Abstract
A collection of Arabidopsis lines with T-DNA insertions in known sites was generated to increase the efficiency of functional genomics. A high-throughput modified thermal asymetric interlaced (TAIL)-PCR protocol was developed and used to amplify DNA fragments flanking the T-DNA left borders from ∼100,000 transformed lines. A total of 85,108 TAIL-PCR products from 52,964 T-DNA lines were sequenced and compared with the Arabidopsis genome to determine the positions of T-DNAs in each line. Predicted T-DNA insertion sites, when mapped, showed a bias against predicted coding sequences. Predicted insertion mutations in genes of interest can be identified using Arabidopsis Gene Index name searches or by BLAST (Basic Local Alignment Search Tool) search. Insertions can be confirmed by simple PCR assays on individual lines. Predicted insertions were confirmed in 257 of 340 lines tested (76%). This resource has been named SAIL (Syngenta Arabidopsis Insertion Library) and is available to the scientific community at www.tmri.org.
INTRODUCTION
The sequenced genome of Arabidopsis contains an estimated 26,000 genes (Arabidopsis Genome Initiative, 2000). Understanding the function of each of these genes is a major challenge that has been taken up by the Arabidopsis research community (Chory et al., 2000).
Sequence homology can be helpful in predicting gene function; however, there are many genes without homology to functionally characterized genes. Furthermore, although sequence homology can reveal general functions, the precise function performed by a specific gene product cannot necessarily be determined from sequence homology alone.
Reverse genetics is a strategy to determine a particular gene's function by studying the phenotypes of individuals with alterations in the gene of interest. Efficient reverse genetics is an essential component of functional genomics programs aimed at the functional characterization of large numbers of genes. Arabidopsis reverse genetics has been aided by the establishment of large insertion mutant collections (Azpiroz-Leehan and Feldmann, 1997; Krysan et al., 1999; Parinov et al., 1999; Speulman et al., 1999; Tissier et al., 1999; Parinov and Sundaresan, 2000; Sussman et al., 2000). Mutations in genes of interest can be identified by PCR screening of pooled mutant populations and confirmed by hybridization (Krysan et al., 1996). Positive pools are deconvoluted sequentially until an individual mutant line is identified (Winkler et al., 1998; Sussman et al., 2000). This screening process is time consuming and laborious and must be repeated for each gene of interest.
An alternative method is to sequence regions flanking insertion sites in individual plants from large insertion mutant populations, thereby determining large numbers of insertion sites in advance (Parinov et al., 1999; Tissier et al., 1999) (http://signal.salk.edu/tabout.html). This approach eliminates the laborious and time-consuming process of PCR-based screening and deconvolution of pools. This work describes the development, analysis, and use of a large collection of insertion site flanking sequences and corresponding seed lots from ∼100,000 T-DNA–mutagenized Arabidopsis plants.
RESULTS
Tissue and Seed Resource
Arabidopsis plants of the Columbia ecotype were transformed using Agrobacterium containing either pCSA110 or pDAP101 by the method of McElver et al. (2001). Approximately 100,000 BASTA-resistant primary transformants were grown in bar-coded trays in a 48-pot format. A small fraction of plants (2.1%) died before tissue harvest. Genomic DNA was extracted from leaf material of each remaining plant, and seeds were harvested and stored in a bar-coded, 96-well format. This produced 1045 bar-coded, 96-well plates of genomic DNA and corresponding T2 seeds. Overall, ∼5% of the plants did not produce seeds as a result of sterility, lethality, insect damage, or underwatering. This resource has been named SAIL (Syngenta Arabidopsis Insertion Library).
Amplification and Sequencing of T-DNA Borders
A modified thermal asymetric interlaced (TAIL)-PCR protocol was developed to amplify sequences flanking T-DNA insertion sites. Standard TAIL-PCR amplifies regions flanking known sequences through three successive amplification reactions using nested primers complementary to known sequences and arbitrary degenerate (AD) primers that hybridize to adjacent sequences (Liu et al., 1995; Liu and Whittier, 1995). Typically, six reactions with a T-DNA border primer and each of six AD primers are used per round of PCR to maximize the likelihood of generating a product. Therefore, 18 PCR procedures are performed to amplify products adjacent to any T-DNA insertion.
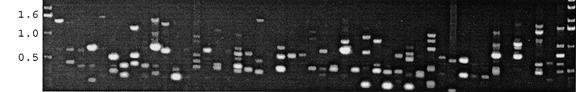
To increase the efficiency of TAIL-PCR, the six AD primers (Liu et al., 1995; Liu and Whittier, 1995) were pooled in various combinations and evaluated for the ability to generate the same products as those produced in reactions with individual AD primers (data not shown). A pool of four AD primers was chosen from various combinations of three to six AD primers because it yielded the most specific products (see Methods). A comparison of two versus three rounds of TAIL-PCR using the pool of four AD primers suggested that, in general, the third round of PCR could be omitted (data not shown). Figure 1 shows typical T-DNA left border secondary TAIL-PCR products produced with the pool of four AD primers. The reactions produced an average of 3.1 products that ranged in size from 100 to 1500 bp. Sequencing of multiple gel-purified products from single reactions demonstrated that distinct TAIL products could be produced from different T-DNAs and that multiple bands could be amplified from a single T-DNA (data not shown). Approximately 4% of the samples failed to produce detectable products in the secondary TAIL-PCR procedure. We refer to this use of two rounds of TAIL amplification with a pool of four AD primers as modified TAIL-PCR (mTAIL-PCR).
Figure 1.
Agarose Gel Analysis of Secondary mTAIL-PCR Products from 48 Lines.
Size standards are shown at left, with band sizes indicated in kb. Note that for individual samples, the lower molecular mass bands generally are present in relatively greater amounts than larger products.
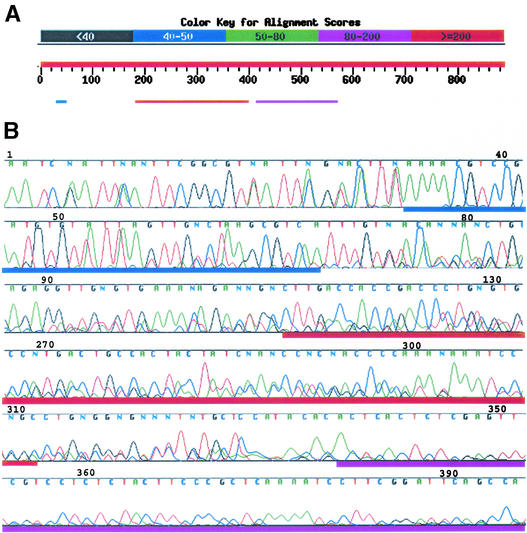
To improve the efficiency of sequencing, all products produced in an individual mTAIL-PCR procedure were sequenced together. Analysis of the resulting sequences revealed that multiple T-DNA left border mTAIL-PCR products could be sequenced together in a single reaction and that gel purification of individual PCR products before sequencing was not necessary. Basic Local Alignment Search Tool (BLAST) comparison of a typical mTAIL-PCR sequence with GenBank identified similarities of distinct regions of the mTAIL-PCR sequencing read to separate and unique regions of the Arabidopsis genome, as shown in Figure 2A. The beginning of the electropherogram of the sequence used for BLAST analysis in Figure 2A is composed of high-amplitude peaks superimposed over low-amplitude peaks, suggesting that it contains more than one mTAIL-PCR product, as shown in Figure 2B. The base-calling software used (Phred; Ewing et al., 1998) calls the base with the highest amplitude peak at each position. This creates a chimeric sequence that begins with shorter mTAIL-PCR products present at higher molar concentrations (Figure 1), producing signal peaks of higher intensity, and ends with sequences of the longer products present at lower molar concentration, producing weaker signals. The boundaries between stretches of sequences from distinct T-DNAs in the chimeric sequencing read are apparent from the results of BLAST comparisons or from inspection of the electropherograms (Figure 2).
Figure 2.
Sequence Analysis of Left Border mTAIL-PCR Products from One Insertion Line.
(A) Scheme of BLAST hits generated for sequence from the plant on plate 1154, well E08, using the NCBI BLAST server (http://www.ncbi.nlm.nih.gov/blast/Blast). The three top hits are color coded for alignment scores, as shown at top. Positions 32 to 68 align with left border T-DNA sequences (green), positions 110 to 315 align with an Arabidopsis chromosome 5 sequence, and positions 338 to 651 align with a chromosome 1 sequence.
(B) Regions of the electropherogram of the sequence in (A), with regions denoted by colored bars as in (A). Note that 5′ sequence has an overall higher signal intensity than 3′ sequence and includes lower amplitude signals from the sequence of an additional mTAIL-PCR fragment. This second sequence is obscured by the higher amplitude sequence until base 340.
Tandem or complex T-DNA insertions result in adjacent T-DNA borders (Jones et al., 1987; Jorgensen et al., 1987; Grevelding et al., 1993; De Neve et al., 1997; Krizkova and Hrouda, 1998; De Buck et al., 1999) and impede the isolation of plant sequences flanking T-DNA inserts. To assess the prevalence of mTAIL-PCR products consisting of only T-DNA sequence, left and right T-DNA borders were amplified from the genomic DNAs of 96 pCSA110 transformants using mTAIL-PCR. Analysis of these border products revealed that 25% of left and 62% of right border products contained only T-DNA sequence. To establish an insertion site database of the entire collection, only sequences flanking the T-DNA left border of each line were amplified. T-DNA left border flanking sequences were amplified from each of the ∼100,000 primary transformants using mTAIL-PCR. The mTAIL-PCR products were sequenced with a distal T-DNA left border primer, analyzed for quality, and in some cases resequenced to generate higher quality reads. These sequences are referred to collectively as SAIL sequences.
Assigning Insertion Sites in the Collection
T-DNA insertions were mapped to the Arabidopsis genome based on homology comparisons followed by additional computational analysis (G. Presting, unpublished data). The Institute for Genomic Research (TIGR) version of the Arabidopsis genome (release date, August 10, 2001; www.tigr.org) was divided into 2-kb segments and used as a query in a BLAST comparison with the collection of SAIL sequences. Based on the resulting high-scoring segment pairs at least 27 bp long, segments of SAIL sequences were mapped to the Arabidopsis genome. Where two or more Arabidopsis genomic regions aligned to the same SAIL sequence segment with the same BLAST score, all sites were recorded as possible insertion sites, leading to a slight overestimation of the total number of insertions. For each high-scoring segment pair between a SAIL sequence segment and a genomic sequence, the coordinates of the aligned bases were used to determine the most likely genomic insertion position for the T-DNAs associated with the SAIL sequence segment. Insertion positions were determined precisely when leading T-DNA left border sequences were present in the SAIL sequence and were estimated when such sequences were absent.
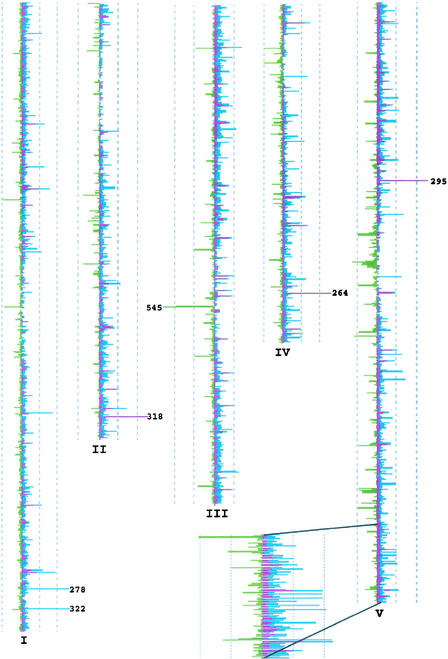
The TIGR Arabidopsis annotation (release date, August 10, 2001; www.tigr.org) then was used to characterize the distribution of insertions in regions of the genome defined as “transcribed regions,” “promoter,” and “intergenic.” Transcribed regions in this analysis were defined as TIGR annotated transcription units, promoters were defined as the lesser of 2 kb upstream or the distance to the next transcription unit, and the remainder of the genome was classified as intergenic. Figure 3 shows the distribution of the three insertion classes in a whole-genome view using 50-kb windows. In this analysis, 85,108 predicted insertions mapped to the genome: 25,710 (30%) mapped to transcribed regions, 37,156 (44%) mapped to promoters, and 22,242 (26%) mapped to intergenic regions. Of the total number, 4064 transcribed region, 6466 promoter, and 6555 intergenic associations were the result of a SAIL sequence segment having more than a single top hit with the highest BLAST score. Thus, in this analysis, the total number of insertions into transcribed, promoter, and intergenic regions was overestimated by 15, 17, and 30%, respectively.
Figure 3.
Chromosomal Distribution of Predicted Insertions.
Transcription unit insertions are shown in maroon, promoter insertions are shown in blue, and intergenic insertions are shown in green. The number of insertions per 50-kb interval is plotted as peaks off of the sides of each chromosome. The scale is indicated by dashed lines that demark intervals of 100 hits, with a maximum of 250. Numbers at peaks greater than 250 indicate value.
Applying the definitions given above, the genome is composed of 48% transcribed regions, 28% promoter, and 24% intergenic. Therefore, the data suggest that there is a bias for insertions outside of the transcribed regions. These 85,108 predicted insertions occur in 52,964 SAIL lines, or ∼53% of the collection. T-DNA–only sequences were produced in 11.4% of the lines, fewer than predicted from the sample test of 96 lines. The remaining 36% almost certainly contain useful insertions. Presumably, insertion sites were not identified in these lines because of mTAIL-PCR failure and/or low-quality sequence from mTAIL-PCR products.
To create a queryable resource to identify all possible insertions in a gene of interest, high-scoring segment pairs that identified SAIL lines containing predicted insertions in promoters and/or transcribed regions were collected. To maximize the identification of all predicted insertions that might disrupt the function of a particular gene, transcribed regions were defined as TIGR annotated transcription units plus 150 bp downstream of the predicted translation stop, and promoters were defined as 2 kb of sequences upstream from a predicted transcription unit regardless of whether the 2 kb extended into an adjacent transcription unit. This analysis was an attempt to detect all possible insertions in promoters and transcribed sequences and differed from that shown in Figure 3 in that once an insertion had been mapped to a transcribed region, it was not excluded from being mapped to a promoter of an adjacent gene. These data were assembled into a table that associates genes and their promoters, as named by their Arabidopsis Gene Index number (www.tigr.org), with mapped insertions detected in individual mutant lines. It also lists sequences of primers useful for the confirmation of insertion sites, as explained below. A sample of this SAIL table is shown in Table 1. The SAIL table includes all duplicate entries from resequenced samples (e.g., line SAIL_504C11 in At5g27100; Table 1) to give the user more confidence in choosing an insertion line. Additionally, the SAIL table lists some predicted insertions as occurring in the promoter and the transcribed region of two adjacent genes when they are separated by less than 2 kb (e.g., SAIL_439B09 in At5g27100 and At5g27110; Table 1).
Table 1.
A Sample of the SAIL Table
| Genea | Insertion Siteb | Gene Lengthc | Insertion Line | e Value | Forward Primerd | Reverse Primerd |
|---|---|---|---|---|---|---|
| At5g27030 | 5824 | 6350 | SAIL_184E07 | 0 | GGAGACACGCGGGTTCAGT | GATTCATGTACCAAACCTTACAA |
| At5g27030 | 5815 | 6350 | SAIL_184E08 | e-137 | GGAGACACGCGGGTTCAGT | CCAATTTCTCATGTTTCAGAAGT |
| At5g27030 | −119 | 6350 | SAIL_583G07 | 0 | CGAATCCGCCACTCTATCTC | CAAAATTCAGCCTCTCACAACA |
| At5g27030 | 5953 | 6350 | SAIL_790B01 | 1.00E-30 | CATCTCTGACCTGGCATGTAT | GGGTGAGCCGCCACAACAA |
| At5g27050 | −756 | 362 | SAIL_1244D06 | 0 | CTTCACCAGCAGCGTTAACC | TGGTTTGTTTGGGCGAAGAAG |
| At5g27050 | −915 | 362 | SAIL_399G12 | 0 | ACTTGACAAGCCCATGTATTTC | GAATGAAAGCAAGCCCCAAATA |
| At5g27050 | −1346 | 362 | SAIL_399H12 | 1.00E-08 | TGCAAATATGACTCCACTTGATA | GGTTGCAAGGCAACCCTAAAT |
| At5g27050 | −547 | 362 | SAIL_759C03 | 1.00E-98 | CTCGGAGAGCTTCCAAGGAA | GCATGTTACATTTGGCTTTGTAT |
| At5g27070 | −159 | 863 | SAIL_281C05 | e-153 | CTTGACACGTGCCACAATCTT | GAAGAAGACAGGGACGATTTC |
| At5g27080 | −644 | 1478 | SAIL_211B11 | 4.00E-52 | GTCGCACAGAGTCGAAAGCT | TGAAAAACCCCAACGGTTAGAT |
| At5g27080 | −577 | 1478 | SAIL_247D04 | 2.00E-14 | TCGTGGTCTTTTTTGCATTGGA | TCCTCTATGATATCAAACATACAA |
| At5g27080 | −573 | 1478 | SAIL_247D04 | 3.00E-37 | TCGTGGTCTTTTTTGCATTGGA | TCCTCTATGATATCAAACATACAA |
| At5g27080 | −875 | 1478 | SAIL_361H04 | 0 | CGTGGAGGATCTGCTTACGT | CGATATGGAAATCGATCGAATTT |
| At5g27080 | −449 | 1478 | SAIL_831F08 | e-127 | CAGTGAAAGTTGAGAGGGTTATA | GCTGAGAGGGAAGCGAATGAT |
| At5g27090 | −120 | 563 | SAIL_211B11 | 4.00E-52 | GTCGCACAGAGTCGAAAGCT | TGAAAAACCCCAACGGTTAGAT |
| At5g27090 | −187 | 563 | SAIL_247D04 | 2.00E-14 | TCGTGGTCTTTTTTGCATTGGA | TCCTCTATGATATCAAACATACAA |
| At5g27090 | −191 | 563 | SAIL_247D04 | 3.00E-37 | TCGTGGTCTTTTTTGCATTGGA | TCCTCTATGATATCAAACATACAA |
| At5g27090 | 111 | 563 | SAIL_361H04 | 0 | CGTGGAGGATCTGCTTACGT | CGATATGGAAATCGATCGAATTT |
| At5g27090 | −315 | 563 | SAIL_831F08 | e-127 | CAGTGAAAGTTGAGAGGGTTATA | GCTGAGAGGGAAGCGAATGAT |
| At5g27100 | 59 | 3151 | SAIL_1290bB10 | e-161 | CACGTATCTCAGTGCATGCAA | TGAGACGAAGTCTCCTCCTTT |
| At5g27100 | −115 | 3151 | SAIL_439B09 | 0 | GAGTAAGCCGTTCCGATATCA | AAACCGGGCTGCAGTTGGAT |
| At5g27100 | −1056 | 3151 | SAIL_504C11 | 0 | AGCATGACCACAAGCTGACAA | GTGAACTGTTCCTTGATTGATC |
| At5g27100 | −1057 | 3151 | SAIL_504C11 | 0 | AGCATGACCACAAGCTGACAA | GTGAACTGTTCCTTGATTGATC |
| At5g27100 | −541 | 3151 | SAIL_856D03 | 2.00E-13 | CATTGACCGGATTCCATGTGA | ACCTGATGGGGTTACTCTTCT |
| At5g27110 | 2221 | 2075 | SAIL_439B09 | 0 | GAGTAAGCCGTTCCGATATCA | AAACCGGGCTGCAGTTGGAT |
| At5g27110 | 1280 | 2075 | SAIL_504C11 | 0 | AGCATGACCACAAGCTGACAA | GTGAACTGTTCCTTGATTGATC |
Arabidopsis Gene Index numbers.
Positive values indicate distance in bp downstream from the first coding base, and negative values indicate distance upstream.
Length of genomic sequence from the first coding base to 150 bp downstream of the predicted stop site.
Forward and reverse primers that flank the predicted insertion that can be used for insert confirmation.
To count the number of genes predicted to be disrupted in the collection, the SAIL table was filtered to remove predicted insertions from resequencing entries and predicted insertions in promoters that also were mapped to an adjacent transcribed region. The filtered SAIL table lists 28,774 insertions within the transcribed regions of 13,878 genes that were detected in 25,909 lines. Additionally, depending on the definition of promoter length, there are between ∼16,000 and ∼36,000 insertions in promoter sequences, as listed in Table 2. In summary, there are between ∼44,000 and 65,000 predicted insertions in the transcribed regions and/or promoters of ∼18,000 to 22,000 genes, depending on the defined length of the promoter. These numbers are likely an overestimation of the identified insertions, given that 15 to 17% of the predicted promoter and transcribed region insertions result from a SAIL sequence segment having more than a single top hit with the highest BLAST score (see above). Subtracting 17% from the listed figures above gives a revised estimate of ∼15,000 to ∼18,000 genes with disruptions in the transcribed regions and/or promoters.
Table 2.
Relationship between the Defined Promoter Size, the Number of Predicted Insertions into Promoters and Coding Regions of Individual Genes, and the Number of Lines the Insertions Derive From
| Hitsb
|
Genesf
|
Lines
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| P sizea | Pc | Cd | Te | P | C | T | P | C | T |
| −2000 | 36,829 | 28,774 | 65,603 | 18,113 | 13,878 | 22,399 | 34,738 | 25,909 | 50,576 |
| −1000 | 26,516 | 28,774 | 55,290 | 13,809 | 13,878 | 20,128 | 23,858 | 25,909 | 45,394 |
| −500 | 16,168 | 28,774 | 44,942 | 9,734 | 13,878 | 18,154 | 15,566 | 25,909 | 39,452 |
Defined size of promoter.
BLAST associations between SAIL sequences and the specified subset of the genome.
Promoters as defined in the text.
Coding sequences as defined in the text.
Total number of unique entries from combined promoter and coding sets.
Number of genes hit in the specified subset of the genome.
Inspection of the SAIL table suggests that there are genes in the Arabidopsis genome in which insertions occur at high frequencies, as listed in Table 3. Twenty-two loci appear to have 20 or more insertions each, suggesting that there are “hot spots” for T-DNA integration. Gene-specific primers were designed to test for the presence of a subset of these insertions using PCR amplification with a T-DNA left border primer. Of the predicted insertion hot spots tested, none appeared to be a true hot spot (Table 3). A subset of insertions was confirmed in two members of a highly conserved multigene family. Because of the high degree of sequence similarity among these genes, the SAIL table analysis could not distinguish them, so an insertion in any one of them was mapped to all of them. None of the genes listed in Table 3 is predicted to contain multiple insertions at the same genomic position. Together, these data suggest that insertion hot spots in coding regions or their associated upstream sequences do not exist.
Table 3.
Genes Predicted to Contain >20 Insertions
| Arabidopsis Gene Index No. |
No. of Hits |
Insertion Sitesa | Gene Lengthb |
No. Confirmed Using PCR of No. Tested |
|---|---|---|---|---|
| At1g02810 | 24 | −1945 to 3818 | 4024 | |
| At1g11680 | 21 | −1358 to −419 | 1808 | |
| At1g24793c | 27 | −1367 to 4766 | 4650 | 6 of 21 |
| At1g24880c | 27 | −502 to 4765 | 4649 | |
| At1g25054c | 26 | −502 to 4766 | 4650 | 3 of 20 |
| At1g25141c | 27 | −502 to 4882 | 4855 | |
| At1g25190c | 24 | −502 to 3493 | 4650 | |
| At1g30950 | 47 | 250 to 282 | 1328 | 0 of 46 |
| At1g51830 | 41 | −830 to 839 | 2985 | |
| At1g62390 | 15 | −233 to −221 | 2529 | |
| At1g69150 | 23 | −1914 to −535 | 1553 | |
| At2g04300 | 20 | −1938 to 1868 | 2367 | |
| At2g05160 | 38 | −1465 to 1888 | 1920 | |
| At3g18680 | 21 | −108 to 1467 | 1987 | |
| At3g33056 | 45 | −1478 to 3781 | 3745 | |
| At3g45780 | 35 | −1395 to 5195 | 5403 | |
| At4g01020 | 20 | 3303 to 7697 | 9023 | |
| At5g11030 | 36 | 38 to 2740 | 4157 | 1 of 26 |
| At5g17330 | 21 | −248 to 3788 | 3698 | |
| At5g22740 | 26 | −551 to 1946 | 4487 | |
| At5g42600 | 26 | −264 to 2495 | 4409 | 0 of 22 |
| At5g44280 | 29 | −408 to 2920 | 2999 |
Predicted within −2 kb upstream of the gene to 150 bp downstream of the translation stop.
Not including the −2-kb region.
Members of a highly conserved multigene family among which the analysis could not distinguish insertions.
To determine the frequency of left border read-through during excision of the T-DNA from the binary vector within Agrobacterium and the resultant transfer of vector backbone into the plant genome (Martineau et al., 1994; Ramanathan and Veluthambi, 1995; van der Graaff et al., 1996), binary vector backbone sequences were compared with SAIL sequences using BLAST. Approximately 4% of the SAIL sequences showed significant homology with vector backbone sequences. This finding agrees with previous reports that agrobacteria regularly transfer binary vector backbone sequences adjacent to the T-DNA left border as a result of improper processing of the T-DNA within the bacterial cell.
Most of the Insertion Sites Predicted in the SAIL Collection Can Be Confirmed Experimentally
To test the utility of the SAIL collection and the SAIL table, lines predicted to have insertions in genes implicated in disease resistance were studied. For the most part, these lines are predicted to contain insertions in genes whose expression increases in response to infection by the bacterial pathogen Pseudomonas syringae. They include transcription factors, protein kinases, enzymes of primary and secondary metabolism, carbohydrate-modifying enzymes, and unknown proteins. They vary widely in gene size and basal expression level. Among 463 genes of interest, the SAIL table predicts that insertions exist in coding sequences of 282 (61%). For 83 genes, the SAIL table predicts more than one allele. In some cases, two lines from adjacent positions on a 96-well plate are predicted to have insertions in identical positions. This is almost certainly attributable to cross-contamination during DNA purification or mTAIL-PCR. These were not counted as cases of two alleles of one gene.
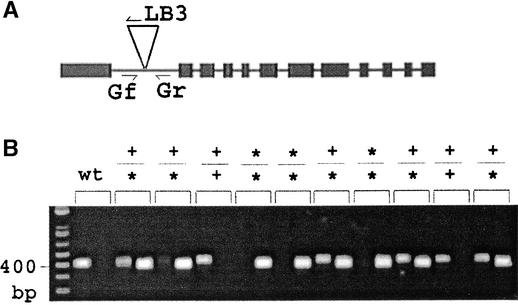
Segregating populations from 340 SAIL lines were tested to determine if the insertions were in the positions predicted in the SAIL table and to isolate plant lines homozygous for T-DNA insertion mutations. As described in Methods, primers flanking each predicted insertion in the SAIL table were designed and listed in the SAIL table. Two PCR procedures were used to test each predicted insertion site. One used the forward and reverse primers to amplify a product from chromosomes without the insertion. The other used the reverse primer and the T-DNA left border primer to amplify the product from a chromosome carrying the insertion. For each SAIL line studied, DNA from each of 20 to 30 plants was used for each of these two reactions. From the results, it was possible to determine if plants were homozygous for the insertion mutation, hemizygous, or lacked the insertion completely. Figure 4 shows an example of the use of this method to confirm an insertion in At4g39030, which encodes the disease resistance gene EDS5 (Nawrath et al., 2002). Among the 340 lines tested, 257 were found to carry an insertion in the predicted position. Thus, the success rate of confirming predicted inserts was ∼76%.
Figure 4.
Insert Confirmation Using PCR with T-DNA Left Border and Gene-Specific Forward and Reverse Primers.
The gene-specific primers were 5′-TTTTCACGATTCTTCTAGAC-3′ (Gf; forward) and 5′-AATAACCTGTTTGGCAAGAG-3′ (Gr; reverse). LB3, left border.
(A) Scheme of At4g39030 (EDS5/SID1) and the predicted insertion in line 1255_E09, which is predicted to lie at position 841 in the first intron.
(B) PCR analysis of the predicted insertion in wild-type (wt) Columbia (negative control) and 10 progeny plants of line 1255_E09. Products of PCR using the primer sets Gf/Gr and Gr/LB3 were run in adjacent lanes for each sample. Plants 1, 3, 6, 8, and 10 are hemizygous for the insertion, whereas plants 4, 5, and 7 are homozygous for the insertion.
DISCUSSION
To obtain 99% confidence of finding a T-DNA insertion in any Arabidopsis gene, ∼280,000 T-DNA inserts would be required (Krysan et al., 1999). To process samples on this scale, an efficient system is required to identify T-DNA insertion sites. To increase the efficiency of characterizing large insertion collections for functional genomics, several important advances were made. Primary transformants, rather than subsequent generations, were characterized directly, thereby saving space and time in plant growth. Plants were grown in a format that facilitated sample collection onto 96-well plates, enabling efficient downstream processing. TAIL-PCR was modified for high throughput and used to amplify sequences flanking T-DNA left borders from each line in only 2 reactions, instead of the typical 12 to 18 reactions. mTAIL-PCR products from each plant were sequenced together, eliminating gel electrophoresis and purification of individual PCR products. Finally, insertions in individual lines were associated with specific genes in a database, allowing users to find insertions in genes of interest by simply looking at a table.
Steps taken to increase efficiency likely decreased the number of sequence tags identified for the following reasons: (1) only sequences flanking the T-DNA left borders were amplified, and not all T-DNA insertions include an intact left border (Zambryski et al., 1982; Jorgensen et al., 1987; Deroles and Gardner, 1988; Gheysen et al., 1991; Mayerhofer et al., 1991; Grevelding et al., 1993); (2) TAIL-PCR will not amplify a T-DNA left border flanking sequence if an AD primer binding site is not available in the appropriate orientation; and (3) sequencing of multiple templates in a single reaction leads to lower quality sequence that reduces the likelihood of identifying a quality BLAST match to the genome.
Only T-DNA left border flanking sequences were chosen for rescue because initial tests showed a lower frequency of T-DNA–only sequences on the left borders compared with the right borders. Previous reports on the characterization of T-DNA integration patterns in various species have found that between 30 and 90% of T-DNAs are arranged in direct or inverted repeats (Jones et al., 1987; Jorgensen et al., 1987; Grevelding et al., 1993; De Neve et al., 1997; Krizkova and Hrouda, 1998; De Buck et al., 1999). Although some reports have found more repeats around the T-DNA right border (Grevelding et al., 1993; Cluster et al., 1996; De Neve et al., 1997), others did not. Analysis of T-DNA borders in lethal mutants from a separate study using the same vectors and transformation protocol (McElver et al., 2001) found evidence for 49 right border inverted repeats in 115 plants (data not shown). Although some researchers have suggested that the tissue transformed, Agrobacterium strain and titer, and/or binary vector system may influence the proportion of tandem and inverted repeat T-DNA integrations (Grevelding et al., 1993; De Neve et al., 1997), it is unclear why there is a higher proportion of repeat structures at the right borders than the left borders in our collection. Analysis of SAIL sequences showed that 11.4% were T-DNA only. This is likely an underestimate of the total number of left border repeats, because inverted repeat structures such as those found at T-DNA borders have proven difficult to amplify by PCR (DeBuck et al., 1999; data not shown).
Approximately 50,000 lines, or 50% of the collection, contain insertions likely to affect gene function and therefore are useful for functional genomics. In addition, there are many insertions in intergenic regions that also may prove useful for elucidating the biological functions of sequences currently thought to be nonfunctional. SAIL sequences from ∼47,000 lines could not be associated with Arabidopsis genomic sequences. This number includes lines that died before tissue collection (2.1% of the collection), mTAIL-PCR failures, T-DNA–only mTAIL-PCR products (11.4% of the collection) resulting from concatenated T-DNAs inserted at a single locus, and low-quality sequences. Although 25% of the predicted insertions could not be verified experimentally, the number of insertions predicted is likely an underestimate of the total mutation coverage in the collection because T-DNAs lacking intact left borders were not rescued and mTAIL-PCR does not amplify every left border. Further efforts to characterize T-DNA right border flanking sequences and to reamplify missing left border flanking regions will identify more mutations within the collection.
Twenty-two genes were predicted to have disproportionately high numbers of insertions, although only a small fraction of predicted insertions in these genes could be confirmed. The repeated amplification of these sequences from independent samples using mTAIL-PCR may represent common artifacts that occur with the mTAIL-PCR protocol. The explanation that seems most likely is that recombination events occurred during some of the TAIL-PCR procedures, resulting in chimeric TAIL products. Taq polymerase is known to switch templates after reaching an end or a damaged region (Paabo et al., 1990). The presence of oligonucleotides hybridized to the template can result in template switching onto the oligonucleotide (Patel et al., 1996). Because our modified TAIL-PCR contained oligonucleotide primers of many different sequences, the frequency of template switching may have been quite high.
Although most of the insertions tested experimentally could be confirmed, 24% of the predicted T-DNA insertions could not. One possible source of these false-positive results could be attributable to cases in which a single SAIL sequence segment had more than a single top hit with the highest BLAST score. In these cases, the insertions predicted but not confirmed could be in a closely related gene. Another likely explanation is template switching during mTAIL-PCR, as described above. Additionally, sample contamination during DNA extraction and mTAIL-PCR, as well as sample tracking errors, could have contributed to the false-positive rate. Finally, because tissue from primary transformants was used for TAIL-PCR and sequencing, it is possible that agrobacteria could have persisted in the tissues of the primary transformants and created somatic insertion events. There are examples in the collection of genes for which more than one allele was identified but only one could be confirmed. Thus, it does not appear that all unconfirmable mutations are gametophytic lethal.
The SAIL collection contains numerous insertions in the transcribed regions and promoters of ∼15,000 to ∼18,000 genes. The ease of screening, and the propagation of the collection as individual lines, will aid Arabidopsis functional genomics efforts and by extension facilitate the assignment of functions to genes from other plant species, including crops. The lines are available to the scientific community. Conditions for seed transfer, the SAIL table, and an interface for performing BLAST queries can be found at www.tmri.org.
METHODS
Binary Vectors, Transformation, and Plant Growth
The T-DNA vectors, Agrobacterium tumefaciens strain (GV3101), transformation methods, selection of transformants, and plant growth were as described by McElver et al. (2001). Two different binary vectors were used to generate a population of ∼100,000 transgenic Arabidopsis thaliana plants, which were collected onto 1045 96-well plates. Lines on plates 1 to 456, 1052 to 1057, 1142 to 1205, and 1206 (rows A to D) are homozygous for a qrt mutation (Preuss et al., 1994; McElver et al., 2001) and were transformed using pCSA110. The T-DNA in pCSA110 is 7541 kb in length and contains from left to right border a BASTA resistance cassette, pBluescript SKII+, and a Lat52 promoter–β-glucuronidase fusion. Lines on plates 500 to 918 and 1206 (rows E to H) to 1307 were transformed using pDAP101. The T-DNA in pDAP101 is 4763 kb and contains from left to right border a BASTA resistance cassette and pBluescript SKII+. Both T-DNAs have the same left border but different right borders. Vector maps can be found at www.tmri.org. Primary transformants were grown in greenhouses in Gilroy, CA, and Research Triangle Park, NC. On the Torrey Mesa Research Institute World Wide Web site and in conference presentations, this resource was referred to previously as the GARLIC collection, for Gilroy Arabidopsis Reverse Lethal Insertion Collection.
DNA Extraction
Approximately 30 mg of leaf tissue was collected from each independent BASTA-resistant plant in two 48-pot flats, transferred to 96-well racks of tubes, and lyophilized overnight. Tissue was disrupted by adding a stainless-steel bead to each tube of the 96-well rack and shaking the rack for 30 s in a clamp attached to a stationary reciprocating saw. DNA was purified using Plant DNeasy 96 DNA Extraction kits from Qiagen (Hilden, Germany).
Modified TAIL-PCR Procedure
The thermal asymetric interlaced (TAIL)-PCR protocol (Liu et al., 1995) was modified for high throughput as follows. A T-DNA border–specific primer and a pool of four arbitrary degenerate (AD) primers (AD1, 5′-NGTCGASWGANAWGAA-3′ [Liu et al., 1995]; AD2, 5′-TGW-GNAGSANCASAGA-3′ [Liu and Whittier, 1995]; AD3, 5′-AGW-GNAGWANCAWAGG-3′ [Liu and Whittier, 1995]; and AD6, 5′-WGTGNAGWANCANAGA-3′ [Liu et al., 1995]) were used per round of TAIL-PCR cycling. The T-DNA border primers used were as follows: for the left border, LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′), LB2 (5′-GCTTCCTATTATATCTTCCCAAATTACCAA-TACA-3′), or LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGAT-ACAC-3′); for the right border in pCSA110, qRB1 (5′-CAAACTAGG-ATAAATTATCGCGCGCGGTGTC-3′), qRB2 (5′-GGTGTCATCTATGTTACTAGATCGGGAATT-GA-3′), or qRB3 (5′-CGCCATGGCATA-TGCTAGCATGCATAATTC-3′); and for the right border of pDAP101, RB1 (5′-ATTAGGCACCCCAGGCTTTACACTTTATG-3′), RB2 (5′-GTATGTTGTGTGGAATTGTGAGCGGATAAC-3′), or RB3 (5′-TAACAATTTCACACAGGAAACAGCTATAC-3′). Two of rounds of mTAIL-PCR cycling were performed. The final concentration of the pooled primers AD1, AD2, AD3, and AD6 were proportional to their level of degeneracy (primary/secondary TAIL primer concentrations were as follows: AD1, 3.0/1.8 μM; AD2, 3.0/1.8 μM; AD3, 3.0/1.8 μM; and AD6, 4.0/1.8 μM). T-DNA border primers were at a final concentration of 0.2 μM.
First and second rounds of mTAIL-PCR cycling were performed on 384-well plates in Applied Biosystems 9700 thermocyclers. Cycling parameters for the first round were as follows: (1) 94°C for 2 min and 95°C for 1 min; (2) 5 cycles of 94°C for 30 s, 62°C for 1 min, and 72°C for 2.5 min; (3) 2 cycles of 94°C for 30 s, 25°C for 3 min (50% ramp), and 72°C for 2.5 min (32% ramp); (4) 15 cycles of 94°C for 10 s, 68°C for 1 min, 72°C for 2.5 min, 94°C for 10 s, 68°C for 1 min, 72°C for 2.5 min, 94°C for 10 s, 44°C for 1 min, and 72°C for 2.5 min; and (5) 72°C for 7 min. Cycling parameters for the second round were as follows: (1) 94°C for 3 min; (2) 5 cycles of 94°C for 10 s, 64°C for 1 min, and 72°C for 2.5 min; (3) 15 cycles of 94°C for 10 s, 64°C for 1 min, 72°C for 2.5 min, 94°C for 10 s, 64°C for 1 min, 72°C for 2.5 min, 94°C for 10 s, 44°C for 1 min, and 72°C for 2.5 min; (4) 5 cycles of 94°C for 10 s, 44°C for 1 min, and 72°C for 3 min; and (5) 72°C for 7 min. Platinum Taq polymerase (Life Technologies, Rockville, MD) was used for all amplifications. The primary mTAIL reaction contained 5 ng of genomic DNA.
Sequencing
Before sequencing, secondary TAIL-PCR products were purified by treatment with exonuclease I (2.5 units; Amersham) and shrimp alkaline phosphatase (0.5 units; Amersham) for 20 min at 37°C, followed by 15 min at 80°C. Sequencing reactions were performed in a 384-well format using the T-DNA LB3 primer and one-eighth of the suggested amount of BigDye terminators and run on Applied Biosystems 3700 sequencers. Sequencing reactions were passed through a Sephadex G-50 matrix to remove salts and unincorporated dye terminators. In total, 116,251 sequencing reactions were performed.
Design of Primers for Testing SAIL Insertion Sites
Two primers were designed to anneal 200 to 300 bp flanking each predicted insertion site. Each 100-bp interval was scanned for possible primer sites, beginning with an 18-nucleotide window. The criteria considered were annealing temperature of 66 to 70°C using the formula temperature = 22 + 1.46 (number of A or T bases + 2× the number of G or C bases), avoidance of direct repeats, low GC content in the six bases closest to the 3′ end, and avoidance of hairpin structure. If fewer than 10 primers satisfied the criteria, then the window was expanded to 19 and the scan was repeated. The window was expanded 1 nucleotide at a time up to 24 nucleotides, until 10 satisfactory primers were obtained. To find compatible primer pairs for each insertion site, all combinations of the 10 satisfactory primers were evaluated for avoidance of primer-dimers and similar annealing temperature. This evaluation was repeated for the primers to be used with the T-DNA left border primer using the left border primer and the set of 10 reverse primers. The sequence of the left border primer used is 5′-TTCATAACCAATCTCGATACAC-3′. Based on these analyses, the best primers were chosen and added to the SAIL table. Occasionally, no primers met the minimal criteria. In these cases, the SAIL table entry is “no good primer” and it is necessary to design primers by another method. Because the existence of highly homologous sequences in the genome was not considered in the primer design, some primer pairs also may amplify fragments from highly homologous sequences. This could result in an apparent hemizygote pattern from a true homozygote in the analysis described for Figure 4.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank University of California at Berkeley, Scripps Research Institute, and Salk Institute professors, post-docs, graduate students, and technicians, and numerous Syngenta scientists, for their generous assistance with leaf sample and seed collection. This work could not have been accomplished without their help.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.004630.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan, R., and Feldmann, K.A. (1997). T-DNA insertion mutagenesis in Arabidopsis: Going back and forth. Trends Genet. 13, 152–159. [DOI] [PubMed] [Google Scholar]
- Chory, J., et al. (2000). Functional genomics and the virtual plant: A blueprint for understanding how plants are built and how to improve them. Plant Physiol. 123, 423–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluster, P., O'Dell, M., Metzlaff, M., and Flavell, R. (1996). Details of T-DNA structural organization from a transgenic Petunia population exhibiting co-suppression. Plant Mol. Biol. 32, 1197–1203. [DOI] [PubMed] [Google Scholar]
- De Buck, S., Jacobs, A., Van Montagu, M., and Depicker, A. (1999). The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 20, 295–304. [DOI] [PubMed] [Google Scholar]
- De Neve, M., De Buck, S., Jacobs, A., Van Montagu, M., and Depicker, A. (1997). T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J. 11, 15–29. [DOI] [PubMed] [Google Scholar]
- Deroles, S., and Gardner, R.C. (1988). Analysis of the T-DNA structure in a large number of transgenic petunias generated by Agrobacterium-mediated transformation. Plant Mol. Biol. 11, 365–377. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M., and Green, P. (1998). Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Gheysen, G., Villarroel, R., and Van Montagu, M. (1991). Illegitimate recombination in plants: A model for T-DNA integration. Genes Dev. 5, 287–297. [DOI] [PubMed] [Google Scholar]
- Grevelding, C., Fantes, V., Kemper, E., Schell, J., and Masterson, R. (1993). Single-copy T-DNA insertions in Arabidopsis are the predominant form of integration in root-derived transgenics, whereas multiple insertions are found in leaf discs. Plant Mol. Biol. 23, 847–860. [DOI] [PubMed] [Google Scholar]
- Jones, J., Gilbert, D., Grady, K., and Jorgensen, R. (1987). T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens C58 derivatives. Mol. Gen. Genet. 207, 478–485. [Google Scholar]
- Jorgensen, R., Snyder, C., and Jones, J. (1987). T-DNA is organized predominantly in inverted repeat structures in plants transformed with Agrobacterium tumefaciens C58 derivatives. Mol. Gen. Genet. 207, 471–477. [Google Scholar]
- Krizkova, L., and Hrouda, M. (1998). Direct repeats of T-DNA integrated in tobacco chromosome: Characterization of junction regions. Plant J. 16, 673–680. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., Tax, F., and Sussman, M.R. (1996). Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci. USA 93, 8145–8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., and Whittier, R.F. (1995). Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Martineau, B., Voelker, T., and Sanders, R. (1994). On defining T-DNA. Plant Cell 6, 1032–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Bakkeren, G., Crameri, A., Angelis, K., Redei, G., Schell, J., Hohn, B., and Koncz, C. (1991). T-DNA integration: A mode of illegitimate recombination in plants. EMBO J. 10, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.-P. (2002). EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paabo, S., Irwin, D.M., and Wilson, A.C. (1990). DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem. 265, 4718–4721. [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W., Kumaran, M., and Sundaresan, V. (1999). Analysis of flanking sequences from Dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11, 2263–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov, S., and Sundaresan, V. (2000). Functional genomics in Arabidopsis: Large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotechnol. 11, 157–161. [DOI] [PubMed] [Google Scholar]
- Patel, R., Lin, C., Laney, M., Kurn, N., Rose, S., and Ullman, E.F. (1996). Formation of chimeric DNA primer extension products by template switching onto an annealed downstream oligonucleotide. Proc. Natl. Acad. Sci. USA 93, 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., Rhee, S.Y., and Davis, R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264, 1458–1460. [DOI] [PubMed] [Google Scholar]
- Ramanathan, V., and Veluthambi, K. (1995). Transfer of non-T-DNA portions of the Agrobacterium tumefaciens Ti plasmid pTiA6 from the left terminus of the Tl-DNA. Plant Mol. Biol. 28, 1149–1154. [DOI] [PubMed] [Google Scholar]
- Speulman, E., Metz, P., van Arkel, G., te Lintel Hekkert, B., Steikema, W.J., and Pereira, A. (1999). A two component Enhancer-Inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell 11, 1853–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124, 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. (1999). Multiple independent defective Suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff, E., Dulk-Ras, A., and Hooykaas, P. (1996). Deviating T-DNA transfer from Agrobacterium tumefaciens to plants. Plant Mol. Biol. 31, 677–681. [DOI] [PubMed] [Google Scholar]
- Winkler, R., Frank, M., Galbraith, D., Feyereisen, R., and Feldmann, K. (1998). Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Plant Physiol. 118, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambryski, P., Depicker, A., Kruger, K., and Goodman, H. (1982). Tumor induction of Agrobacterium tumefaciens: Analysis of the boundaries of T-DNA. J. Mol. Appl. Genet. 1, 361–370. [PubMed] [Google Scholar]