Short abstract
Two recent papers describe how arrays of oligosaccharide and polysaccharide molecules can be used to investigate the interactions between carbohydrates and proteins more fully.
Abstract
Interactions between carbohydrates and proteins mediate intracellular traffic, cell adhesion, cell recognition and immune system function. Two recent papers describe how arrays of oligosaccharide and polysaccharide molecules can be used to investigate these interactions more fully.
Protein-carbohydrate interactions occupy a small but important niche in the repertoire of molecular interactions that underlie the development and function of multicellular organisms [1]. Glycans, chains of sugar units attached to proteins or lipids, serve as sorting tags for the intracellular and extracellular trafficking of glycoproteins as a result of their interactions with sugar-binding receptors. In this way, oligosaccharides can direct the movements of the glycoproteins through intracellular compartments and into and out of the circulation. Other protein-glycan interactions result in cellular adhesion, particularly in the case of transient interactions involving cells of the immune system. In addition, mammalian carbohydrate receptors can detect differences between the types of sugars on mammalian cells and those on microorganisms. Recognition of the surfaces of potential pathogens often initiates innate immune protective responses. Conversely, bacteria, viruses and toxins often bind to oligosaccharides on mammalian surfaces as a means of gaining entry into target cells.
In order to understand the biology of protein-glycan interactions, it is necessary to identify the sugar-binding receptors, also known as lectins, and to characterize the glycan ligands that they bind. Two recent papers describe strategies for characterizing the specificity of lectins by creating and screening libraries of immobilized poly- and oligosaccharides [2,3]. Up to now, most sugar-binding activity has been analyzed using material-intensive assays, for example in hapten inhibition studies in which soluble oligosaccharides compete with a reporter ligand for binding to immobilized lectins. The ability to test arrays of glycans in a single experiment would clearly increase the rate of lectin characterization. The oligosaccharide array method is an outgrowth of previous studies in which libraries of oligosaccharides from individual glycoproteins were created and probed with lectins (Figure 1) [4].
Figure 1.
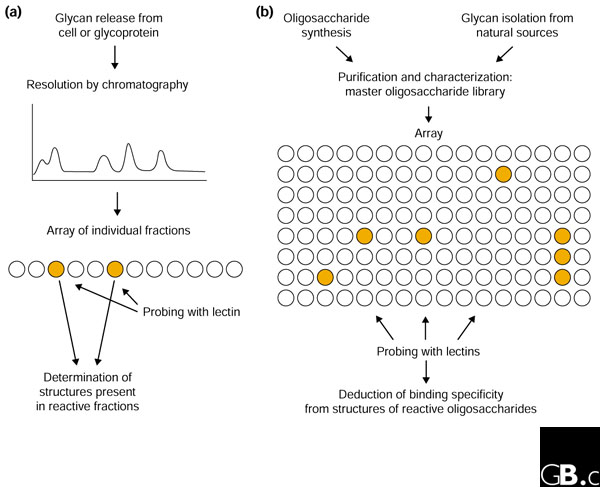
Two conceptual approaches to screening glycan libraries. (a) When specific target cell types or glycoprotein ligands are known for a lectin, oligosaccharides released specifically from these sources can be separated by chromatography and examined by probing a relatively small array. (b) In the alternative 'glycomic' approach, a generic library of purified glycans produced synthetically and/or from natural sources can be screened with various different lectins.
The need for more efficient screening methods reflects recent increases in the numbers of known or potential lectins and the growing pool of possible glycan ligands. In line with the concepts of the genome and the proteome, the term 'glycome' is used to denote the complement of glycans in a cell or organism. If one is interested in studying a novel sugar-binding receptor that mediates cell-cell adhesion in a model organism such as the mouse, the magnitude of the screening task depends on the size of the mouse glycome. Unfortunately, it is difficult to estimate the size of typical mammalian glycomes. Any two monosaccharides may be linked through various combinations of hydroxyl groups, and many glycans are branched, so a large number of glycans can be imagined to be in the 'theoretical' glycome. It is important to remember, by analogy, that the number of possible proteins of 100 amino acids (10400) vastly exceeds the number of proteins encoded in the human genome (fewer than 105). In both proteomics and glycomics, the focus needs to be on what is actually found in nature, rather than on what could theoretically exist.
Projects to catalog the structures of all the glycans associated with particular cells are just getting underway, but databases containing published glycan sequences can provide a clue to the scale of glycomes. Examination of a curated database containing mostly protein-linked oligosaccharides suggests that there are probably around 200 well-characterized structures in the mouse, which is the best studied model organism in this respect [5]. A database that includes more glycolipids contains in excess of 100 structures [6]. Allowing for structures missing from the databases and for others that have not yet been examined in detail, it seems reasonable to estimate that there may be about 500 endogenous mammalian glycan structures in glycoproteins and glycolipids.
A second way to gauge glycan complexity is to examine the complement of glycosyltransferases that are present in genomes. Genes encoding transferases can be identified with reasonable confidence, leading to estimates that there are roughly 300 potential transferases in mouse and humans [7]. Although some of these enzymes are not well studied, the actions of the known transferases can be accounted for by the structures in the glycan databases, and most enzymes needed to make the known structures have been described. Thus, it seems likely that we are approaching a complete description of the glycan biosynthetic machinery and the resulting glycome, and that the total magnitude of the glycome is likely to be in the range of 500 unique structures.
From these considerations, it is plausible to imagine an array containing a complete set of endogenous mammalian glycans. A major use of such an array would be the identification of ligands for mammalian lectins. The human genome sequence provides information about the number of potential lectins. There are about 100 lectins that fall into the known structural categories, and the binding specificities of about half of these are known with considerable precision [8]. Glycan arrays would be useful for confirming the existing specificity data and for characterizing the remaining lectins. Glycan arrays will not be used exclusively for testing interactions of known types of mammalian lectins, however. Because sugar-binding sites are often relatively shallow, they have evolved independently in the context of many different protein folds [9]. There may well be additional structural classes of lectins remaining to be described, which we cannot yet identify because we do not know what to look for. Probing glycan arrays could provide a way to demonstrate sugar-binding activity in novel proteins as well as probing the specificity of the interactions.
Of course, glycan arrays need not be used only to probe interactions of mammalian lectins with mammalian oligosaccharides. Appropriately constructed arrays could be used to test for interactions of mammalian lectins with sugar structures from microorganisms and for testing interactions between novel glycans and lectins in other organisms, including plants. This raises the issue of whether generic glycan arrays will be most useful or whether custom-made arrays designed to answer specific questions will be needed. Generic arrays from mammals and from panels of microorganisms will probably suffice for many purposes.
The format described by Fukui et al. [3] is one of several currently being put forward. They have built on previously described chemistry to attach oligosaccharides to lipid tails, and the resulting neoglycolipids are spotted onto nitrocellulose for probing with peroxidase-conjugated lectins. Alternative technologies use other ways of immobilizing oligosaccharides in microwells. One microwell format uses streptavidin to capture biotinylated glycosides [10], and a broadly inclusive array using this format is being developed by the Consortium for Functional Glycomics [11]. An array of relatively simple mono-, di-, tri- and tetra-saccharides chemically coupled to microwells is commercially available (the GlycoChip® from GlycoMinds [12]). Chemical coupling of monosaccharides to gold surfaces has also been reported [13].
So what kind of results can one envision coming from experiments using these arrays? Model proteins, such as the selectin cell-adhesion molecules that operate to attach lymphocytes to blood vessel walls at sites of inflammation, would be expected to recognize a very restricted set of glycans with closely related structures (Figure 2a). In contrast, molecules such as the macrophage mannose receptor, which detects a range of oligosaccharides typical of the extracellular matrix, intracellular glycoproteins and microbial surfaces, will probably bind to a much broader spectrum of compounds in the array (Figure 2b). Both types of result are informative.
Figure 2.
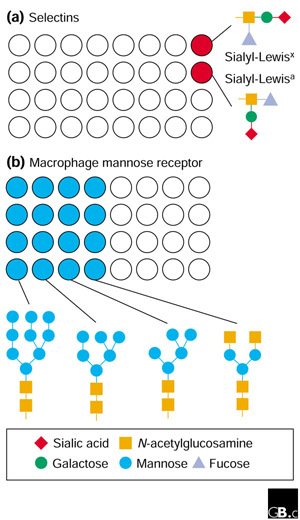
Possible outcomes of screening glycan libraries. (a) Some lectins, such as the selectin cell-adhesion molecules, will recognize a few specific endogenous mammalian glycans with related structures, such as the sialyl-Lewis glycans shown. (b) Other lectins, such as the macrophage mannose receptor, will bind broader classes of endogenous and microbial glycans, such as those shown below the array; the receptor is shown recognizing structures that bear terminal mannose or N-acetylglucosamine residues.
A key outstanding issue in the glycan-screening field is the importance of the context in which glycans are normally seen by lectins. Oligosaccharides are usually present on the surfaces of glycoproteins and lipid bilayers rather than free in solution or on synthetic chemical surfaces. Conjugation of glycans to proteins and lipids can have at least three different effects on their interactions with lectins. First, lectins are usually oligomeric and they bind with highest avidity when they make multiple interactions with appropriately spaced oligosaccharides [14]. The tightest binding might be achieved when multiple glycans are present on the surface of a specific glycoprotein at appropriate spacings. Second, some lectins actually bind to protein-carbohydrate co-determinants rather than to glycans alone. The best studied example of such a situation is the simultaneous interaction of P-selectin with a glycan and with adjacent sulfotyrosine residues in the P-selectin glycoprotein ligand 1 [11]. Finally, the proximity to protein or polysaccharide surfaces can increase the affinity of binding of lectins to glycoproteins or to terminal structures on large oligosaccharides without the need for extended, highly specific binding sites. These considerations suggest that glycan arrays will be useful tools for defining potential ligands, but characterization of natural ligands will often require analysis of glycoconjugates.
Despite these issues, the development of panels of oligosaccharides and methods that allow rapid screening of lectin binding is clearly an important advance and represents a tangible step toward studying functional aspects of the glycome.
References
- Taylor ME, Drickamer K. Introduction to Glycobiology. New York: Oxford University Press; 2002. [Google Scholar]
- Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- Feizi T, Stoll MS, Yuen C-T, Chai W, Lawson AM. Neoglycolipids: probes of oligosaccharide structure, antigenicity and function. Methods Enzymol. 1994;230:484–519. doi: 10.1016/0076-6879(94)30030-5. [DOI] [PubMed] [Google Scholar]
- GlycoSuiteDB http://www.glycosuite.com/
- SweetDB http://www.dkfz-heidelberg.de/spec/sweetdb/
- CAZy - Carbohydrate-Active enZYmes http://afmb.cnrs-mrs.fr/CAZY/
- A genomics resource for animal lectins http://ctld.glycob.ox.ac.uk/
- Weis WI, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- Leppanen A, Penttila L, Renkonen O, McEver RP, Cummings RD. Glycosulfopeptides with O-glycans containing sialylated and polyfucosylated polylactosamine bind with low affinity to P-selectin. J Biol Chem. 2002;277:39749–39759. doi: 10.1074/jbc.M206281200. [DOI] [PubMed] [Google Scholar]
- Consortium for Functional Glycomics http://glycomics.scripps.edu/
- Glycominds http://www.glycominds.com/
- Houseman BT, Mrksich M. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- Drickamer K. C-type lectin-like domains. Curr Opin Struct Biol. 1999;9:585–590. doi: 10.1016/s0959-440x(99)00009-3. [DOI] [PubMed] [Google Scholar]


