Abstract
The geminivirus Tomato golden mosaic virus (TGMV) replicates in differentiated plant cells using host DNA synthesis machinery. We used 5-bromo-2-deoxyuridine (BrdU) incorporation to examine DNA synthesis directly in infected Nicotiana benthamiana plants to determine if viral reprogramming of host replication controls had an impact on host DNA replication. Immunoblot analysis revealed that up to 17-fold more BrdU was incorporated into chromosomal DNA of TGMV-infected versus mock-infected, similarly treated healthy leaves. Colocalization studies of viral DNA and BrdU demonstrated that BrdU incorporation was specific to infected cells and was associated with both host and viral DNA. TGMV and host DNA synthesis were inhibited differentially by aphidicolin but were equally sensitive to hydroxyurea. Short BrdU labeling times resulted in some infected cells showing punctate foci associated with host DNA. Longer periods showed BrdU label uniformly throughout host DNA, some of which showed condensed chromatin, only in infected nuclei. By contrast, BrdU associated with viral DNA was centralized and showed uniform, compartmentalized labeling. Our results demonstrate that chromosomal DNA is replicated in TGMV-infected cells.
INTRODUCTION
Geminiviruses are small, single-stranded DNA viruses that replicate through double-stranded DNA intermediates in the nuclei of their plant hosts (reviewed by Hanley-Bowdoin et al., 1999; Gutierrez, 2000). Tomato golden mosaic virus (TGMV), a member of the begomovirus genus, infects a wide variety of differentiated cell types (Rushing et al., 1987; Nagar et al., 1995; Bass et al., 2000). Other geminiviruses, including Beet curly top virus, Maize streak virus, and Bean dwarf mosaic virus, also localize to quiescent, differentiated cells (Esau, 1977; Lucy et al., 1996; Sudarshana et al., 1998). By contrast, Abutilon mosaic virus and Squash leaf curl virus are con-fined to vascular tissue (Horns and Jeske, 1991; Sanderfoot and Lazarowitz, 1996) and may be restricted to provascular and cambial cells that can support DNA replication (Hemerly et al., 1993). Independent of their tissue specificity, all geminiviruses rely on host DNA synthesis machinery to replicate their genomes.
Only one viral protein, AL1 (also known as AC1 or Rep), is required for TGMV DNA replication. AL1 is a multifunctional protein that binds the origin of replication, nicks DNA to initiate rolling-circle replication, forms oligomers (Orozco et al., 1997), and localizes to infected nuclei (Nagar et al., 1995). AL3 (also known as AC3 or Ren) interacts with AL1 (Settlage et al., 1996), increases viral DNA accumulation (Sunter et al., 1990), and, like AL1, localizes to infected nuclei (Nagar et al., 1995). Both AL1 and AL3 also interact with pRBR, a plant homolog of the retinoblastoma tumor suppressor protein (pRb) (Ach et al., 1997; Settlage et al., 2001).
TGMV-infected cells also accumulate proliferating cell nuclear antigen (PCNA), the processivity factor of DNA polymerase δ (Nagar et al., 1995). Because PCNA functions in both replicative and repair DNA synthesis (Bravo et al., 1987; Kelman, 1997), its induction does not directly reveal the nature of the interaction between host and virus that leads to DNA synthesis. The involvement of DNA repair machinery would be consistent with the fact that most TGMV-infected cells in a plant are differentiated and no longer contain detectable levels of replication enzymes (Rushing et al., 1987; Coello et al., 1992; Nagar et al., 1995). However, the involvement of host DNA synthesis enzymes associated with cell cycle activity is supported by the finding that many infected nuclei contain condensed chromatin (Bass et al., 2000).
Mammalian DNA tumor viruses establish permissive conditions for DNA replication by inactivating tumor suppressor proteins, such as pRb and p53, and upregulating DNA replication-associated proteins, such as PCNA (Nevins, 1992; Jansen-Durr, 1996). TGMV may use a similar strategy because the TGMV AL1 protein, which interacts with pRBR (Ach et al., 1997), also causes PCNA accumulation in differentiated plant cells (Nagar et al., 1995; Egelkrout et al., 2001). TGMV AL1 mutants impaired for pRBR binding are confined to vascular tissue (Kong et al., 2000), suggesting that efficient pRBR binding is required for viral DNA replication in mesophyll and epidermal cells. The altered tissue specificity of the mutant virus demonstrates that differentiated plant cells can vary in their ability to reenter the cell cycle and provides a rationale for examining viral DNA replication in planta.
Some mammalian DNA tumor viruses induce both host and viral DNA replication in quiescent cells (Cheng et al., 1995; Morin et al., 1996). Because both geminiviruses and mammalian DNA viruses interact with conserved cell cycle proteins, it is possible that plant chromosomal DNA also is replicated in TGMV-infected cells. Active DNA synthesis in cells of pea and tobacco root meristems has been visualized using antibodies to the thymidine analog 5-bromo-2-deoxyuridine (BrdU) (Levi et al., 1987; Suzuki et al., 1992). We used a combination of immunolocalization and immunoblotting to separate host and viral DNA synthesis in plants. This combination demonstrated that host DNA replication is upregulated significantly in differentiated TGMV-infected cells and that host chromatin could be labeled uniformly with anti-BrdU antibodies.
RESULTS
BrdU Incorporation into TGMV and Chromosomal DNA
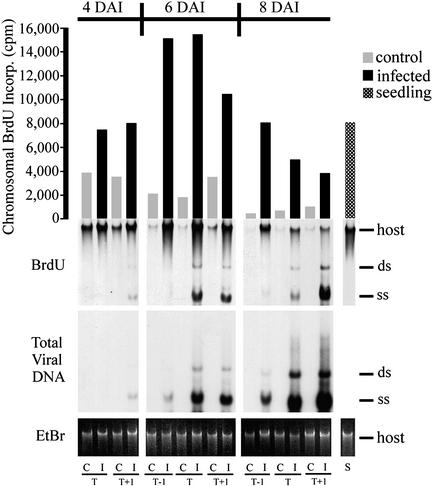
The findings that TGMV infection induces PCNA accumulation in terminally differentiated cells (Nagar et al., 1995) and that geminivirus proteins interact with cell cycle regulators (Hanley-Bowdoin et al., 1999; Gutierrez, 2000) suggested that TGMV alters cell cycle regulation to support DNA synthesis. To determine if host DNA also is replicated during TGMV infection, we examined the incorporation of BrdU into host and viral DNA at different time points during TGMV infection of Nicotiana benthamiana. Fully expanded leaves matched by size and position on the plants were excised from paired mock-infected control and TGMV-infected plants at different days after infection (DAI) and placed in a solution of 100 μM BrdU for 24 h. Leaves were designated by position as primary target (T), which showed the most bombardment damage, the leaf immediately below T (T−1), and the leaf immediately above T (T+1). Total DNA then was isolated from individual leaves and analyzed by immunoblotting with anti-BrdU antibodies (Figure 1), which detect only nascent DNA. These experiments were performed a total of six times on different plants and included a range of time points. The results presented here are representative of the patterns we observed in each experiment.
Figure 1.
BrdU Is Incorporated into Both Chromosomal and Viral DNA of Mature Leaves.
Pairs of TGMV-infected (I) and control (C) leaves were labeled with 100 μM BrdU for 24 h. Total DNA was separated on agarose, blotted, and incubated with anti-BrdU antibodies followed by chemiluminescent detection. Photon counting was used to quantify the BrdU signal (in cpm) for chromosomal DNA (bar graph). The positions of double-stranded (ds) and single-stranded (ss) TGMV and host chromosomal DNA are marked. Total viral DNA was detected with a 32P-labeled TGMV A DNA probe. EtBr, ethidium bromide; S, DNA from healthy seedlings treated with 100 μM BrdU for 24 h; T, DNA from the leaf intercepting most of the microprojectiles (target); T−1, the leaf immediately below T; T+1, the leaf immediately above T.
High-level viral DNA accumulation was not detected until 6 DAI (Figure 1). At 4 DAI, the T leaf was asymptomatic and contained no detectable viral DNA, whereas the T+1 leaf had only a few isolated chlorotic spots (1 to 2 mm) and contained low amounts of viral DNA (Figure 1). The T and T+1 leaves at 6 DAI showed moderate symptoms, including chlorotic areas and leaf curling, and contained higher levels of TGMV DNA. At 8 DAI, the T and T+1 leaves showed severe symptoms of extensive yellow mottling and puckering and contained the highest levels of total viral DNA. Only a few isolated yellow areas were observed on T−1 leaves at 6 and 8 DAI. At all three time points, total viral DNA and BrdU incorporation into viral DNA were similar within each leaf, with the exception of T−1 leaves. Together, these results demonstrated that there is a strong correlation between active viral DNA accumulation and symptom development in TGMV-infected plants and established that high-level TGMV replication coincides with symptom formation in N. benthamiana.
We also detected BrdU incorporation into chromosomal DNA. In every case, more BrdU (up to 17-fold) was incorporated into the host DNA of TGMV-infected leaves than in identically treated, mock-inoculated control leaves (Figure 1). TGMV-infected tissues incorporated similarly high levels of BrdU as seedlings, based on equivalent amounts of total DNA (Figure 1). Chromosomal BrdU incorporation decreased over time in control leaves (cf. 4, 6, and 8 DAI). However, in TGMV-infected leaves, BrdU incorporation increased over time and peaked at 6 DAI. Because lower levels of BrdU incorporation also were seen in excised leaves of nonbombarded plants (data not shown), this finding may reflect an initial response of the plant to the presence of BrdU. The large differences in BrdU incorporation between chromosomal DNA of infected and mock-inoculated plants suggest that chromosomal DNA is replicated in infected plants.
Aphidicolin Inhibits Viral DNA Synthesis
BrdU incorporation into chromosomal and TGMV DNA suggested that similar enzymes were used. To test this idea, we examined BrdU incorporation in the presence of two well-characterized S-phase inhibitors, aphidicolin and hydroxyurea. Aphidicolin inhibits mammalian DNA polymerase α and, to a lesser extent, DNA polymerase δ and blocks the elongation phase of DNA replication (Wang, 1991; Wright et al., 1994). Hydroxyurea inhibits ribonucleotide reductase, a key enzyme for the synthesis of deoxyribonucleotides. Hydroxyurea prevents DNA synthesis by depleting nucleotide pools (Young and Hodas, 1964). Because adequate nucleotide pools must be available before DNA elongation can begin, hydroxyurea should block S-phase at an earlier time point than aphidicolin.
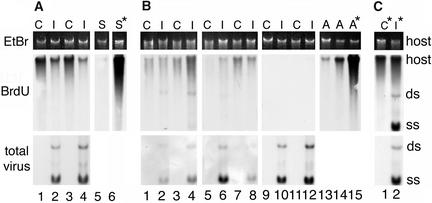
Excised TGMV-infected and mock-inoculated leaves were incubated with BrdU in the presence of either hydroxyurea or aphidicolin, and nascent DNA was analyzed as in Figure 1. Aphidicolin blocked the synthesis of single-stranded and double-stranded forms of TGMV DNA and partially reduced BrdU incorporation into chromosomal DNA of infected leaves (Figure 2A, lanes 2 and 4) but did not affect BrdU incorporation in control leaves (Figures 2A, lanes 1 and 3, and 2C, lane 1). Reduction of BrdU incorporation into healthy N. benthamiana seedling chromosomal DNA (Figure 2A, lanes 5 and 6) suggested that there are differences in the nature of DNA synthesis between seedlings and TGMV-infected mature leaves and healthy control leaves. By contrast, hydroxyurea treatment inhibited both host and viral DNA synthesis, dramatically reducing single-stranded DNA accumulation, in a concentration-dependent manner (Figure 2B, lanes 1 to 12). The presence of viral DNA synthesized before hydroxyurea and aphidicolin treatments was confirmed by hybridization with a TGMV-specific DNA probe. These inhibitor experiments were performed three times and yielded similar results each time. We conclude that hydroxyurea inhibits both viral and chromosomal DNA synthesis in healthy and infected tissues. Aphidicolin did not significantly affect BrdU incorporation into DNA of control, mature leaves but greatly inhibited seedling uptake. Like seedling DNA, aphidicolin also inhibited viral DNA uptake to levels below the detection limits of our system. Chromosomal DNA in TGMV-infected tissue showed significantly lower levels of BrdU incorporation than matched control leaves, but some incorporation persisted.
Figure 2.
DNA Synthesis Is Inhibited Differentially by Aphidicolin and Hydroxyurea.
(A) Matched pairs of TGMV-infected (I) and control (C) leaves at position T+1 were placed in 100 μM aphidicolin (lanes 1 to 4) for 14.5 h and then transferred to the same inhibitor solution containing 100 μM BrdU for 9.5 h (lanes 1–5). Healthy seedlings (S) were treated with and without 100 μM aphidicolin. Total viral DNA (lower panel) was detected on the same blot as BrdU incorporation. *, no inhibitor. EtBr, ethidium bromide; shows host DNA levels before transfer to a nitrocellulose membrane.
(B) Similar sets of control and infected leaves were placed in 10 (lanes 1–4), 50 (lanes 5–8), or 100 mM (lanes 9–12) hydroxyurea (HU) for 16 h and then hydroxyurea + BrdU for 9 h. The apical portions of healthy N. benthamiana plants were incubated with or without 50 mM hydroxyurea (lanes 13 to 15). BrdU incorporation was analyzed as described for Figure 1. Viral DNA was detected by chemifluorescence using a digoxigenin-labeled AL1 probe.
(C) Lanes 1 and 2 show control and infected leaf DNA, respectively, treated with BrdU and no inhibitor. Double-stranded (ds) and single-stranded (ss) TGMV DNA and host DNA are shown.
TGMV-Mediated Induction of DNA Synthesis Is Cell Autonomous
High levels of BrdU incorporation into chromosomal DNA of TGMV-infected plants (Figure 1) may be a systemic pathogen response that results in a general increase in DNA repair. Alternatively, TGMV may act at the cellular level by interfering with transcriptional and cell cycle controls, thereby stimulating cells to begin DNA synthesis. To distinguish between these possibilities, we colocalized newly incorporated BrdU with viral DNA visualized by fluorescent in situ hybridization.
Figures 3A and 3B show BrdU incorporation into nuclei of a healthy seedling leaf containing actively cycling cells. The heterogeneous pattern of BrdU incorporation was consistent with cells in various stages of the cell cycle, with only some in S-phase. In mature tissue of healthy plants, high levels of BrdU incorporation were not seen (Figures 3C2, 3G1, and 3G2). Examination of systemically infected leaf, root, and stem sections showed high levels of BrdU incorporation into nuclei of a variety of differentiated cell types, including epidermal, mesophyll, vascular, cortical, and pith cells (Figure 3). The asynchronous and widespread nature of systemic TGMV infection in N. benthamiana made it possible to find large groups of infected cells at different stages of infection in multiple tissue types. Comparison of BrdU, TGMV DNA, and 4′,6-diamidino-2-phenylindole (DAPI) images showed that almost every nucleus that had incorporated significant amounts of BrdU also contained TGMV DNA. Colocalization of TGMV DNA with BrdU in the same nuclei of differentiated cell types confirmed that TGMV DNA replicates in a variety of mature cells that normally are inactive in DNA replication.
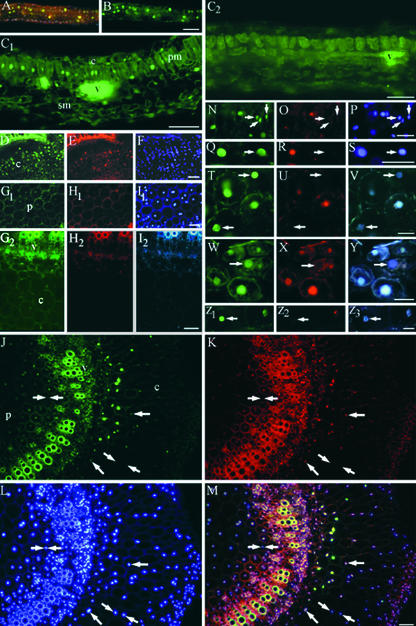
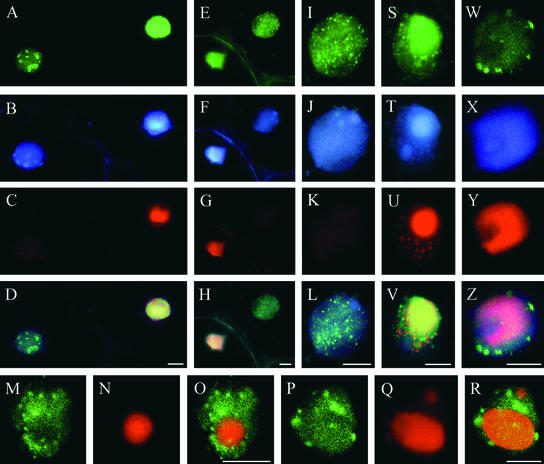
Figure 3.
Induction of BrdU Incorporation by TGMV Is Cell Autonomous.
Systemically infected tissues were incubated with 100 μM BrdU for 24 h as described for Figure 1, fixed, and Vibratome sectioned. BrdU was detected using anti-BrdU antibodies and Alexa 488–conjugated secondary antibodies (green in [A] to [D], [G1], [G2], [J], [M], [N], [Q], [T], [W], and [Z1]). Sections also were hybridized with Texas Red–labeled oligonucleotide probes to detect TGMV DNA (red in [E], [H1], [H2], [K], [M], [O], [R], [U], [X], and [Z2]) and stained with DAPI (blue in [A], [F], [I1], [L], [M], [P], and [S]). TGMV AL1 was immunolocalized with anti-AL1 antibodies and Marina Blue–labeled secondary antibodies (blue in [I2], [V], [Y], and [Z3]).
(A) and (B) BrdU incorporation (green) in an immature healthy leaf is confined to isolated nuclei throughout the lamina. A triple-wavelength excitation filter was used in (A) to show BrdU-incorporating nuclei among nuclei with no BrdU (blue).
(C1) BrdU incorporation into nuclei of a TGMV-infected mature leaf. Nuclei of adjacent palisade mesophyll cells and some spongy mesophyll cells show abundant BrdU labeling.
(C2) Mock-inoculated control mature leaf section. Bright autofluorescence of vascular tissue is visible.
(D) to (F) TGMV-infected root section. Viral DNA and BrdU are evident in cortical cells.
(G1) to (I2) Mock-inoculated control stem cross-sections. Double nuclei are common in N. benthamiana pith. Low levels of background BrdU incorporation are observed in some nuclei.
(J) to (M) TGMV-infected stem cross-section. BrdU (green) and viral DNA (red) are colocalized. (M) shows a three-color overlay of (J) to (L), demonstrating colocalization of BrdU and viral DNA in DAPI-stained nuclei. Single arrows show nuclei with low amounts of viral DNA and correspondingly lower BrdU signals. Nuclei between double arrows lack viral DNA and BrdU.
(N) to (S) TGMV-infected stem sections. Arrows show nuclei that have high BrdU but minimally detectable viral DNA. Blue indicates DAPI.
(T) to (Z3) TGMV-infected stem sections. Arrows show nuclei that have BrdU and minimally detectable viral DNA but contain TGMV AL1. Blue indicates AL1-specific signal.
c, cortex; e, epidermis; pm, palisade mesophyll; sm, spongy mesophyll; v, vascular tissue. Bars = 100 μm for (A) to (M), (V), and (Z3) and 50 μm for (P), (S), and (Y).
We observed variation in the levels of BrdU and viral DNA among nuclei in a given section (Figures 3J to 3M), illustrating the asynchronous nature of TGMV infection. Although most nuclei with BrdU label contained high levels of viral DNA, some had minimally detectable viral DNA (Figures 3N to 3S, arrows), raising the possibility that BrdU was incorporated into host chromatin in the absence of TGMV. To address this discrepancy, we colocalized BrdU, viral DNA, and the TGMV replication protein AL1. In every BrdU-labeled nucleus that had minimally detectable viral DNA, high levels of AL1 were detected (Figures 3T to 3Z3, arrows), demonstrating the presence of TGMV. We interpret the presence of AL1 but only minimal amounts of viral DNA in BrdU-labeled cells to reflect an early stage of infection, before high levels of viral DNA had accumulated. In some cells, the incorporation of BrdU into cellular DNA may precede significant viral DNA accumulation.
BrdU Can Be Distributed Differentially between TGMV and Host DNA
The observation of heterogeneous BrdU labeling patterns prompted us to wonder whether we could distinguish further how BrdU was distributed within nuclei. Laser scanning confocal microscopy was used in addition to normal epifluorescence microscopy to compare BrdU incorporation into viral and host DNA after a 24-h BrdU treatment. The most common patterns, accounting for ∼95% of all nuclei with BrdU and TGMV signals, were nuclei showing colocalization of BrdU exclusively with viral DNA inclusions (Figures 3 and 4A to 4H) and nuclei in which BrdU signals were localized throughout the nucleus (Figures 3 and 4I to 4P). Up to 60% of all TGMV-infected nuclei exhibited condensed chromatin during later stages of infection, based on the presence of large viral DNA inclusions. In ∼5 to 10% of these nuclei, BrdU was associated exclusively with condensed host chromatin and not with TGMV DNA (Figures 4Q to 4X). The detection of uniform BrdU labeling throughout the length of condensed chromosomal DNA provides additional evidence that host DNA is replicated during infection. Figures 4A to 4H and 4Q to 4X also demonstrate that host and viral DNA can replicate at different times during infection.
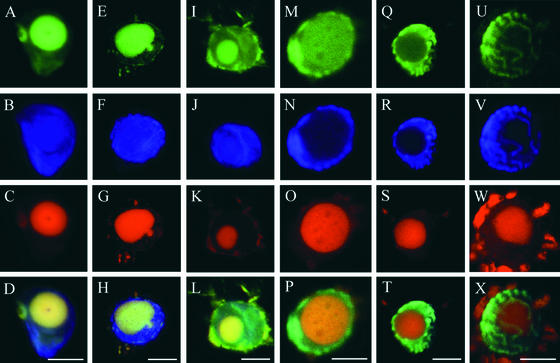
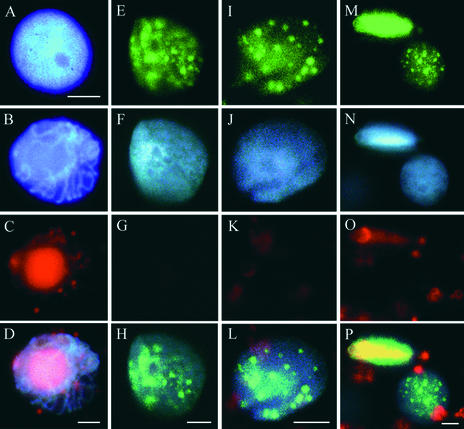
Figure 4.
BrdU Can Be Incorporated Preferentially into Viral or Host DNA of an Infected Nucleus.
TGMV-infected leaf and stem tissues were processed as described for Figure 3 and analyzed using laser scanning confocal microscopy. BrdU (green) is shown in the top row, DAPI (blue) is shown in the second row, TGMV DNA (red) is shown in the third row, and three-color overlays are shown in the bottom row.
(A) to (H) Preferential BrdU incorporation into viral DNA. BrdU signals colocalize with viral DNA inclusions.
(I) to (P) BrdU incorporation into both viral and host DNA. The BrdU signal at the nuclear periphery in (M) colocalizes with marginalized host chromatin (N).
(Q) to (X) Preferential BrdU incorporation into chromosomal DNA. BrdU is associated with condensed chromatin.
Red objects outside of the nuclei in (G), (K), (O), (S), and (W) are autofluorescent chloroplasts. Bars = 10 μm.
Nuclei labeled for 24 h with BrdU most often showed a diffuse, uniform distribution of BrdU associated with both TGMV DNA compartments and whole nuclei. To determine if BrdU could be incorporated in distinct foci, infected tissue was pulse-labeled with BrdU for 4 h. Stem sections were chosen because cortical and pith cells are large and contain fewer autofluorescent organelles. Figure 5 shows that BrdU incorporation was punctate in some nuclei, whereas in adjacent cells, BrdU labeling remained uniform throughout nuclei (Figures 5A to 5H). Punctate labeling of BrdU was never observed in healthy control sections (data not shown). BrdU foci were detected in only ∼10% of nuclei in a given TGMV-infected section, probably as a result of the relatively short labeling period used, and were most prevalent in nuclei containing AL1 but only low amounts of viral DNA (Figures 5A to 5L). However, BrdU foci also were seen occasionally in nuclei that contained large TGMV DNA inclusions (Figures 5M to 5Z). In these nuclei, BrdU foci did not colocalize with the larger viral DNA inclusions, which consistently showed nonpunctate BrdU labeling (Figures 5S to 5V). BrdU foci also were observed at the margins of TGMV-infected nuclei (Figures 5W to 5Z). Previous studies demonstrated that host DNA redistributes to the nuclear margins, whereas viral inclusions are located centrally in infected cells (Rushing et al., 1987; Bass et al., 2000). Thus, the location of punctate BrdU foci at nuclear margins is consistent with host DNA replication.
Figure 5.
BrdU Foci Are Not Associated with TGMV DNA.
TGMV-infected stem tissues were processed and imaged as described for Figure 3, except that tissues were administered BrdU for only 4 h. BrdU (green) is shown in the top row, TGMV AL1 (blue) is shown in the second row, TGMV DNA (red) is shown in the third row, and three-color overlays are shown in the fourth row.
(A) to (H) BrdU foci are prevalent in nuclei with AL1 but minimally detectable in viral DNA that are adjacent to nuclei with high viral DNA, AL1, and BrdU signals. BrdU association with viral DNA is uniform, not punctate.
(I) to (L) Same as (E) to (H), showing details of BrdU foci in one nucleus.
(M) to (R) TGMV-infected nuclei with BrdU foci that do not colocalize with large TGMV DNA inclusions. (O) and (R) show overlays of BrdU and viral DNA signals.
(S) to (V) A nucleus with BrdU foci that do not colocalize with large or small areas of viral DNA. Viral DNA shows uniform (nonpunctate) BrdU labeling.
(W) to (Z) BrdU foci are localized at the nuclear periphery with marginalized chromatin, not with viral DNA.
Bars = 10 μm.
Most nuclei at advanced stages of infection had large areas of viral DNA accumulation, and many had condensed chromatin, which is characteristic of cells in early mitosis (Figures 6B to 6D) (Bass et al., 2000). By contrast, all nuclei with punctate BrdU labeling (Figures 6E, 6I, and 6M) and undetectable levels of viral DNA (Figures 6G, 6K, and 6O) had evenly dispersed chromatin throughout their interiors (Figures 6F, 6J, and 6N), similar to healthy interphase nuclei (Figure 6A) and more specifically to early S-phase chromatin organization. The presence of host-associated BrdU foci in nuclei with no detectable TGMV DNA and diffuse chromatin, as well as in nuclei with large viral inclusions but no detectable BrdU (Figure 5), is consistent with the idea that host DNA replication can occur both before and after viral DNA replication, depending on the particular cell.
Figure 6.
Host Chromatin of TGMV-Infected Nuclei Is Distributed Evenly and Resembles S-Phase Chromatin Organization.
Stem tissues were pulse labeled with BrdU for 4 h and then processed and imaged as described for Figure 5. For (E) to (P), BrdU (green) is shown in the top row, DAPI-stained host chromatin (blue) is shown in the second row, TGMV DNA (red) is shown in the third row, and three-color overlays are shown in the fourth row except for (D), which was imaged using a triple cube.
(A) Healthy interphase nucleus stained with DAPI showing typical dispersed chromatin.
(B) to (D) Advanced-stage TGMV-infected nucleus showing high viral DNA accumulation and condensed chromatin.
(E) to (L) TGMV-infected nuclei showing BrdU foci and no detectable viral DNA. Host DNA in these nuclei is dispersed evenly.
(M) to (P) A nucleus with BrdU foci but no detectable viral DNA adjacent to a nucleus with viral DNA and uniform BrdU label.
Red objects outside of the nuclei in (C), (K), (L), (O), and (P) are autofluorescent chloroplasts. Bars = 5 μm.
DISCUSSION
TGMV Activates Host DNA Synthesis Machinery in Differentiated Cells
TGMV encodes seven proteins, two of which interact with the plant cell cycle regulatory protein, pRBR, and only one of which is required for induction of the host DNA replication factor PCNA (Egelkrout et al., 2001). Because of the small number of viral proteins and the complexity of DNA replication, it is likely that the virus targets critical cell cycle regulatory components. We used BrdU labeling in mature plant tissues to analyze how interactions between the virus and the cell cycle affect host DNA synthesis in infected cells. Our results suggest that TGMV induces conditions favorable for the replication of both the viral genome and chromosomal DNA in a variety of mature cell types. However, replica-tion of chromosomal DNA was confined to cells containing viral DNA and/or AL1, suggesting that replication competence must be established in each infected cell. Based on the different patterns of BrdU labeling, host and viral DNA can replicate at different times and areas within each nucleus, and there may be fundamental differences between the organization of viral and host DNA replication centers. The punctate labeling of host DNA after a 4-h treatment and the uniform incorporation of BrdU into condensed chromatin are not consistent with DNA repair. Punctate BrdU labeling of host DNA has been reported to be associated with DNA replication factories in onion cells (Samaniego et al., 2002) and animal cells (Cossmann et al., 2000). These results establish TGMV infection of N. benthamiana plants as a useful system for analyzing the molecular basis for how plants control DNA replication and demonstrate that host DNA replication can be caused by a plant virus.
Previous studies of TGMV infection did not examine active TGMV DNA synthesis directly (Rushing et al., 1987; Nagar et al., 1995; Bass et al., 2000). The use of BrdU incorporation in intact TGMV-infected N. benthamiana organs allowed us to demonstrate that TGMV replicates its DNA in a variety of differentiated cells, including mesophyll, epidermal, vascular, pith, and cortical cells of the leaf, stem, and root (Figure 3). The variation in BrdU incorporation patterns (Figure 3) compared with viral DNA accumulation illustrates the heterogeneous nature of TGMV infection and cellular interaction. The finding that host DNA also was replicated was unexpected, because chromosomal replication is tightly regulated. Further supporting the heterogeneity in cellular interactions, we found that viral and host DNA could incorporate BrdU in the same cell at different times (Figure 4). The molecular basis for the variation in BrdU labeling patterns is not clear, but this variation illustrates the importance of in planta analyses of viral DNA replication in addition to studies using cultured cells.
Host DNA Is Replicated in TGMV-Infected Cells
Analysis of BrdU-labeled DNA from TGMV-infected and control leaves (Figure 1) suggested that host DNA replication was activated in infected tissues. Because developmentally identical leaves were treated similarly, except for the presence of TGMV, the higher level of BrdU incorporation into chromosomal DNA of infected leaves was most likely the result of the stimulation of host DNA replication by TGMV. The alternative explanation, that increased BrdU incorporation into chromosomal DNA reflects a general increase in DNA repair, is not consistent with the cell-autonomous character of BrdU incorporation into TGMV-infected nuclei (Figures 3 and 4).
The distribution of BrdU in host DNA of TGMV-infected cells provided additional evidence for replication. Nuclei with uniform BrdU incorporation into condensed chromatin were seen (Figures 4Q and 4U), consistent with the incorporation of BrdU during S-phase, transit through G2, and arrest in prophase. Similar uniform BrdU labeling of condensed chromatin has been observed in dividing cultured soybean cells (Wang et al., 1989). Recent results showing that the TGMV AL1-interacting kinesin, GRIMP, colocalizes with mitotic chromosomes in healthy cells (Kong and Hanley-Bowdoin, 2002), possibly preventing progression through mitosis in infected cells, may account for the finding of many cells with condensed chromatin.
Some mammalian DNA viruses, such as Human papilloma virus (HPV), cause host DNA replication in differentiated cells (Cheng et al., 1995). In other virus infections, including those by Human cytomegalovirus (HCMV), host DNA is not replicated (Morin et al., 1996). HCMV, a much larger and perhaps more sophisticated virus than HPV and TGMV, activates cell cycle progression and nucleotide synthesis but blocks cells at late G1, thereby inhibiting cellular DNA synthesis and allowing exclusive viral access to nucleotide pools (Bresnahan et al., 1996). Our data suggest that TGMV, like the similarly sized HPV, stimulates differentiated cells to enter S-phase but does not prevent cellular DNA synthesis. However, cell division apparently is prevented, because enations, such as those found in other geminivirus-host combinations (Esau and Hoefert, 1978; Latham et al., 1997), and morphological evidence of mitotic events are not found in TGMV-infected N. benthamiana cells.
BrdU Foci Are Associated with Host DNA Synthesis
Punctate BrdU foci, observed only during the 4-h BrdU labeling period (Figure 5), did not colocalize with TGMV DNA. These foci were found most frequently in nuclei that contained no detectable viral DNA but did contain TGMV AL1 protein (Figure 5). Similar patterns of BrdU labeling have been observed in onion, tobacco, and barley (Suzuki et al., 1992; Sanathkumar et al., 1996; Jasencakova et al., 2001) and are associated with replication factories in mammalian cells (Hozak et al., 1994; Cossmann et al., 2000), suggesting that the punctate areas may represent sites of host chromatin replication during TGMV infection. The observation of diffuse S-phase–like chromatin associated with BrdU foci in nuclei with no detectable viral DNA (Figure 6) also supports the idea that TGMV can induce host replication early in infection after cell cycle reactivation, before high-level viral replication begins and chromatin condenses in the later stages of infection. The fact that we never observed punctate BrdU label in TGMV DNA compartments suggests that there may be differences between the size and/or structure of viral and host replication factories that our methods could not resolve. Alternatively, newly replicated viral DNA may be displaced from sites of BrdU incorporation, resulting in diffuse BrdU signals, whereas host replication factories support processive synthesis of long segments of DNA and can be visualized as discrete foci.
Viral Induction of DNA Replication Is Cell Autonomous
BrdU was detected only in nuclei possessing viral DNA and/or TGMV AL1 (Figures 3 to 5). The presence of AL1 in nuclei with BrdU incorporation but no detectable TGMV DNA may be related to AL1's essential role in altering cell cycle controls before significant viral DNA replication (Ach et al., 1997). Different AL1 functions are mediated by different AL1 and AL3 oligomerization states (Kong et al., 2000; Orozco et al., 2000; Settlage et al., 2001). One possibility is that early in infection, AL1 forms protein complexes that bind to pRBR, inducing cells to reenter the cell cycle, but do not support efficient initiation of viral DNA synthesis (Settlage et al., 2001). Nonetheless, host DNA replication may proceed as induced host DNA replication factors, such as PCNA, are integrated at chromosomal replication initiation complexes (Samaniego et al., 2002). Later in infection, AL1 forms replication-competent oligomers and may “compete” with host DNA to recruit the available replication factors. Consistent with our data, this model predicts that chromosomal and viral DNA replication are cell autonomous but not necessarily simultaneous events.
TGMV DNA Synthesis Is Dependent on Host DNA Polymerase α– or δ–Like Activity
TGMV does not encode its own polymerase and instead induces the expression of host DNA synthesis machinery. Other than PCNA and minichromosome maintenance (Sabelli et al., 1996; Holding and Springer, 2002), specific components of the replication machinery have not been identified. However, because the encapsidated form of TGMV DNA is single stranded, DNA polymerase α activity is likely to be required for the production of double-stranded DNA replication templates during initial infection (Bryant et al., 2001). Aphidicolin inhibits mammalian polymerase α and polymerase δ (Wang, 1991; Wright et al., 1994), has been shown to inhibit plant polymerase α–like activity (Sala et al., 1980), and is used routinely to synchronize tobacco cells at S-phase (Nagata et al., 1982). We showed that the synthesis of both single-stranded and double-stranded TGMV DNA is inhibited by aphidicolin at concentrations that still allow incorporation into host DNA to proceed (Figure 2). The absence of BrdU incorporation into seedlings treated with aphidicolin, along with the persistence of incorporation into DNA of mature cells, suggest that repair, or endoreduplication, is active in uninfected cells. The lower level of BrdU incorporation into infected tissue may be explained by the heterogeneity of cellular states—not all cells were infected, and some showed repair similar to controls—or by endoreduplication, which is not as sensitive to aphidicolin.
TGMV Infection May Cause Endoreduplication
Endoreduplication occurs naturally in a variety of differentiated cell types in plants (Larkins et al., 2001). Root knot nematode infection causes increases in both nuclear DNA content (Wiggers et al., 1990) and nuclear volume (de Almeida Engler et al., 1999). We showed previously (Bass et al., 2000) that the nuclear volume of TGMV-infected cells increased as much as 24-fold, which, together with the BrdU incorporation data presented here, is consistent with endoreduplication of host DNA. The unexpected results of host DNA replication occurring both before and after significant viral DNA replication (Figures 1 and 3 to 6) may be partly explained by the induction of additional rounds of DNA replication by TGMV.
The persistence of aphidicolin-resistant BrdU incorporation into chromosomal DNA of TGMV-infected leaves, but not into seedling DNA (Figure 2), might be explained by a DNA polymerase β–like enzyme. In animal cells, polymerase β is associated with endoreduplication and repair and is resistant to aphidicolin inhibition (Nealon et al., 1996; Burgers, 1998; Dianov et al., 1999). Enzymes with polymerase β–like activities have been isolated from plants (Sanathkumar et al., 1996; Luque et al., 1998). Recent studies using tobacco mesophyll cultures induced to undergo endoreduplication by cytokinin deprivation demonstrated that cells continued to synthesize DNA when treated with aphidicolin (Quelo et al., 2002), suggesting the involvement of an enzyme with polymerase β–like activity. Further studies will be required to identify the specific enzymes that function in plant DNA replication, repair, and endoreduplication and to understand the biochemical basis of TGMV-induced DNA replication.
METHODS
Plant Growth and Inoculation
Nicotiana benthamiana plants and excised tissues were grown at 25°C and 65% RH with a 14-h-light/10-h-dark photoperiod. Plants at the four– to six–expanded leaf stage (leaves averaging 5 cm × 4 cm) were inoculated with Tomato golden mosaic virus (TGMV) by microprojectile bombardment as described previously (Nagar et al., 1995). Control plants at the same stage of development were mock infected with microprojectiles lacking viral DNA.
DNA Gel Blot Analysis of 5-Bromo-2-Deoxyuridine Incorporation
TGMV-infected and control leaves were excised, and their petioles were placed in vials containing 6 mL of 0.5 × Hoagland solution without micronutrients (Hoagland and Arnon, 1950) containing 100 μM 5-bromo-2-deoxyuridine (BrdU) (Calbiochem, La Jolla, CA) for 24 h. Seedlings were treated similarly by immersing their roots in BrdU solution. Total DNA was isolated as described (Kjemtrup et al., 1998). Uncut DNA (250 ng) was electrophoresed on 1.0% agarose gels, stained with ethidium bromide, and photographed. DNA was blotted (Sambrook et al., 1989) to nitrocellulose membranes and UV cross-linked.
All detection steps were performed at 25°C on an orbital shaker at 50 rpm. Membranes were blocked for 1 h in Tris-buffered saline containing 0.5% Tween 20 (TBS-T), 5% powdered milk, and 50 μg/mL heparin. After one 15-min and two 5-min washes in TBS-T, membranes were incubated for 1 h with murine anti-BrdU antibodies (Roche Biochemicals, Indianapolis, IN) diluted 1:1000 in TBS-T, washed in TBS-T as described above, and incubated for 1 h in goat anti-mouse horseradish peroxidase conjugate (Amersham Pharmacia Biotech, Piscataway, NJ) diluted 1:5000 in TBS-T. After washes in TBS-T, BrdU was detected using an enhanced chemiluminescence kit (Amersham Pharmacia Biotech). Chemiluminescent signals were collected using a Hamamatsu photon imager (Bridgewater, NJ) during a 5-min count time. Membranes were reprobed with a 32P-dCTP–labeled TGMV A DNA probe that included the TGMV common region and exposed to film (Sambrook et al., 1989).
Inhibitor Analysis
Mock-infected control and TGMV-infected leaves were excised at 8 days after infection (DAI) and placed in 0.5 × Hoagland solution containing 100 μM aphidicolin (Biomol, Plymouth Meeting, PA) for 12 h and then transferred to the same inhibitor solution containing 100 μM BrdU for 14 h. For hydroxyurea (Calbiochem), leaves were excised at 12 DAI and incubated in 10, 50, or 100 mM hydroxyurea alone for 16 h and then in the same concentration of hydroxyurea plus 100 μM BrdU for 24 h. Seedling controls were incubated in Hoagland solution containing 100 μM aphidicolin for 14.5 h and then in 100 μM aphidicolin plus 100 μM BrdU or 100 μM BrdU alone for 9.5 h. BrdU incorporation was analyzed as described above except that viral DNA was detected by reprobing the membrane with a 280-bp digoxigenin-labeled AL1 probe (DIG High Prime DNA Labeling Kit I; Roche Biochemicals). The chemifluorescent product of the substrate Vistra ECF (Amersham Pharmacia Biotech) was detected using a STORM gel blot analysis system (Molecular Dynamics, Sunnyvale, CA).
BrdU Labeling, in Situ Hybridization, and Immunolocalization
Systemically infected tissues harvested at 7 to 11 DAI were placed in 0.5 × Hoagland solution containing 100 μM BrdU for 4 or 24 h. For leaf and stem samples, whole plants were cut above the roots and placed in BrdU solution. For roots, whole plants were placed in small beakers of BrdU solution. Tissues were cut into 0.5- to 1.0-cm pieces and fixed for 3 to 5 h at 25°C in chromatin-preserving buffer (Bass et al., 2000). Tissues then were washed for 30 min in several changes of buffer and stored at 4°C. Fixed tissues were embedded in 5% low-gelling-temperature agarose (type XI; Sigma, St. Louis, MO) in distilled water, and 50- to 60-μm sections were cut using a Vibratome 1000 (Technical Products International, St. Louis, MO) and collected in 1 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate).
All hybridizations and washes were performed at 37°C. Sections were prehybridized for 3 h in hybridization solution containing 47% deionized formamide, 4 × SSC, 1 mM EDTA, 100 μg/mL denatured salmon sperm DNA, 100 μg/mL yeast tRNA, and 5 × Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA). Texas Red end-labeled oligonucleotide probes (Bass et al., 2000) then were diluted in hybridization solution to 5 ng/μL and hybridized with sections for 16 h. Posthybridization washes consisted of one 30-min wash in hybridization solution, two 10-min washes in 2 × SSC, and two 10-min washes in 1 × SSC.
All immunolocalization steps were performed at 25°C. After in situ hybridization and washes, sections were transferred to 1 × PBS, pH 7.4, containing 0.1% BSA (PBS-BSA). Sections were blocked in 10% goat serum (Sigma) in PBS-BSA for 1 h. After a 5-min wash in PBS-BSA, sections were incubated for 1 h with anti-BrdU monoclonal antibodies (Roche Biochemicals) diluted 1:17 in Incubation Buffer from the BrdU Labeling and Detection Kit I (Roche Biochemicals) with the addition of 300 units/mL exonuclease III and 15 units/mL Sau3A. For experiments involving colocalization of AL1 with BrdU, samples were washed after the anti-BrdU antibody step with three 10-min PBS-BSA washes and then incubated for 1 h with polyclonal rabbit anti-AL1 antiserum (Settlage et al., 1996) diluted 1:100 in PBS-BSA. After three 10-min washes in PBS-BSA, sections were incubated for 1 h with an Alexa 488 goat anti-mouse conjugate (Molecular Probes, Eugene, OR) diluted 1:250 in PBS-BSA for BrdU detection alone or with the addition of a Marina Blue (Molecular Probes) goat anti-rabbit conjugate diluted 1:250 in the same Alexa 488/PBS-BSA solution to colocalize AL1 with BrdU. After three 10-min washes in PBS-BSA, sections were mounted on slides in 1 × PBS containing 90% glycerol. 4′,6-Diamidino-2-phenylindole (1 μg/mL) was included in the mounting medium only for samples not colabeled for AL1. Images were recorded on Kodak Elite ASA 400 color slide film using a Nikon Eclipse E800 fluorescence microscope and a Nikon U-III camera system (Tokyo, Japan). For confocal analyses, a Leica DMRXA upright microscope with an attached TCS SP scanner head was used in conjunction with Leica NTS image-analysis software (Wetzlar, Germany) to obtain triple-wavelength images of TGMV-infected nuclei. A UV laser was used at 360 nm for 4′,6-diamidino-2-phenylindole images, an argon laser was used at 488 nm for BrdU/Alexa 488 images, and a krypton laser was used at 568 nm for TGMV DNA/Texas Red images. Images were analyzed and three-color overlays were made using Adobe Photoshop (Mountain View, CA). Quantitative analyses were performed by microscopic examination of tissues from different leaves and plants as well as by inspection of at least 100 sections for each treatment.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank David Collings, Amie Scott, and Nina Allen for help with confocal microscopy, which was performed at the Cell and Molecular Imaging Facility at North Carolina State University. This work was supported by National Science Foundation Grant MCB-9601893 to D.R. and U.S. Department of Agriculture Grant 96-35301-3177 to L.H.-B. and D.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005777.
References
- Ach, R.A., Durfee, T., Miller, A.B., Taranto, P., Hanley-Bowdoin, L., Zambriski, P.C., and Gruissem, W. (1997). RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and a geminivirus replication protein. Mol. Cell. Biol. 17, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, H.W., Nagar, S., Hanley-Bowdoin, L., and Robertson, D. (2000). Chromosome condensation induced by geminivirus infection of mature plant cells. J. Cell Sci. 113, 1149–1160. [DOI] [PubMed] [Google Scholar]
- Bravo, R., Frank, R., Blundell, P.A., and MacDonald-Bravo, H. (1987). Cyclin/PCNA is the auxiliary protein of DNA polymerase δ. Nature 326, 515–517. [DOI] [PubMed] [Google Scholar]
- Bresnahan, W.A., Boldogh, I., Thompson, E.A., and Albrecht, T. (1996). Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224, 150–160. [DOI] [PubMed] [Google Scholar]
- Bryant, J.A., Moore, K., and Aves, S.J. (2001). Origins and complexes: The initiation of DNA replication. J. Exp. Bot. 52, 193–202. [PubMed] [Google Scholar]
- Burgers, P.M. (1998). Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107, 218–227. [DOI] [PubMed] [Google Scholar]
- Cheng, S., Schmidt-Grimminger, D.-C., Murant, T., Broker, T.R., and Chow, L. (1995). Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9, 2335–2349. [DOI] [PubMed] [Google Scholar]
- Coello, P., Rodriguez, R., Garcia, E., and Vazquez-Ramos, J.M. (1992). A DNA polymerase from maize axes: Its purification and possible role. Plant Mol. Biol. 20, 1159–1168. [DOI] [PubMed] [Google Scholar]
- Cossmann, P.H., Eggli, P.S., and Kurz, H. (2000). Three-dimensional analysis of DNA replication foci: A comparative study on species and cell type in situ. Histochem. Cell Biol. 113, 195–205. [DOI] [PubMed] [Google Scholar]
- de Almeida Engler, J., de Vleesschauwer, V., Burssens, S., Celenza, J.L., Inzé, D., Van Montagu, M., Engler, G., and Gheysen, G. (1999). Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11, 793–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov, G.L., Prasad, R., Wilson, S.H., and Bohr, V.A. (1999). Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J. Biol. Chem. 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- Egelkrout, E.M., Robertson, D., and Hanley-Bowdoin, L. (2001). Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau, K. (1977). Virus-like particles in the nuclei of phloem cells in spinach leaves infected with the curly top virus. J. Ultrastruct. Res. 61, 78–88. [DOI] [PubMed] [Google Scholar]
- Esau, K., and Hoefert, L. (1978). Hyperplastic phloem in sugar beet leaves infected with the beet curly top virus. Am. J. Bot. 65, 772–783. [Google Scholar]
- Gutierrez, C. (2000). DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 19, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Settlage, S.B., Orozco, B.M., Nagar, S., and Robertson, D. (1999). Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18, 71–106. [PubMed] [Google Scholar]
- Hemerly, A.S., Ferreira, P., de Almeida Engler, J., Van Montagu, M., Engler, G., and Inzé, D. (1993). cdc2A expression in Arabidopsis is linked with competence for cell division. Plant Cell 5, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland, D.R., and Arnon, D.I. (1950). The water culture method for growing plants without soil. Calif. Agric. Ext. Serv. Circ. 347, 1–32. [Google Scholar]
- Holding, D.R., and Springer, P.S. (2002). The Arabidopsis gene PROLIFERA is required for proper cytokinesis during seed development. Planta 214, 373–382. [DOI] [PubMed] [Google Scholar]
- Horns, T., and Jeske, H. (1991). Localization of abutilon mosaic virus (AbMV) DNA within leaf tissue by in situ hybridization. Virology 181, 580–588. [DOI] [PubMed] [Google Scholar]
- Hozak, P., Jackson, D.A., and Cook, P.R. (1994). Replication factories and nuclear bodies: The ultrastructural characterization of replication sites during the cell cycle. J. Cell Sci. 107, 2191–2202. [DOI] [PubMed] [Google Scholar]
- Jansen-Durr, P. (1996). How viral oncogenes make the cell cycle. Trends Genet. 12, 270–275. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Meister, A., and Schubert, I. (2001). Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley. Chromosoma 110, 83–92. [DOI] [PubMed] [Google Scholar]
- Kelman, Z. (1997). PCNA: Structure, functions and interactions. Oncogene 14, 629–640. [DOI] [PubMed] [Google Scholar]
- Kjemtrup, S., Sampson, K.S., Peele, C., Long, L.V., Conkling, M.A., Thompson, W.F., and Robertson, D. (1998). Gene silencing from plant DNA carried by a geminivirus. Plant J. 14, 91–100. [DOI] [PubMed] [Google Scholar]
- Kong, L.-J., and Hanley-Bowdoin, L. (2002). A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L.J., Orozco, B.M., Roe, J.L., Nagar, S., Ou, S., Feiler, H.S., Durfee, T., Miller, A.B., Gruissem, W., Robertson, D., and Hanley-Bowdoin, L. (2000). A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19, 3485–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins, B.A., Dilkes, B.P., Dante, R.A., Coelho, C.M., Woo, Y.M., and Liu, Y. (2001). Investigating the hows and whys of DNA endoreduplication. J. Exp. Bot. 52, 183–192. [PubMed] [Google Scholar]
- Latham, J.R., Saunders, K., Pinner, M.S., and Stanley, J. (1997). Induction of plant cell division by beet curly top virus gene C4. Plant J. 11, 1273–1283. [Google Scholar]
- Levi, M., Sparvoli, E., Sgorbati, S., and Chiatante, D. (1987). Rapid immunofluorescent determination of cells in S-phase in pea root meristems: An alternative to autoradiography. Physiol. Plant. 71, 68–72. [Google Scholar]
- Lucy, A.P., Boulton, M.I., Davies, J.W., and Maule, A.J. (1996). Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant-Microbe Interact. 9, 22–31. [Google Scholar]
- Luque, A.E., Benedetto, J.P., and Castroviejo, M. (1998). Wheat DNA polymerase CI: A homologue of rat DNA polymerase beta. Plant Mol. Biol. 38, 647–654. [DOI] [PubMed] [Google Scholar]
- Morin, J., Johann, S., O'Hara, B., and Gluzman, Y. (1996). Exogenous thymidine is preferentially incorporated into human cytomegalovirus DNA in infected human fibroblasts. J. Virol. 70, 6402–6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar, S., Pedersen, T.J., Carrick, K.M., Hanley-Bowdoin, L., and Robertson, D. (1995). A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, T., Okada, K., and Takebe, I. (1982). Mitotic protoplasts and their infection with tobacco mosaic virus RNA encapsulated in liposomes. Plant Cell Rep. 1, 250–252. [DOI] [PubMed] [Google Scholar]
- Nealon, K., Nicholl, I.D., and Kenny, M.K. (1996). Characterization of the DNA polymerase requirement of human base excision repair. Nucleic Acids Res. 24, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins, J. (1992). E2F: A link between the Rb tumor suppressor protein and viral oncoproteins. Science 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Orozco, B.M., Kong, L.-J., Batts, L.A., Elledge, S., and Hanley-Bowdoin, L. (2000). The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275, 6114–6122. [DOI] [PubMed] [Google Scholar]
- Orozco, B.M., Miller, A.B., Settlage, S.B., and Hanley-Bowdoin, L. (1997). Functional domains of a geminivirus replication protein. J. Biol. Chem. 272, 9840–9846. [DOI] [PubMed] [Google Scholar]
- Quelo, A.H., Bryant, J.A., and Verbelen, J.P. (2002). Endoreduplication is not inhibited but induced by aphidicolin in cultured cells of tobacco. J. Exp. Bot. 53, 669–675. [DOI] [PubMed] [Google Scholar]
- Rushing, A.E., Sunter, G., Gardiner, W.E., Dute, R.R., and Bisaro, D.M. (1987). Ultrastructural aspects of tomato golden mosaic virus infection in tobacco. Phytopathology 77, 1231–1236. [Google Scholar]
- Sabelli, P.A., Burgess, S.R., Kush, A.K., Young, M.R., and Shewry, P.R. (1996). cDNA cloning and characterization of a maize homologue of the MCM proteins required for the initiation of DNA replication. Mol. Gen. Genet. 252, 125–136. [DOI] [PubMed] [Google Scholar]
- Sala, F., Parisi, B., Burroni, D., Amileni, A.R., Pedrali-Noy, G., and Spadari, S. (1980). Specific and reversible inhibition by aphidicolin of the α-like DNA polymerase of plant cells. FEBS Lett. 117, 93–98. [DOI] [PubMed] [Google Scholar]
- Samaniego, R., de la Torre, C., Moreno Diaz de la Espina, S. (2002). Dynamics of replication foci and nuclear matrix during S phase in Allium cepa L. cells. Planta 215, 195–204. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanathkumar, M., Ghosh, B., and SenGupta, D.N. (1996). Isolation of mammalian pol beta type DNA polymerase in shoot tips of germinated seedlings of IR-8 rice (Oryza sativa L.). Biochem. Mol. Biol. Int. 39, 117–126. [DOI] [PubMed] [Google Scholar]
- Sanderfoot, A.A., and Lazarowitz, S.G. (1996). Getting it together in plant virus movement: Cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 6, 353–358. [DOI] [PubMed] [Google Scholar]
- Settlage, S.B., Miller, A.B., Gruissem, W., and Hanley-Bowdoin, L. (2001). Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279, 570–576. [DOI] [PubMed] [Google Scholar]
- Settlage, S.B., Miller, A.B., and Hanley-Bowdoin, L. (1996). Interactions between geminivirus replication proteins. J. Virol. 70, 6790–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarshana, M.R., Wang, H.L., Lucas, W.J., and Gilbertson, R.L. (1998). Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol. Plant-Microbe Interact. 11, 277–291. [Google Scholar]
- Sunter, G., Hartitz, M.D., Hormuzdi, S.G., Brough, C.L., and Bisaro, D.M. (1990). Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179, 69–77. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Kawano, S., Sakai, A., Fujie, M., Kuroiwa, H., Nakamura, H., and Kuroiwa, T. (1992). Preferential mitochondrial and plastid DNA synthesis before multiple cell divisions in Nicotiana tabacum. J. Cell Sci. 103, 831–837. [Google Scholar]
- Wang, H., Cutler, A.J., Saleem, M., and Fowke, L.C. (1989). Immunocytochemical detection of DNA synthesis. J. Plant Physiol. 135, 15–20. [Google Scholar]
- Wang, T.S. (1991). Eukaryotic DNA polymerases. Annu. Rev. Biochem. 60, 513–552. [DOI] [PubMed] [Google Scholar]
- Wiggers, R.J., Starr, J.L., and Price, H.J. (1990). DNA content and variation in chromosome number in plant cells affected by Meloidogyne incognita and M. arenaria. Phytopathology 80, 1391–1395. [Google Scholar]
- Wright, G.E., Hubscher, U., Khan, N.N., Focher, F., and Verri, A. (1994). Inhibitor analysis of calf thymus DNA polymerases alpha, delta and epsilon. FEBS Lett. 14, 128–130. [DOI] [PubMed] [Google Scholar]
- Young, C.W., and Hodas, S. (1964). Hydroxyurea: Inhibitory effect of DNA metabolism. Science 146, 1172–1174. [DOI] [PubMed] [Google Scholar]