Short abstract
Recent work in the fission yeast Schizosaccharomyces pombe reveals that RNA is an integral component of silent heterochromatin.
Abstract
Repeated DNA elements and region-specific protein modifications combine within chromosomes to form a transcriptionally silent chromatin structure called heterochromatin. Recent work in the fission yeast Schizosaccharomyces pombe reveals that RNA is also an integral component of silent heterochromatin, providing a new perspective on how heterochromatin is organized and maintained in eukaryotic cells.
The heterochromatin of eukaryotic genomes is concentrated at and around centromeres and plays a central role in the maintenance of genome integrity through regulation of both gene expression and chromosome segregation [1,2]. Centric heterochromatin is required for cohesion between sister centromeres, which ensures proper chromosome segregation at mitosis [3,4]. Additional heterochromatin at telomeres protects the DNA at chromosome ends from inappropriate fusions and recombination. If genes are inserted into heterochromatin they are transcriptionally silenced; this silencing is maintained epigenetically (independent of DNA sequence) and can be propagated through multiple cell divisions. Interestingly, transposable elements and repeated DNA arrays are abundant within heterochromatin [1,5], suggesting that the molecular nature of repeated DNA sequences is related to assembly of this specialized chromatin structure. Two recent studies have implicated a new and unexpected player in heterochromatin assembly: RNA interference (RNAi; [6,7]).
The specification of a region for heterochromatin assembly results from a 'histone code' that modifies the tails of histone proteins within nucleosomes in heterochromatin differentially from those in euchromatin [8,9]. The fission yeast Schizosaccharomyces pombe provides an important model for heterochromatin assembly in eukaryotic cells, because the regions of heterochromatin have been molecularly defined (the mating-type locus, telomeres, and inverted repeats surrounding centromeres), and the evolutionary conserved proteins required for heterochromatin formation have been characterized. Several steps in the pathway of heterochromatin assembly have been identified in S. pombe. The histone deacetylase (HDAC) Clr3 deacetylates one specific residue, lysine 9 of histone H3; lysine 14 of histone H3 is deacetylated by other HDACs, potentially Clr6 and/or Hda1 [10]. Deacetylation of histone H3 lysine 9 allows this residue to be methylated by the Clr4 methyltransferase (the homolog of Drosophila Su(var)3-9), which acts together with Rik1, a protein that contains WD40 repeats, domains predicted to be involved in protein-protein interactions [10,11]. The heterochromatin protein 1 (HP1) homolog Swi6 then binds to the methylated lysine 9 of histone H3 [10] and spreads the silent heterochromatin structure (with methylated lysine 9) to surrounding sequences [12]. Spreading is ultimately blocked by boundary elements that define the extent of the heterochromatic region [11,12,13]. Interestingly, both the Clr4 methyltransferase and Swi6 contain chromodomains, protein motifs that are capable of binding RNA as well as methylated histone tails [14,15]. Once established, transcriptionally silent heterochromatin can be passed to successive generations of cells epigenetically through mitosis and meiosis [16].
Two recent studies have revealed a surprising involvement for RNAi in heterochromatin assembly [6,7]. RNAi was first identified as a mechanism of gene silencing in Caenorhabditis elegans and other animals; it also acts in the related processes of quelling in Neurospora and post-transcriptional gene silencing in plants [17]. RNAi involves a conserved group of proteins: Dicer, Argonaute, and RNA-dependent RNA polymerase (RdRP) [17,18]. Double-stranded RNA (dsRNA) amplified by RdRP is processed by the Dicer enzyme to form small interfering RNAs (siRNAs) of 21-25 nucleotides, which bind to an RNA-induced silencing complex (RISC) containing Argonaute. The siRNAs then target the mRNA homologous to the siRNA for destruction. The RISC complex is thought to promote methylation of the DNA [17,18], which further inhibits transcription in the region.
The two recent studies [6,7] implicate these same RNAi enzymes specifically in the initiation of chromatin silencing and heterochromatin assembly in S. pombe. These findings additionally indicate that maintenance of silencing requires other mechanisms, including Swi6-dependent spreading of heterochromatin. This allows the definition of a pathway of heterochromatin assembly that differentiates between factors required for the establishment and the propagation of a heterochromatin domain (Figure 1). Homologs of the genes encoding Dicer (dcr1+), Argonaute (ago1+) and RdRP (rdp1+), identified by the fission yeast genome project [19], were cloned and disrupted. The mutants are viable but defective in chromosome transmission and centromeric silencing. A mechanism underlying the role for RNAi in these processes was suggested by the accumulation of RNA transcripts in the mutants that correspond to heterochromatic centromere repeat sequences. This implies that processing of RNAs derived from these sequences is required to silence centromeric heterochromatin. Consistent with this view, the RdRP protein associates with the centromeric repeats in vivo, raising the possibility that dsRNA arises from RdRP activity on a single transcribed strand of RNA [6]. Repeated rounds of this process would lead to amplification of the RNAs, which presumably target other factors to the heterochromatin to initiate silencing.
Figure 1.
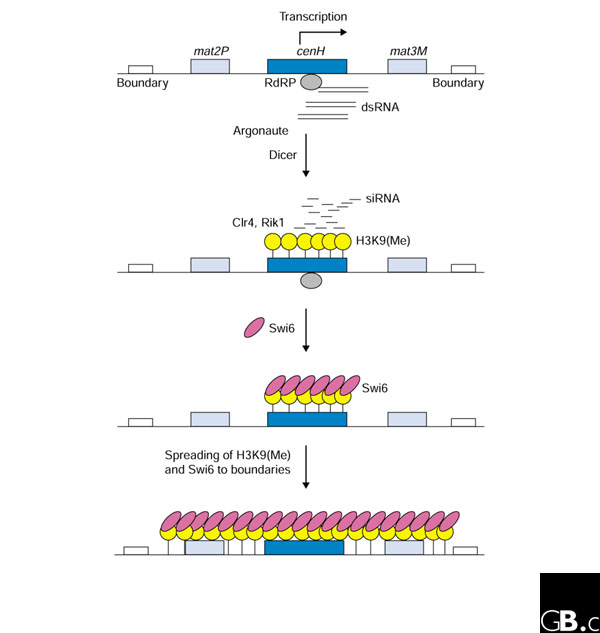
A model for heterochromatin assembly at the S. pombe mating-type locus. RNA transcription, presumably from repeated sequences within the locus, produces dsRNA that is rapidly cleaved by Dicer to form siRNAs. The siRNAs target enzymes to the chromatin so as to methylate histone H3 at lysine 9 - H3K9(Me) on the figure. Histone methylation then recruits the Swi6 protein to the heterochromatin nucleation site. The binding of Swi6 to methylated lysine 9 of histone H3 in this nucleation region then promotes spreading of the silent heterochromatin structure into adjacent sequences by further methylation of H3 at lysine 9 and its association with Swi6, throughout the mating-type locus. Broadly similar mechanisms are likely to work in the centromere. Additional factors are discussed further in the text.
Curiously, few RNA transcripts from the centromere were observed in wild-type cells [6]. But in an independent study, a search for potential Dicer cleavage products in S. pombe revealed sequences homologous to the centromeric repeats even in wild-type cells [20], suggesting that the mechanism is finely tuned and requires relatively little RNA. Because sequences homologous to telomeres and the mating-type locus were not revealed by these studies, it will be important to identify the RNA transcripts that act at these non-centric sites of heterochromatin.
To determine at what stage of the pathway RNAi components act, Volpe and colleagues [6] used chromatin immunoprecipitation to demonstrate that RNAi is required for efficient methylation of lysine 9 of histone H3. An obvious model is that siRNAs or the RNAi protein components directly target the appropriate enzymes to histones, promoting deacetylation and methylation. Indirect support for this model is provided by the observation that chromodomain proteins such as Clr4 and Swi6 can bind both methylated histone tails and RNA [14,15]. Direct evidence that the dsRNA or siRNA binds these proteins remains to be determined.
Interestingly, assembly of the silent heterochromatin at the mating-type locus requires a sequence, cenH, that is related to the centromere repeats [7,21]. A region containing this sequence is sufficient to promote silencing at an ectopic site in an RNAi-dependent fashion [7]. Because epigenetic inheritance of the silent heterochromatin at the mat (mating-type) locus is extremely robust, Hall and colleagues [7] used the cenH site at the mat locus to further analyze the role of RNAi proteins in the establishment of silent heterochromatin. The histone methylation required for the assembly of silent heterochromatin can be blocked by inhibiting the upstream histone deacetylase with Trichostatin A or by deleting the Clr4 methyltransferase itself. Both of these treatments erase the epigenetic mark for silencing [22,23]. Significantly, in the RNAi mutants, methylation and silencing at the mating-type locus could not be re-established when Trichostatin A was removed or Clr4 was restored [7]. In striking contrast, a previously established, epigenetically inherited silent chromatin state could be maintained in the absence of RNAi proteins, suggesting that RNAi is required specifically for heterochromatin assembly but is dispensable for heterochromatin maintenance. This suggests a linear pathway from RNAi activity to histone H3 lysine 9 methylation to Swi6 binding, consistent with prior observations that Swi6 association with heterochromatin requires prior methylation of histone H3 lysine 9 [10,11].
Unexpectedly, although Swi6 appears to act downstream of histone methylation, Hall and colleagues [7] found that Swi6 itself is required to maintain the methylated state and to promote spreading of heterochromatin. In a swi6 mutant, methylation of histone H3 lysine 9 within the mating-type locus is restricted to cenH rather than distributed throughout a broad region as in wild-type cells. This indicates that binding of Swi6 to methylated histone H3 lysine 9 at cenH is required for further methylation outside this region, resulting in a positive feedback mechanism that spreads silent chromatin. This observation is consistent with the ability of cenH to nucleate silencing at an ectopic chromosomal site, and defines two steps in heterochromatin assembly: the initial methylation event triggered by RNAi, and the Swi6-mediated spreading of the silent chromatin state toward the boundaries with euchromatin.
This result is consistent with a similar study by Partridge and colleagues [24]. Using silencing at an ectopic site as an assay, they define a sequence from the centromere repeats - related to cenH - that is sufficient to promote histone H3 lysine 9 methylation and silencing. Histone methylation of this site itself is independent of Swi6, similar to the situation for cenH. But methylation at the centromere sequence requires another chromodomain protein, Chp1 [24]. Thus, there appear to be different requirements for the two chromodomain proteins Chp1 and Swi6 during heterochromatin nucleation, although both proteins subsequently display a similar localization pattern across the centromere [12].
These new studies [6,7], as well as recent work from a number of other laboratories, have uncovered important details about the formation and function of heterochromatin in fission yeast. Although heterochromatin commonly contains repeated DNA arrays, this work demonstrates that RNA components also act in coordination with region-specific histone modification to drive the association of heterochromatin-specific factors and spreading of a silent chromatin structure. Notably, this work emphasizes the mechanistic similarities in diverse cellular processes. In plants and C. elegans, RNAi is known to function in gene silencing, probably by promoting DNA methylation [17]. Maintenance of male fertility in Drosophila requires the RNAi-like silencing of germline transcripts [25]. In Tetrahymena, a strong link has been uncovered between heterochromatin formation and programmed DNA elimination during cell growth [26]. During X-chromosome inactivation in mammals, methylation of histone H3 lysine 9 at a specific site targets an RNA, Xist, to spread its silent epigenetic mark along the length of the chromosome [27]. Furthermore, a pericentromeric RNA component has recently been shown to be required for histone methylation and heterochromatin formation in mouse [28]. It is also important to note, however, that in at least one eukaryote, Saccharomyces cerevisiae, there are no apparent homologs of the RNAi machinery, Swi6/HP1 proteins or methyltransferases; transcriptional silencing is achieved through a pathway that employs different factors, the 'silent information regulator' (SIR) proteins [29].
The identification of RNAi as a core component of the chromatin silencing mechanism increases the known complexity of formation of higher-order chromatin structure. Many details of this process, such as the initiation of RNA transcription from specific heterochromatin sequences and the mechanism by which methyltransferases are targeted to heterochromatin by siRNAs, remain to be determined. In addition, it is not known how histone H3 lysine 9 methylation and Swi6 cooperate to promote spreading of the silent chromatin state or whether this process involves other chromosomal factors. And there are likely to be subtle differences in the pathways that establish heterochromatin at different sites in the genome. While Chp1 is required to silence centromere sequences, it does not appear to affect silencing at the mating-type locus (see [24] and references therein). In addition, although RNAi mutants lose silencing at centromeres, the mutants do not display obvious defects in mating or in mating-type silencing, suggesting that the mat locus is better able to maintain its epigenetic imprint [6,7]. This strongly suggests that there will prove to be region-specific modifiers to the common pathway of heterochromatin assembly. Although many of the molecular components of the pathway are now defined, and linked mechanistically to other cellular processes, it is clear that there are still tantalizing puzzles to solve in the silence of heterochromatin.
Acknowledgments
Acknowledgements
Work in S.L.F.'s laboratory is supported by the NIH and the American Cancer Society. S.L.F. is a Stohlman Scholar of the Leukemia and Lymphoma Society. J.M.B. is a fellow of the Damon Runyon Cancer Research Foundation.
References
- Henikoff S. Heterochromatin function in complex genomes. Biochem Biophys Acta. 2000;1470:1–8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- Dillon N, Festenstein R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 2002;18:252–258. doi: 10.1016/S0168-9525(02)02648-3. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Avramova ZV. Heterochromatin in animals and plants. Similarities and differences. Plant Physiol. 2002;129:40–49. doi: 10.1104/pp.010981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Thon G, Bjerling P, Bunner CM, Verhein-Hansen J. Expression-state boundaries in the mating-type region of fission yeast. Genetics. 2002;161:611–622. doi: 10.1093/genetics/161.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1a. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Matzke M, Matzke AJ, Kooter JM. RNA: guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146:1221–1238. doi: 10.1093/genetics/146.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors Clr4+ and Rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12:1652–1660. doi: 10.1016/S0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/S0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]


