot analysis using a standard protocol.
Analysis of Monoallelic Expression.
cDNA synthesis was performed by using 5 μg of total RNA by oligo(dT) and superScript TM II reverse transcriptase (GIBCO/BRL) according to the manufacturer’s instructions. PCR reactions were performed with either 2 μl of reverse transcription products or 50 ng of genomic DNA by a set of primers, which amplify a 326-bp fragment encompassing the coding +231 A/G polymorphism. A portion (10 μl) of the PCR product was digested with 15 units of HhaI by incubation at 37°C overnight and then was analyzed on a 2% agarose gel.
Assay of Allele-Specific Methylation.
Allele-specific methylation was assayed by using a modification of the method of Issa et al. (17). To study imprinting methylation, DNA from different families was genotyped by using the NOEY2 TA repeat marker. Informative offspring’s genomic DNA was digested with BstUI and was PCR amplified by using primer methy F1/LOH R1, which encompasses sequence −160 to 2680 and is ≈2.94 kb. Only the methylated allele is amplified after digestion. Allele typing was confirmed by nested PCR using primer LOH F1/LOH R1.
RESULTS
A Member of the ras Superfamily.
Most ovarian and breast cancers are clonal neoplasms that arise through multiple mutations in normal epithelial cells. The ability to isolate and maintain normal ovarian and breast epithelial cells in culture (18, 19) has facilitated the identification of molecular alterations in ovarian and breast cancer cells. We have used differential display PCR (20) to identify several sequences that were expressed in three specimens of normal ovarian surface epithelial (OSE) cells maintained in culture but not expressed in six ovarian cancer cell lines. One of these sequences (NOEY2) was extended by using rapid amplification of cDNA ends. A cDNA library from OSE cells constructed in Lambda ZAPII (Stratagene) was screened by using the extended NOEY2 cDNA as probe. With this strategy, a full length NOEY2 cDNA sequence and ORF were obtained. The nucleotide sequence contains a 5′ untranslated region of 149 bp, an ORF of 687 bp encoding a 26-kDa protein of 229 amino acids, and 660 bp of 3′ untranslated sequence with a poly(A) tail. NOEY2 is a member of the Ras superfamily of small G proteins. Starting from its N-terminal amino acid 35, NOEY2 shares 56% amino acid homology with Rap1A, 56% with Rap1B, 58% with Rap 2A, 62% with Rap2B, 59% with c-K-Ras, and 54% with H-Ras. The NOEY2 gene ORF contains three motifs typical of Ras/Rap family members (21): (i) a highly conserved GTP binding domain, (ii) a putative effector domain YLPTIENTY, and (iii) the membrane localizing CAAX motif (where C is cysteine, A is an aliphatic amino acid, and X is any amino acid) at the COOH terminus (Fig. 1). Within the effector domain, however, NOEY2 differs both from Ras and Rap family members where the sequence YDPTIEDSY is found in all Ras and Rap proteins. NOEY2 instead has YLPTIENTY. In addition, when compared with the sequence of p21ras, NOEY2 has substitutions of alanine for glycine at amino acid 12 and glycine for glutamine at amino acid 61, consistent with constitutive activation of a small G protein.
Figure 1.
Comparison of NOEY2 with Ras and Rap family members. The four conserved GDP/GTP binding domains and the CAAX motif are underlined. Bold type indicates residues conserved in nearly all GTPases. Amino acids are designated according to the single letter code.
Loss of NOEY2 Expression in Ovarian and Breast Carcinomas.
An NOEY2 message of 1.9 kb was detected by Northern blot analysis in all 17 primary normal OSE cultures and 14 primary normal breast epithelial cultures but was not detected in 8 of 9 ovarian cancer cell lines (See Fig. 2A for representative samples) and 7 of 8 breast cancer cell lines (See Fig. 2B for representative samples). One ovarian cancer cell line (CAOv3) and one breast cancer cell line (MDA-MB-468) expressed NOEY2, but at lower levels than normal epithelial cells. NOEY2 expression was not detected in any of nine primary ovarian cancer cell preparations that were separated and purified from patients’ ascites (Fig. 2C). By using multiple tissue blots containing poly(A)+ RNA from 16 different normal human tissues, the expression of NOEY2 was detected in several other normal tissues, including heart, liver, pancreas, and brain, but the highest expression occurred in normal ovarian tissue (Fig. 2D). Murine mAbs were prepared against an NOEY2–glutathione S-transferase fusion protein. On Western blot analysis, a 26-kDa protein was expressed by normal ovarian and breast epithelial cells but could not be detected in ovarian and breast cancer cell lines (Fig. 2E).
Figure 2.
NOEY2 expression in cells and tissues. (A) Northern blot analysis of OSE cells (right three lanes) and ovarian cancer cell lines (remaining lanes). (B)Northern blot analysis of normal breast epithelial (NBE) cells (right four lanes) and breast cancer cell lines (remaining lanes). (C) Northern blot analysis of OSE cells (left three lanes) and ovarian cancer cells purified from patients’ ascites fluid (remaining lanes). (D) Northern blot of multiple human tissues. Poly(A)+ RNA (2 μg) from multiple human tissues was supplied in a blot prepared by CLONTECH. (E) Western blot analysis of positive control (NIH 3T3 transiently transfected with NOEY2 cDNA), OSE cells (left lanes 2–4), and ovarian cancer cell lines (remaining lanes).
NOEY2 as a Growth Inhibitor in Cultured Cells.
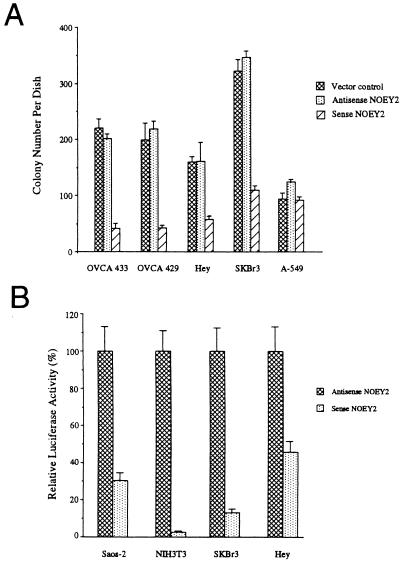
Sense and antisense constructs of NOEY2 were transfected with lipofectamine into three ovarian cancer cell lines (OVCA433, OVCA429, Hey) and one breast cancer cell line (SKBr3) that did not express NOEY2 as well as one lung cancer cell line that did express NOEY2 (A-549). All four ovarian and breast cancer cell lines were growth inhibited clonogenically by transfection of NOEY2 in a sense but not in an antisense orientation (Fig. 3A). This growth inhibition also was found with other cell lines that did not express NOEY2, such as Hela, Saos-2, NIH 3T3, 293, and Cos7. However, NOEY2 transfection did not significantly affect the growth of the A-549 lung cancer cell line that exhibited higher expression of NOEY2 (Fig. 3A).
Figure 3.
NOEY2-induced growth inhibition of ovarian and breast cancer cell lines. (A) Colony formation after NOEY2 cDNA transfection. NOEY2 in the sense orientation inhibited growth of OVCA433, OVCA429, Hey, and SKBr3 (P < 0.001). (B) Inhibition of cyclin D1 promoter activity in Saos-2, NIH 3T3, SKBr3, and Hey cells. Luciferase activity in cells transfected with the sense construct was expressed as a percentage of the activity in cells transfected with the antisense vector. The results are from representative experiments performed in triplicate.
We have examined the effects of NOEY2 on other growth regulatory molecules. In Saos-2 and NIH 3T3 as well as ovarian and breast cancer cell lines whose growth could be inhibited, an NOEY2 sense construct, but not an antisense construct, strongly inhibited cyclin D1 promoter activity when cotransfected with a plasmid containing the luciferase gene under the control of the cyclin D1 promoter (Fig. 3B). Because cyclin D1 is required for G1-S progression (14), the potent inhibition of the cyclin D1 promoter activity by NOEY2 could contribute to the observed growth inhibition. Cyclin D1 expression is up-regulated in 25–30% of ovarian cancers in the absence of gene amplification (22) and also is overexpressed in as many as 49% of invasive breast carcinomas, consistent with the possible impact of loss of NOEY2 function in these tumors.
Multiple growth factors, including epidermal growth factor, insulin, hydrocortisone, and bovine pituitary extract can stimulate growth of primary breast and ovarian epithelial cells. Increased growth rates were associated with down-regulation of both NOEY2 and P21WAF1/CIP1 in Northern blot analysis. To examine the interaction between NOEY2 and P21WAF1/CIP1, an NOEY2 fragment that included the entire ORF was fused in frame with a plasmid that contained a triple hemagglutinin repeat, and this hemagglutinin–NOEY2 construct was transfected into NIH 3T3 cells. Two-color immunofluorescent staining demonstrated that hemagglutinin–NOEY2 transfectants had higher P21WAF1/CIP1 protein expression than did nontransfected cells or cells transfected with a hemagglutinin–Erk2 cDNA (data not shown). The P21WAF1/CIP1 protein has been shown to arrest cell growth by inhibition of cyclin-dependent kinases (23). Introduction of NOEY2 as a transgene into FVB/N mice has produced small body size associated with up-regulation of P21WAF1/CIP1 (data not shown). These observations collectively suggest that NOEY2 functions as a negative regulator of cell growth, probably through interaction with components of cell cycle control.
Localization of NOEY2 to Chromosome 1p31 at a Site of LOH.
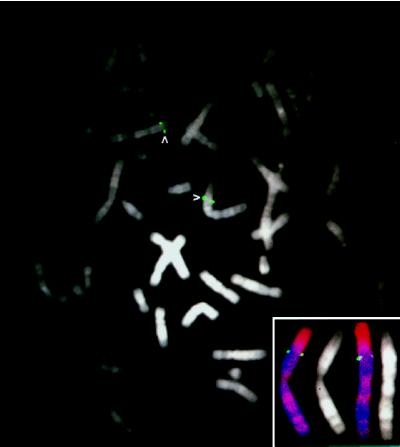
The NOEY2 gene was mapped to chromosome 1p31 by screening a P1 genomic library (DuPont) with two sets of primers that encompass the entire coding and 3′ untranslated region. Clones positive with either or both of the primer sets were mapped to 1p31 by fluorescence in situ hybridization (Fig. 4). The chromosomal location was confirmed with the fractional length measurement (0.27) of the hybridized signal from the p terminus of the chromosome 1, as described (24). As LOH has been reported in breast cancers in this region, we have determined whether LOH affected the NOEY2 locus in both ovarian and breast carcinomas by studying 49 cancer samples for which paired samples of normal DNA were available. PCR-LOH analysis was performed by using an intragenic TA repeat marker within NOEY2. Of 22 informative cases, 9 (41%) showed LOH at this gene. Frequent LOH for NOEY2 supports its potential importance in these neoplasms and prompted further studies to identify mechanisms by which the function of the contralateral allele might be lost.
Figure 4.
Metaphase mapping of NOEY2 by fluorescence in situ hybridization. A P1 clone containing NOEY2 was mapped to 4′,6-diamidino-2-phenylindole-banded human chromosome 1p31 in normal human lymphocytes. (Inset) Localization of NOEY2 (green) on chromosome 1 counterstained with propidium iodide and 4′,6-diamidino-2-phenylindole compared with 4′,6-diamidino-2-phenylindole-banded chromosome 1.
DNA Sequence Abnormalities.
The genomic sequence of NOEY2 contains two exons and one intron. We have screened the entire coding region by single-strand conformation polymorphism sequencing analysis and sequenced 1.9 kb upstream to identify putative mutations in NOEY2 by using DNA from 18 different cell lines and 4 normal controls. Genetic analysis reveals one germline mutation in the promoter region (with a C to G change at position −19) but lack of DNA mutation in coding sequence. Two polymorphisms (a G to A substitution at coding +231 encoding an Ala to Thr change and A to G substitution at −750 promoter region) were found and confirmed in 110 normal DNA samples.
Methylation and Monoallelic Expression of NOEY2.
To find whether aberrant methylation plays a role in inactivation of NOEY2 gene, methylation status of NOEY2 was studied by Southern blot analysis. Hypermethylation was found in two of eight breast cancer cell lines, indicating an alternative mechanism of down-regulation of NOEY2 gene in a fraction of breast cancer. Hypomethylation was observed in three of eight breast cancer cell lines, reflecting either LOH or the actual hypomethylation of two alleles. Surprisingly, all normal and most cancer cells exhibited partial methylation at these sites, consistent with the possibility that the gene is expressed only monoallelically.
Taking advantage of the coding +231 G/A polymorphism, which eliminates an HhaI restriction site, a total of 43 normal samples were screened to detect additional heterozygous individuals. Aside from two cancer cell lines (BT20 and CAOv3), two more informative samples (OSE031 and normal breast epithelial 024) were found. Only one allele of NOEY2, either the A allele or the G allele, was expressed in all four informative samples, as assessed by HhaI digestion of a 326-bp transcript from reverse transcription–PCR cDNA (Fig. 5A) and confirmed by sequencing. These data verified that NOEY2 is indeed a monoallelically expressed gene.
Figure 5.
Monoallelic expression and imprinting of NOEY2. (A) Monoallelic expression. Shown is analysis of coding +231 G/A allele expression by using HhaI digestion of a reverse transcription–PCR transcript (lanes 2, 4, 6, 8) and genomic DNA as controls (lanes 1, 3, 5, 7) from homozygote G allele in OSE001 and heterozygote G/A alleles in BT20, CAOv3, and OSE 031. Arrowheads: a, 326 bp for an HhaI-uncut fragment of A allele; b, 206 bp; and c, 120 bp for HhaI-cut two fragments of G allele. (B) Maternal imprinting of NOEY2. Genotype of TA repeat length polymorphism of three families (F1, F2, and F3) was shown as peaks. Maternal (lane 1); paternal (lane 2); offspring from the same families (lanes 3 and 5; D, daughter; S, son). Maternal origin of methylated allele is demonstrated in each offspring (lanes 4 and 6). (C) DNA fragments analysis of LOH and methylated retained allele in ovarian cancer patients. Genotype of TA repeat length polymorphism of normal DNA from two ovarian cancer patients (lane 1): one allele is lost in tumor DNA (lane 2); the retained allele is methylated (lane 3). (D) The genomic structure of NOEY2 and the location of the primers and polymorphisms.
Maternal Imprinting and Loss of the Functional Allele.
To determine whether NOEY2 is imprinted, 3 informative families with TA repeat length polymorphism and one family with −750 A/G polymorphism were identified from a total of 10 families assessed. As shown in Fig. 5B, only the maternal allele can be amplified after genomic DNA is digested by the methylation-sensitive restriction enzyme BstUI in all six offspring from three families. In a family with the −750 polymorphism, the mother is A/G heterozygous, and the father is A homozygous. Four of eight offspring are A/G heterozygous. Only the maternal G allele could be amplified after genomic DNA was digested with BstUI in all four heterozygous offspring. Moreover, we also have demonstrated that NOEY2 is paternally expressed in a coding +231 G/A informative family (NBE024). Two normal G/A heterozygous daughters were found to express only the paternal G allele at the transcript level. These results confirmed that NOEY2 is a maternally imprinted gene.
Loss of NOEY2 expression in a fraction of ovarian and breast cancers could result from LOH of the nonimprinted allele. To test this possibility, we have examined five informative cases in which both LOH and imprinting could be evaluated. By comparing the TA repeat genotype from BstUI- cut DNA with the genotype of normal and tumor DNA from same patient, we have found that the retained allele is the methylated allele in four of five patients with NOEY2 LOH. Two representative examples are displayed in Fig. 5C. In one case, the methylated allele was lost, suggesting that loss of other genes at 1p31 may also be important in this instance.
DISCUSSION
NOEY2 is a member of the ras superfamily sharing 54 to 62% amino acid homology with ras and rap. The presence of a distinctive effector domain suggests, however, that the function of NOEY2 may differ from that of the ras and rap proteins. As in the case of ras and rap, preliminary immunohistochemical data suggest that the protein is located both in the cytoplasm and in the cell membrane. Earlier studies have demonstrated that certain members of the ras superfamily, particularly rap-related genes, can inhibit ras activation and signal transduction in certain types of cells. Our unpublished data suggest that expression of NOEY2 may truncate signaling through ras/mitogen-activated protein kinase induced by growth factors. Data presented above suggest that introduction of NOEY2 can inhibit the growth of transformed cells that have lost expression of the endogenous gene. Whether this relates to a direct impact on signaling through ras remains to be determined. The role of NOEY2 may depend on the cellular context in which the protein is expressed.
NOEY2 has been mapped to a region of chromosome 1p31 that has been found deleted in a substantial fraction of breast cancers. Our study has shown that 1p31 is an area in which LOH is observed frequently in ovarian cancers. Previous LOH studies on chromosome 1p in breast cancer have found two well defined deleted regions (1p36 and 1p31) (5, 6). By comparing the rate of allelic loss for an NOEY2 intragenic marker with rates of LOH in four flanking 1p31 markers, we found that the NOEY2 marker exhibited the highest rate of LOH, indicating that NOEY2 is the most frequently deleted locus at 1p31 in breast and ovarian cancers (H.P., F.X., J.W.G., R.C.B., and Y.Y., unpublished data). To the extent that the association of growth regulatory genes with LOH has signaled the presence of tumor suppressor loci, NOEY2 is a reasonable candidate as a tumor suppressor gene at 1p31 in these malignancies.
In recent years, evidence has accumulated to suggest that genomic imprinting and monoallelic gene expression appear to play an important role in tumorigenesis (25). Sapienz was the first to incorporate genetic imprinting into Knudson’s two-“hit” tumorigenesis model (26). He noted that, if an imprinted gene were involved, the first hit might actually be explained by nonexpression of one of the alleles because of the imprinting process, leaving a hemizygous phenotype. The second hit may be mutational or may result from a loss of all or part of chromosome carrying the remaining functional suppressor allele. Only the paternal allele of NOEY2 is expressed because of maternal imprinting. The second somatic hit in NOEY2 could occur through multiple mechanisms. LOH has been detected in 40% of ovarian and breast cancers, and the functional allele has been lost preferentially in informative cases. The germline mutation with a C to G change at position −19, which is located in the cis-regulatory sequences that control methylation, also could act through a cis-acting control pathway. Either hyper- or hypomethylation was detected in five of eight breast cancer cell lines in the current study. Cis- or trans-acting defects could underlie such aberrant methylation. In addition to genetic regulation of NOEY2, our unpublished data have demonstrated repressed NOEY2 promoter activity in ovarian and breast cancer cells. Down-regulation in transformed cells could result from a loss of transcriptional activators or from the expression of transcriptional repressors through both cis- and trans-control pathways. The observation that NOEY2 expression can be down-regulated by the medium enriched in multiple growth factors and hormones suggests that some of these growth factors and hormones might regulate the expression of trans-acting factors for NOEY2.
In summary, we have identified an imprinted growth regulatory gene on chromosome 1p31 whose loss may play an important role in the development of ovarian and breast cancers. Other imprinted genes may map to a cluster in this area. NOEY2 may play an important role physiologically in regulating cell growth through regulating expression of the cyclins and cyclin dependent kinase inhibitors. In addition to its fundamental importance, further evaluation of NOEY2 should elucidate its potential prognostic significance and its possible value for gene therapy in breast and ovarian cancer.
Acknowledgments
We thank Dr. Elvio G. Silva, Dr. Kelly K. Hunt, Dr. Andrew Berchuck, and Dr. Marsha L. Frazier for providing normal and malignant tissue samples. Dr. Elvio G. Silva and Dr. Carmen S. Tornos aided with pathologic analysis. We thank Rashmi Pershad in DNA Core Facility for sequence and DNA fragment analysis. We thank Dr. J. P.-J. Issa and Dr. Cherry Li for advice on methylation analysis. This work was supported by the Ovarian Cancer Research Fund, the Breast Cancer Research Program of the University of Texas M. D. Anderson Cancer Center, and National Institutes of Health Grants P01 CA64602, CA16672, and CA79003.
ABBREVIATIONS
- LOH
loss of heterozygosity
- OSE
ovarian surface epithelial
- kb
kilobase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U96750).
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett L M, Ding W, et al. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Nature (London) 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 3.Malkin D, Li F P, Strong L C, Fraumeni J F, Jr, Nelson C E, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, et al. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 4.Nagai H, Negrini M, Carter S L, Gillum D R, Rosenberg A L, Schwartz G F, Croce C M. Cancer Res. 1995;55:1752–1757. [PubMed] [Google Scholar]
- 5.Loupart M L, Armour J, Walker R, Adams S, Brammar W, Varley J. Genes Chromosomes Cancer. 1995;12:16–23. doi: 10.1002/gcc.2870120104. [DOI] [PubMed] [Google Scholar]
- 6.Hoggard N, Brintnell B, Howell A, Weissenbach J, Varley J. Genes Chromosomes Cancer. 1995;12:24–31. doi: 10.1002/gcc.2870120105. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y H, Shields T, Crenshaw T, Hao Y, Moulton T, Tycko B. Am J Hum Genet. 1993;53:113–124. [PMC free article] [PubMed] [Google Scholar]
- 8.Caron H, Peter M, Sluis P, Speleman F, Kraker J, Laureys G, Michon J, Brugieres L, Voute P A, Westerveld A, et al. Hum Mol Genet. 1995;4:535–539. doi: 10.1093/hmg/4.4.535. [DOI] [PubMed] [Google Scholar]
- 9.Patten S E, Martin M L, Nelsen L L, Fang X, Mills G B, Bast R C, Jr, Ostrowski M C. Cancer Res. 1998;58:2253–2259. [PubMed] [Google Scholar]
- 10.Cook S J, Rubinfeld B, Albert I, McCormick F. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 12.York R D, Yao H, Dillon T, Ellig C L, Eckert S P, McCleskey E W, Stork P S. Nature (London) 1998;392:622–626. doi: 10.1038/33451. [DOI] [PubMed] [Google Scholar]
- 13.Altschuler D, Ribeiro-Neto F. Proc Natl Acad Sci USA. 1998;95:7475–7479. doi: 10.1073/pnas.95.13.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 15.Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick R B, Aaltonen L A, Chapelle A. Cancer Res. 1996;56:3331–3337. [PubMed] [Google Scholar]
- 16.Peng H Q, Hogg D, Malkin D, Bailey D, Gallie B L, Bulbul M, Jewett M, Buchanan J, Goss P E. Cancer Res. 1993;53:3574–3578. [PubMed] [Google Scholar]
- 17.Issa J P, Vertino P M, Boehm C D, Newsham I F, Baylin S B. Proc Natl Acad Sci USA. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruk P A, Maines-Bandiera S L, Auersperg N. Lab Invest. 1990;63:132–136. [PubMed] [Google Scholar]
- 19.Stampfer M R. J Tissue Cult Methods. 1985;9:107–115. [Google Scholar]
- 20.Liang P, Pardee A B. Science. 1992;257:967–970. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 21.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 22.Worsley S D, Ponder B A J, Davies B R. Gynecol Oncol. 1997;64:189–195. doi: 10.1006/gyno.1996.4569. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashl R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 24.Sakamoto M, Pinkel D, Mascio L, Peter D, Kuo W-L, Yamakawa K, Nakamura Y, Drabkin H, Jericevic Z, Smith L, et al. Cytometry. 1995;19:60–69. doi: 10.1002/cyto.990190108. [DOI] [PubMed] [Google Scholar]
- 25.Barlow D P. Science. 1995;270:1610–1613. doi: 10.1126/science.270.5242.1610. [DOI] [PubMed] [Google Scholar]
- 26.Sapienza C. Biochim Biophys Acta. 1991;1072:51–61. doi: 10.1016/0304-419x(91)90006-7. [DOI] [PubMed] [Google Scholar]