Abstract
Selective ubiquitin-mediated proteolysis through the cell cycle controls the availability, and therefore the activity, of several cell proliferation proteins. E2F transcription factors play distinct roles in both proliferating and differentiated cells by regulating gene expression. Here, we report that Arabidopsis AtE2Fc is regulated by a balance between gene expression and ubiquitin-proteasome proteolysis. AtE2Fc degradation implicates the function of the E3 ubiquitin-ligase Skp1, Cullin, F-box (SCFAtSKP2) complex and seems to be dependent on cyclin-dependent kinase phosphorylation. In addition, we found that AtE2Fc degradation is triggered by light stimulation of dark-grown seedlings. Interestingly, the auxin response mutant axr1-12, in which RUB1 modification of the SCF component CUL1 is impaired, shows increased AtE2Fc protein levels, suggesting a dysfunction in the control of AtE2Fc stability. Likewise, overexpression of a stable form of the AtE2Fc protein negatively affects cell division and increases cell size. These effects are mediated, at least in part, by downregulating the cell cycle gene AtCDC6. The negative role of AtE2Fc in gene expression is further supported by the fact that AtE2Fc interacts with plant retinoblastoma-related protein, suggesting that AtE2Fc might form part of a repressor complex. We propose that AtE2Fc might play a role in cell division and during the transition from skotomorphogenesis to photomorphogenesis.
INTRODUCTION
The retinoblastoma E2F pathway plays an important role in the regulation of the cell cycle and in different differentiation processes (Harbour and Dean, 2000; Müller et al., 2001), although its role in differentiated cells remains poorly understood. E2F proteins interact with dimerization proteins to form the active transcription factors (E2F factors) that modulate gene expression. E2F activity, in turn, is modulated by the retinoblastoma family members that contribute to the formation of repressor complexes. The retinoblastoma proteins mediate this repression by blocking the activation domain of the E2F proteins and/or by recruiting chromatin-remodeling factors to the E2F sites of the gene promoters (for review, see Müller and Helin, 2000). This repression is relieved when the retinoblastoma protein is phosphorylated by cyclin-dependent kinase (CDK) activities (Cooper and Shayman, 2001).
In plants, the retinoblastoma-related (RBR)-E2F pathway components, as well as the majority of the cell cycle regulators, also are present, but very little is known about their function and regulation during plant growth and development (Gutierrez, 1998; Meijer and Murray, 2000; Vandepoele et al., 2002). E2F family proteins have been identified in different plant species (Ramirez-Parra et al., 1999; Sekine et al., 1999; Albani et al., 2000), and in some cases, plant E2Fs have been shown to transactivate E2F-regulated markers in planta (Egelkrout et al., 2001; Kosugi and Ohashi, 2002a). In the model plant Arabidopsis, six different E2Fs and two dimerization proteins (DPa and DPb) have been identified (de Jager et al., 2001; Vandepoele et al., 2002). Three Arabidopsis E2Fs (E2Fa, E2Fb, and E2Fc, also named E2F3, E2F1, and E2F2, respectively) share their domain organization with human E2F1, E2F2, and E2F3. Recently, De Veylder et al. (2002) showed that overexpression of AtE2Fa/DPa produces ectopic cell division in differentiated cells and also extra DNA replication rounds, depending on the competence of the cells. However, data regarding the role, activity, regulation, or availability of other AtE2Fs are lacking.
The ubiquitin-proteasome pathway is emerging as a widespread mechanism to control developmental programs, hormone signaling, and stress responses, among others processes, by targeted proteolysis of key regulatory proteins (Patton et al., 1998; Deshaies, 1999; Callis and Vierstra, 2000; Hellmann and Estelle, 2002). In particular, ubiquitin-mediated degradation plays a crucial role in the control of cell cycle progression and exit (Bartek and Lukas, 2001; Vodermaier, 2001). Ubiquitin is attached covalently to the target protein in a sequential reaction that involves the activity of the E1, E2, and E3 enzymes (Deshaies, 1999). The Skp1, Cullin, F-box (SCF) complex is one type of E3 enzyme that is composed of a structural core (SKP1, Cullin/Cdc53, and RBX/ROC1 proteins) and an F-box protein, which contains an F-box motif that is required to interact with SKP1 (Deshaies, 1999; Schulman et al., 2000). Different F-box proteins can assemble into the core and are responsible for target recognition, conferring specificity to the complexes. The majority of the F-box proteins contain WD40 repeats or Leu-rich repeats that are involved in specific target interaction (Deshaies, 1999).
In this work, we focused our studies on the regulation of AtE2Fc, also known as AtE2F2 (At1g47870). We found that AtE2Fc is cell cycle regulated and is expressed in meristematic areas and early-differentiated cells during plant development. Overexpression of a stable form of AtE2Fc affects cell division and size and reduces the expression of the cell cycle gene AtCDC6. In vitro data indicate that AtE2Fc activity is likely regulated by plant RBR interaction and by AtCDKA;1 phosphorylation. In addition, we found that AtE2Fc is regulated by proteolysis through the ubiquitin-proteasome pathway, implicating the function of the SCFAtSKP2 complex. Interestingly, AtE2Fc is degraded during the transition from dark to light, suggesting a novel role of this protein in this process. Finally, we have found that efficient AtE2Fc degradation requires the modification of AtCUL1, a component of the SCF, with RUB.
RESULTS
AtE2Fc Is Cell Cycle Regulated and Is Expressed in Dividing and Differentiated Cells
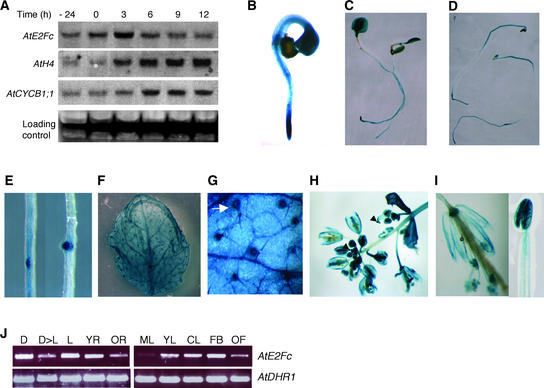
To gain knowledge of AtE2Fc function, we first decided to analyze the expression pattern of this gene. We analyzed Arabidopsis cultured cells partially synchronized with aphidicolin, a drug that blocks cell proliferation in G1/S. We found that AtE2Fc mRNA accumulated during early S-phase, with a peak at 3 h after aphidicolin removal (Figure 1A). To assess how aphidicolin-treated cells progressed through the cell cycle after drug removal, we hybridized the blot with the histone H4 probe, as a marker of S-phase cells (Reichheld et al., 1998), and with the cyclin B1;1, as a marker of G2/M (Hemerly et al., 1992). The histone H4 mRNA accumulated soon after drug removal, and the AtCYCB1;1 mRNA accumulated later, suggesting that the cells were at least partially synchronized, as has been shown recently (Menges and Murray, 2002). Our results are in agreement with previous results showing that AtE2Fc is cell cycle regulated (de Jager et al., 2001; Mariconti et al., 2002).
Figure 1.
AtE2Fc mRNA Accumulated during S-Phase as Well as in Meristems and Specialized Cells during Plant Development.
(A) Two-day-old cells (−24 h) were treated with aphidicolin (8 μg/mL) for 24 h (0 h). After aphidicolin removal, the cells were grown for 3, 6, 9, and 12 h. Total RNA from these samples was analyzed by RNA gel blotting using AtE2Fc, histone H4, and AtCYCB1;1 probes. As a loading control, a portion of gel stained with ethidium bromide is shown. PNE2Fc:N-GUS seedlings were analyzed for GUS activity at different stages of development. Several independent lines were analyzed, and representative images are shown.
(B) PNE2Fc:N-GUS seedling grown in continuous light for 1.5 days.
(C) PNE2Fc:N-GUS seedling grown in continuous light for 5 days.
(D) 5-day-old dark-grown PNE2Fc:N-GUS seedling.
(E) GUS staining of lateral roots of an auxin-treated PNE2Fc:N-GUS seedling.
(F) Mature rosette leaf of a 2-week-old PNE2Fc:N-GUS plant stained for GUS activity.
(G) Magnification of a mature leaf showing GUS staining in the base of trichome cells. The arrow indicates a trichome cell.
(H) Flower bud of an PNE2Fc:N-GUS plant stained for GUS activity. Arrowheads indicate young flower buds.
(I) Mature flower stained for GUS activity. A stamen is shown in detail at right.
(J) AtE2Fc expression was analyzed by semiquantitative reverse transcriptase–mediated PCR. As a control, AtDHR1 was used. Samples examined were as follows: D, seedlings grown for 3 days in the dark; D>L, seedlings grown for 3 days in the dark and then transferred to the light for 7 h; L, seedlings grown for 3 days in the light; YR, young roots from 3-day-old seedlings; OR, old roots from 10-day-old seedlings; ML, mature leaves from 2-week-old plants; YL, youngest leaves from 2-week-old plants; CL, cauline leaves; FB, young flower buds; OF, old flowers.
To gain insight into the possible role of AtE2Fc during plant growth and development, we studied the spatial expression pattern of this gene. First, we tried to generate transgenic plants harboring the 1.2-kb 5′ region upstream from the ATG fused to the reporter gene β-glucuronidase (GUS). We did not recover any plant that was positive for GUS activity. Because it has been suggested that some promoters must contain the first intron to be functional (Gidekel et al., 1996; Morello et al., 2002), we generated a construct that contained the 1.2 kb upstream from the ATG and the genomic region from the ATG to the second in-frame ATG (PNE2Fc:N-GUS), which includes the first intron. We recovered several lines, and five independent lines, all of them showing a similar staining pattern, were analyzed in detail for GUS activity. Histochemical analysis of PNE2Fc:N-GUS transgenic seedlings showed that GUS activity was high in actively dividing cells but also in differentiated and specialized cells, such as root cells, dark-grown hypocotyl cells, and trichomes. GUS activity was strikingly high in young seedlings, especially in the cotyledons and the shoot and root meristems (Figure 1B).
When we analyzed GUS activity in older seedlings, we found that the activity was restricted to the meristematic areas, the vascular tissue, and the apical one-third of the root, with high levels in the root tip (Figure 1C). In dark-grown seedlings, GUS activity was high in cotyledons, along the root, and in the upper region of the hypocotyl (Figure 1D). AtE2Fc also was expressed in the early stages of lateral root development (Figure 1E). When we analyzed 2-week-old rosette leaves, we found slight GUS activity in the leaves, with high staining in the vascular tissue and in the specialized trichome cells (Figures 1F and 1G). We also found activity of the reporter gene during flower development, and in particular in young and developing flower buds (Figure 1H). In mature flowers, however, GUS activity was restricted to pollen grains and to a lesser extent to the vascular tissue (Figure 1I). The expression pattern of AtE2Fc was analyzed further at the mRNA level in different organs by semiquantitative reverse transcriptase–mediated PCR (Figure 1J). We found that AtE2Fc mRNA accumulated to high levels in young dark- and light-grown seedlings and was reduced when the seedlings were transferred from the dark to the light. However, the levels of AtE2Fc mRNA were lower in mature leaves than in young leaves or cauline leaves. We also found higher levels in young flower buds than in old, mature flowers. This expression pattern seems to correlate with the pattern found for GUS activity.
AtE2Fc Is Regulated by the Ubiquitin-Proteasome Pathway
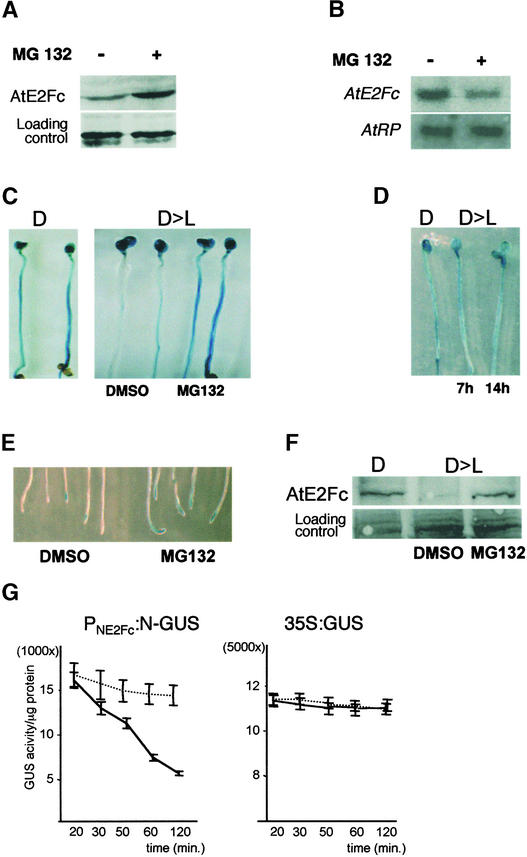
Several factors are regulated by a balance between gene expression and ubiquitin-mediated proteolysis during the cell cycle (Bartek and Lukas, 2001; Vodermaier, 2001). Thus, we studied whether AtE2Fc protein might be regulated through this pathway. To address this issue, we analyzed AtE2Fc protein levels in cultured cells after treatment with the proteasome inhibitor MG132, which is known to block the degradation of ubiquitin-labeled proteins in plants (Callis and Vierstra, 2000; Gray et al., 2001). Addition of the proteasome inhibitor MG132 provoked a striking increase in AtE2Fc protein levels (Figure 2A). This increase in AtE2Fc protein levels was not attributable to an increase in gene transcription, as shown by RNA gel blot hybridization (Figure 2B). These results suggest that AtE2Fc stability is regulated by ubiquitin-dependent degradation.
Figure 2.
AtE2Fc Is Regulated through the Ubiquitin-Proteasome Pathway.
(A) After Arabidopsis cultured cells were synchronized with aphidicolin, the cells were treated with the proteasome inhibitor MG132 or with the solvent DMSO for 3 h. Total protein extracts from these samples were analyzed by protein gel blot hybridization using affinity-purified anti-AtE2Fc IgGs. A portion of the membrane, stained with Ponceau-S dye, is shown as a loading control.
(B) RNA gel blot of total RNA from Arabidopsis cultured cells treated with (+) or without (−) MG132 at 3 hours after aphidicolin synchronization. The blot was hybridized with AtE2Fc and loading control (AtRP) probes.
(C) PNE2Fc:N-GUS seedlings were grown for 3 days in the dark (D) and then stained for GUS activity (left). The image at right shows GUS staining of dark-grown PNE2Fc:N-GUS seedlings that were transferred to light for 7 h (D>L) in the presence of DMSO or the proteasome inhibitor MG132 in the medium.
(D) 35S:GUS seedlings were grown as described above (D and D>L) in the presence of DMSO and then stained for GUS activity. In this case, seedlings were transferred to the light for 7 or 14 h.
(E) Roots of 3-day-old PNE2Fc:N-GUS seedlings that were incubated with DMSO or MG132 for 8 h and stained afterward for GUS activity.
(F) Wild-type seedlings were grown for 3 days in the dark (D) and then transferred to the light (D>L) in the presence of DMSO or MG132 for 7 h. Total protein was extracted from excised hypocotyls and analyzed by protein gel blot hybridization using anti-E2Fc. A region of the Coomassie blue–stained blot is shown as a loading control.
(G) Time course of NE2Fc-GUS degradation. Total protein extracts from PNE2Fc:N-GUS or 35S:GUS calli were incubated in the presence of DMSO or MG132 in the degradation assay. Values for remnant GUS activity after the incubation time represent means of three independent assays. Continuous line, DMSO; broken line, MG132.
The N-terminal region of the human E2F1 protein has been implicated in the control of ubiquitin-mediated degradation (Marti et al., 1999). To determine whether the N-terminal region of AtE2Fc was capable of mediating its ubiquitin-mediated proteolysis, we analyzed the stability of the NE2Fc-GUS fusion protein, which contained the first 100 amino acids of AtE2Fc fused to the GUS protein, expressed under the control of the AtE2Fc promoter (PNE2Fc:N-GUS). We observed that the GUS staining in the hypocotyls of dark-grown seedlings was reduced dramatically when these seedlings were transferred to light for 7 h (Figure 2C). This phenotype was observed in at least five independent PNE2Fc:N-GUS lines. To determine whether this degradation occurred in a ubiquitin-dependent manner, we added the proteasome inhibitor MG132 to the medium during light stimulation. Under these conditions, we found that MG132, but not the DMSO solvent, blocked the reduction of GUS staining in the hypocotyl (Figure 2C). Consistent with the described stability of the GUS protein, we found that GUS activity in the hypocotyls of 35S:GUS transgenic seedlings did not decrease in response to the light stimulation (Figure 2D), indicating that the degradation of the NE2Fc-GUS protein was dependent on the AtE2Fc moiety. Therefore, these results suggest that the NE2Fc-GUS fusion protein was degraded through the ubiquitin-proteasome pathway in response to changes in light. Ubiquitin-dependent degradation was not restricted to the hypocotyl, because stabilization of the NE2Fc-GUS protein by MG132 also occurred in the root tip (Figure 2E). To confirm these results, total hypocotyl protein extracts were analyzed with affinity-purified anti-AtE2Fc IgGs. AtE2Fc protein levels were reduced in the hypocotyls of light-stimulated seedlings but not when the MG132 inhibitor was added to the medium (Figure 2F).
To analyze the stability of the NE2Fc-GUS fusion protein in actively dividing cells, we generated calli lines from PNE2Fc:N-GUS and 35S:GUS transgenic seedlings. Cell-free protein extracts from these calli lines were prepared and used to perform a time-course assay of GUS activity in the presence or in the absence of the proteasome inhibitor MG132. Figure 2G shows that GUS activity in the PNE2Fc:N-GUS extract decreased with time, with an estimated half-life of activity of ∼60 min, and that this reduction was largely prevented by the addition of MG132 to the reaction. Control assays performed with cell-free protein extracts from 35S:GUS calli showed that, in this time-course assay, GUS activity did not change and was independent of MG132 treatment.
Overexpression of AtE2Fc Affects Cell Division and Cell Morphogenesis
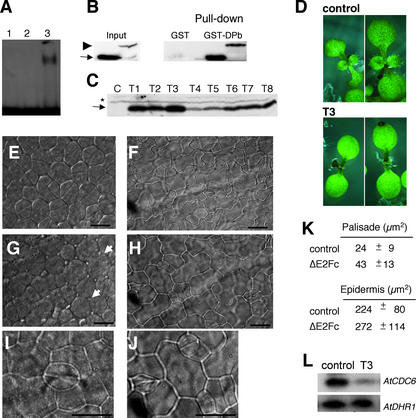
To date, a T-DNA knockout mutant for AtE2Fc is not available in the different collections. Therefore, to gain insight into the cellular role of AtE2Fc, we decided to generate Arabidopsis plants that overexpress this gene. We were unable to recover transgenic lines that accumulated detectable levels of the full-length AtE2Fc protein. Thus, we chose to express a truncated form of AtE2Fc (ΔE2Fc) that lacked the N-terminal region that seems to be involved in regulating AtE2Fc protein stability. However, this ΔE2Fc protein retained other activities, such as binding to a DNA sequence that contains a canonical E2F site (Figure 3A) and its ability to interact with AtDPb protein (Figure 3B) and with AtCDKA;1 protein (Figure 4C). These data indicate that ΔE2Fc is likely a functional protein in vivo. We isolated >20 independent kanamycin-resistant lines that expressed the ΔE2Fc transgene to different extents (Figure 3C). When we analyzed these ΔE2Fc-overexpressing lines, we found no severe changes in morphology with respect to the control plants, except a slight delay in the development of the primary true leaves (Figure 3D). However, microscopic analysis revealed that, in contrast to control plants (Figure 3E), cotyledon palisade parenchyma cells of dark-grown ΔE2Fc transgenic seedlings (Figure 3G) were more irregular in shape and, in many cases, lacked the division plane (corresponding to the last division in most cells). In addition, these ΔE2Fc palisade cells were almost double the size of control cells (Figure 3K). We also analyzed the size and shape of the epidermal cells of seedlings grown in the dark. In this case, ΔE2Fc epidermal cells were larger than the control cells (Figures 3F, 3H, and 3K). Furthermore, the development of the guard cells seemed to be delayed by the overexpression of ΔE2Fc (Figures 3I and 3J). These observations suggest a negative effect of ΔE2Fc overexpression on cell division and might explain the delayed development of the primary true leaves in the transgenic plants (Figure 3D).
Figure 3.
Overexpression of a Stable AtE2Fc Protein Affects Cell Division and Cell Shape.
(A) Electrophoretic mobility shift assay was performed with purified His-ΔE2Fc and GST-DPb using an oligonucleotide containing the consensus E2F motif as described (Ramirez-Parra et al., 1999). Lane 1, GST-DPb alone; lane 2, His-ΔE2Fc alone; lane 3, GST-DPb and His-ΔE2Fc.
(B) Purified His-E2Fc or His-ΔE2Fc were incubated with GST-DPb bound to beads. E2Fc proteins were detected with the IgGs against the AtE2Fc protein. The arrow points to the His-ΔE2Fc protein, and the arrowhead points to the His-E2Fc protein.
(C) Protein gel blot analyses of several independent plants that overexpress the ΔE2Fc protein using the affinity-purified IgGs against AtE2Fc. Control (C) and ΔE2Fc-overexpressing lines (T1 to T8) are indicated. The arrow points to the truncated ΔE2Fc protein, and the asterisk indicates the wild-type AtE2Fc protein. Additional microscopy analyses were performed on the transgenic T3 line.
(D) Five-day-old light-grown control and ΔE2Fc seedlings.
(E) Palisade parenchyma cells from the cotyledons of 3-day-old control dark-grown seedlings. Bar = 10 μm.
(F) Epidermal cells from the cotyledons of 3-day-old control dark-grown seedlings. Bar = 50 μm.
(G) Palisade parenchyma cells from the cotyledons of 3-day-old ΔE2Fc dark-grown seedlings. Arrows indicate cells that showed an uncompleted division plane. Bar = 10 μm.
(H) Epidermal cells from the cotyledons of 3-day-old ΔE2Fc dark-grown seedlings. Bar = 50 μm.
(I) Typical guard cell found in the cotyledons of control dark-grown seedlings. Bar = 10 μm.
(J) Guard cells found in the cotyledons of ΔE2Fc dark-grown seedlings. Bar = 10 μm.
(K) Mean values of the areas of palisade and epidermal cells from 3-day-old control and ΔE2Fc dark-grown seedlings.
(L) Semiquantitative reverse transcriptase–mediated PCR of AtCDC6 using RNA extracted from 3-day-old dark-grown control and ΔE2Fc seedlings. As a control, we used the constitutively expressed AtDHR1 gene.
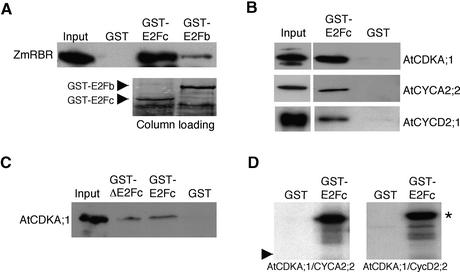
Figure 4.
AtE2Fc Interacts with RBR and with AtCDKA;1, CYCA2;2, and CYCD2;1.
(A) Purified recombinant His-ZmRb protein was incubated with GST, GST-AtE2Fc, or GST-AtE2Fb bound to beads. ZmRb was detected with IgGs against the ZmRb protein. A region of the Coomassie blue–stained blot is shown as a loading control in the GST-E2Fc and GST-E2Fb columns. Note that the purified proteins contained degradation products of these fusion proteins.
(B) Insect cell extracts containing AtCDKA;1, His-AtCYCA2;2, or His-AtCYCD2;1 were incubated with purified GST or GST-AtE2Fc proteins bound to beads. AtCDKA;1 was detected by protein gel blot analysis using the anti-PSTAIRE antibody, and CYCA2;2 and CYCD2;1 were detected with anti-His antibody.
(C) Protein extracts from 3-day-old light-grown wild-type seedlings were incubated with purified GST, GST-E2Fc, or GST-ΔE2Fc bound to beads. Arabidopsis AtCDKA;1 was detected using the anti-PSTAIRE antibody.
(D) Purified GST or GST-AtE2Fc was incubated in a kinase reaction with the AtCDKA;1/CYCA2;2 or AtCDKA;1/CYCD2;1 complex in the presence of γ-32P-ATP. The asterisk indicates the phosphorylated form of AtE2Fc. The arrowhead indicates the mobility of the GST.
To analyze the effect of ΔE2Fc overexpression at the molecular level, we analyzed by semiquantitative reverse transcriptase–mediated PCR the expression of the cell division marker AtCDC6, which contains an E2F element in its promoter (Castellano et al., 2001). Figure 3L shows that dark-grown ΔE2Fc-overexpressing seedlings expressed lower levels of AtCDC6 mRNA than control seedlings. Together, our data indicate that AtE2Fc may act as a repressor of genes required for normal proliferation and therefore negatively affects cell division.
AtE2Fc Interacts with RBR and Is Regulated by CDK Phosphorylation
The ability of E2F/dimerization protein factors to modulate gene expression is regulated, in part, by the interaction with retinoblastoma family proteins through binding to a C-terminal motif in E2F and blocking its transactivation domain. Analysis of the AtE2Fc sequence revealed that it lacks a putative transactivation domain; therefore, it is conceivable that it may act as a repressor rather than an activator. Although AtE2Fc showed a putative retinoblastoma binding motif at the C-terminal region, it has been reported that AtE2Fc did not interact with ZmRb in the two-hybrid system (de Jager et al., 2001). To evaluate the possible interaction between these proteins in a different system, we used recombinant purified proteins. Under these conditions, we found that AtE2Fc and a plant RBR protein (ZmRb) can interact directly (Figure 4A). By contrast, AtE2Fb (also named AtE2F1), which interacts with ZmRb in the yeast two-hybrid system (de Jager et al., 2001), bound with less efficiency to ZmRb (Figure 4A). These results suggest that AtE2Fc might form a repressor complex in cooperation with RBR.
The transition from G1- to S-phase is tightly regulated by CDK activities. Computer assisted analysis of the AtE2Fc sequence (http://www.cbs.dtu.k/services/NetPhos/) revealed that it contains several putative CDK phosphorylation sites. The majority of these sites are located in the N-terminal region, although some of them are distributed at the heterodimerization and retinoblastoma binding motifs (data not shown). To address whether Arabidopsis CDK activity might regulate AtE2Fc, we first analyzed the interaction between Arabidopsis AtCDKA;1, cyclin A, and cyclin D2 proteins with AtE2Fc. We performed pulldown experiments using purified glutathione S-transferase (GST)-AtE2Fc or GST bound to beads and AtCDKA;1, AtCYCA2;2, or CYCD2;1 proteins expressed in baculovirus. In vitro binding assays showed that AtE2Fc interacted physically with these three proteins (Figure 4B). Furthermore, we found that AtE2Fc, and also the truncated form AtΔE2Fc, interacted with AtCDKA;1 from plant extracts (Figure 4C), indicating that these proteins might interact in vivo. Likewise, the GST-AtE2Fc protein was phosphorylated in vitro by both AtCDKA;1/CYCA2;2 and AtCDKA;1/CYCD2;1 complexes (Figure 4D).
The F-Box SKP2 Forms Part of an SCF Complex and Recruits AtE2Fc
Because AtE2Fc seems to be regulated through the ubiquitin pathway, we investigated the mechanisms that underlie this degradation, in particular, identifying the F-box protein responsible for targeting AtE2Fc. Using human SKP2, the F-box protein that recruits E2F1, as a query to search the Arabidopsis database, we retrieved several proteins with significant homology, but two of them, which we named AtSKP2;1 (At1g21410) and AtSKP2;2 (At1g77000), showed the highest homology with the substrate recognition C-terminal Leu-rich repeat region of human SKP2. The two AtSKP2 proteins shared 83% identity in their primary amino acid sequences and 44% identity within their putative promoter DNA sequences, suggesting that they might be duplicated genes. In fact, a search of the MIPS database (http://mips.gsf.de/proj/thal/db/main.html) revealed that both AtSKP2;1 and AtSKP2;2 were located in a duplicated region of chromosome 1. These proteins contain an F-box motif at the N-terminal region and a Leu-rich repeat domain along the C-terminal portion of the protein. Here, we focused our studies on AtSKP2;1 (which we refer to as AtSKP2 throughout).
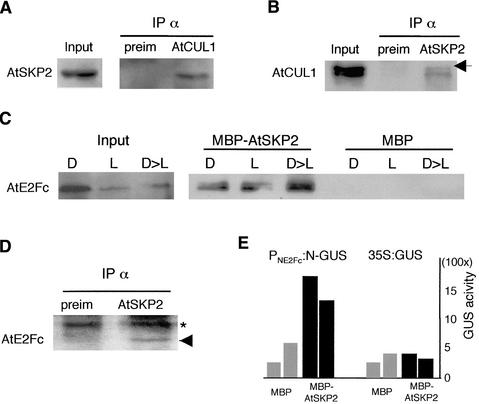
In vitro assays and crystallization studies have demonstrated that the F-box proteins assemble into the SCF core through direct interaction with the SKP1 protein, which interacts with the CUL1 protein (Schulman et al., 2000; Zheng et al., 2002). Therefore, to determine whether AtSKP2 is integrated into an SCF complex, we used an AtCUL1 serum (Gray et al., 1999) to perform immunoprecipitation assays. Figure 5A shows that AtSKP2 coimmunoprecipitated with AtCUL1, suggesting that AtSKP2 is part of an SCF complex. Likewise, when the immunoprecipitation was carried out using the anti-AtSKP2 serum, we found that AtSKP2 interacted with both the RUB1-modified and the nonmodified isoforms of AtCUL1 (Figure 5B). Because Arabidopsis AtSKP2 forms part of an SCF complex and AtE2Fc seems to be regulated by the ubiquitin-proteasome pathway, we wondered whether this F-box protein would be responsible for targeting AtE2Fc for ubiquitylation. To address this possibility, we analyzed the interaction between both proteins using pulldown or immunoprecipitation assays. First, we performed pulldown experiments using protein extracts from 3-day-old seedlings grown in the dark, grown in the light, or grown in the dark and then stimulated with light for 7 h. We found that recombinant maltose binding protein (MBP)-AtSKP2, but not MBP alone, recruited AtE2Fc in these three conditions, but the interaction between MBP-AtSKP2 and AtE2Fc was favored in the light-stimulated protein extract (Figure 5C). This interaction also was detected in vivo by coimmunoprecipitation analysis using the anti-AtSKP2 serum. Figure 5D shows that AtSKP2 serum, but not the preimmune serum, immunoprecipitated AtE2Fc, suggesting that the F-box AtSKP2 might be responsible for targeting AtE2Fc for specific degradation. Furthermore, using extracts of PNE2Fc:N-GUS callus, we found that MBP-AtSKP2 pulled down higher GUS activity than the MBP control (Figure 5E). However, MBP-AtSKP2 did not pull down significant GUS activity when 35S:GUS extracts were used (Figure 5E), suggesting that the N-terminal region of AtE2Fc is important for both AtSKP2 binding and ubiquitin-mediated degradation.
Figure 5.
AtSKP2, an F-Box Protein, Recruits AtE2Fc.
(A) Total protein extracts from 3-day-old light-grown seedlings were immunoprecipitated (IP) with preimmune or anti-AtCUL1 sera and analyzed by protein gel blotting using affinity-purified anti-AtSKP2 IgGs.
(B) Total protein extracts were immunoprecipitated with preimmune or anti-SKP2 sera. Precipitated proteins were analyzed using anti-AtCUL1 serum. The arrow points to the RUB1-modified form of AtCUL1 (del Pozo and Estelle, 1999).
(C) Total protein extracts from 3-day-old dark-grown seedlings (D), light-grown seedlings (L), or seedlings transferred from the dark to the light for 7 h (D>L) were used in pulldown experiments in which MBP or MBP-AtSKP2 was bound to beads and then analyzed by immunoblotting using anti-E2Fc IgGs.
(D) Coimmunoprecipitation assays using rabbit preimmune or anti-AtSKP2 sera. The precipitated proteins were analyzed by protein gel blotting using affinity purified anti-AtE2Fc IgGs. The arrowhead points to the AtE2Fc protein, and the asterisk indicates the IgG band.
(E) PNE2Fc:N-GUS or 35S:GUS calli protein extracts were incubated with MBP and MBP-AtSKP2 proteins bound to beads. GUS activity of the pulled down proteins was quantified in two independent experiments.
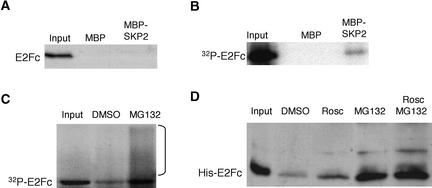
AtSKP2 Recruits Phosphorylated AtE2Fc
The SCF paradigm predicts that the F-box protein recruits phosphorylated targets. Because AtE2Fc is phosphorylated by the AtCDKA;1/CYC complexes in vitro, we wanted to investigate the importance of AtE2Fc phosphorylation for AtSKP2 recruitment. First, we analyzed the interaction between recombinant AtE2Fc and AtSKP2 proteins expressed in bacteria. Under this condition, in the absence of any CDK activity, we detected no interaction between these proteins (Figure 6A). However, when GST-AtE2Fc was phosphorylated by the AtCDKA;1/CYCA2;2 complex, we found that 32P–GST-AtE2Fc was recruited by AtSKP2 (Figure 6B). Similar results were obtained when AtE2Fc was phosphorylated with the AtCDKA;1/CYCD2;1 complex (data not shown). These results strongly suggest that CDKA-dependent phosphorylation is a limiting step in the targeting of AtE2Fc by the SCFAtSKP2 complex. To determine whether phosphorylation of AtE2Fc is required for its ubiquitin-mediated degradation, we used phosphorylated AtE2Fc in a cell-free degradation assay. We found that 32P–His-AtE2Fc was degraded efficiently (Figure 6C) and that the addition of MG132 blocked 32P–His-AtE2Fc degradation, concomitant with the appearance of high molecular mass species that likely correspond to ubiquitylated forms of AtE2Fc (Figure 6C). In addition, and supporting these results, ubiquitin-mediated degradation of the recombinant His-AtE2Fc protein in a cell-free assay was reduced when roscovitine, a CDK activity inhibitor, was added to the extract and was blocked efficiently by the addition of the proteasome inhibitor MG132 (Figure 6D).
Figure 6.
AtSKP2 Recruits Phosphorylated AtE2Fc.
(A) Recombinant purified GST-AtE2Fc was incubated with MBP or MBP-AtSKP2 bound to beads. The pulled down proteins were analyzed by protein gel blot hybridization with anti-E2Fc IgGs.
(B) Phosphorylated 32P–GST-AtE2Fc was incubated with MBP or MBP-AtSKP2 in pulldown experiments.
(C) 32P–His-AtE2Fc was incubated in a degradation assay for 10 min in the presence of DMSO or MG132 (50 μM). The bracket indicates high molecular mass species of radiolabeled 32P–His-AtE2Fc.
(D) Recombinant His-AtE2Fc was incubated in a degradation assay for 20 min. Where indicated, the cell-free protein extract was preincubated for 5 min with 50 μM roscovitine (Rosc), 50 μM MG132, or both before adding the recombinant His-AtE2Fc protein.
RUB1 Modification of AtCUL1 Is Required for AtE2Fc Degradation
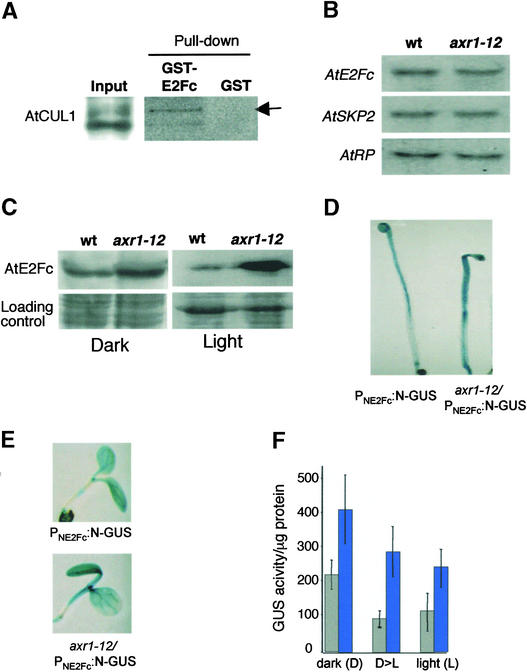
SCF complex activity is regulated through different pathways. In plants, RUB modification of the structural component of the SCF AtCUL1, which is dependent on AXR1 protein function (del Pozo et al., 2002), is required for normal response to the hormone auxin (del Pozo et al., 1998; Gray and Estelle, 2000). Previously, we showed that AtSKP2 interacted with both the RUB-modified and nonmodified isoforms of AtCUL1 (Figure 5B). However, when we performed pulldown experiments using GST-AtE2Fc, we found that the RUB1-modified isoform of AtCUL1 was pulled down preferentially (Figure 7A). Because it is well known that auxin controls cell division, cell expansion, and several aspects of plant development, and that the auxin response mutant axr1 affects SCF complex activity, we determined whether RUB modification of AtCUL1 might have an effect on AtE2Fc stability. To address this possibility, we first compared the mRNA and protein levels of AtE2Fc in wild-type and axr1-12 seedlings, in which RUB modification is impaired (del Pozo et al., 2002). AtE2Fc and AtSKP2 mRNA levels were similar in both wild-type and axr1-12 seedlings (Figure 7B). However, AtE2Fc protein levels were strikingly higher in axr1-12 than in wild-type seedlings grown in the light for 3 days, whereas these levels were only slightly higher when the seedlings were grown in the dark (Figure 7C). Together, these results indicate that AXR1-dependent modification of AtCUL1 with RUB is required for the efficient degradation of AtE2Fc. Because the NE2Fc-GUS fusion protein is regulated through the ubiquitin pathway, we wondered whether this fusion protein also was more stable in the axr1-12 mutant background. We found that GUS staining levels of dark- or light-grown axr1-12/PNE2Fc:N-GUS were higher than those in PNE2Fc:N-GUS seedlings (Figure 7F), especially in the hypocotyl (Figures 7D and 7E), indicating that the axr1-12 mutation affects the degradation of AtE2Fc.
Figure 7.
RUB1 Modification of AtCUL1 Is Necessary for Efficient AtE2Fc Degradation.
(A) Total protein extracts from 3-day-old light-grown seedlings were incubated with GST or GST-AtE2Fc bound to beads. The precipitated proteins were analyzed by protein gel blotting using anti-AtCUL1 serum. The arrow points to the RUB-modified form of AtCUL1. Note that the gel showing the pulled down proteins was exposed five times longer than the input gel.
(B) Wild-type (wt) and axr1-12 seedlings were germinated for 3 days in continuous light. Total RNA was extracted and analyzed by RNA gel blot hybridization using the AtE2Fc or AtSKP2 probe. The AtRP probe was used as a loading control.
(C) Wild-type and axr1-12 seedlings were germinated in the dark or in the light for 3 days. Total protein was extracted and analyzed by protein gel blotting using affinity purified anti-AtE2Fc IgGs. A portion of these blots stained with Ponceau-S solution are shown as a loading control.
(D) PNE2Fc:N-GUS and axr1-12/PNE2Fc:N-GUS seedlings were grown for 3 days in the dark and then stained for GUS activity.
(E) PNE2Fc:N-GUS and axr1-12/PNE2Fc:N-GUS seedlings were grown for 5 days in the light and then stained for GUS activity.
(F) Total protein was extracted from PNE2Fc:N-GUS and axr1-12/PNE2Fc:N-GUS seedlings grown for 3 days in the dark (D), 5 days in the light (L), or 3 days in the dark before transfer to the light for 7 h (D>L), and GUS activities were quantified. Gray bars, PNE2Fc:N-GUS; blue bars, axr1-12/PNE2Fc:N-GUS.
DISCUSSION
The components of the RBR-E2F pathway have been identified in plants. The recent completion of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) has allowed the identification of six different E2F family members. AtE2Fc shows a domain organization similar to that of human E2F1, E2F2, and E2F3 containing a DNA binding motif, a heterodimerization domain, a retinoblastoma binding motif, and a marked box (de Jager et al., 2001; Mariconti et al., 2002; Vandepoele et al., 2002). Although AtE2Fa and AtE2Fb interact with ZmRb in the two-hybrid system, AtE2Fc does not (de Jager et al., 2001). Nevertheless, using recombinant proteins, we detected a more efficient interaction between ZmRb and AtE2Fc than between ZmRb and AtE2Fb. This discrepancy might be explained by an AtE2Fc-mediated repression of the two-hybrid markers. In fact, as with the repressor Drosophila E2F2 (Trimarchi et al., 2001), AtE2Fc does not show a typical transactivation domain and does not transactivate in yeast (de Jager et al., 2001), suggesting a repressor function of AtE2Fc. Although the overexpression of AtE2Fc in cultured cells slightly transactivates E2F-regulated markers (Kosugi and Ohashi, 2002a), it is not clear whether AtE2Fc activates the E2F markers directly or whether it has a dominant-negative effect, titrating E2F-interacting proteins, such as RBR, and indirectly promoting the function of other E2Fs. It also has been reported that the E2F sites located in the PCNA promoter mediate transcriptional activation in proliferating cells and repression in differentiated tissues (Egelkrout et al., 2001; Kosugi and Ohashi, 2002b). These reports indicate that different E2F activities (activators and/or repressors) function in relation to cell status and plant development.
AtE2Fc is expressed in dividing cells, with high expression in the shoot and root meristems, and also is regulated through the cell cycle, with a peak of expression during S-phase. In addition, AtE2Fc also is expressed in the specialized trichome cells and in differentiated cells throughout plant development. This expression pattern suggests that AtE2Fc might play a role in both dividing and differentiated cells. It is remarkable that the expression pattern of AtE2Fc is similar to that of AtCDC6, an E2F-regulated gene that is implicated in cell proliferation (Castellano et al., 2001). Furthermore, we found that overexpression of AtE2Fc reduces the expression levels of AtCDC6, contrary to the overexpression of AtE2Fa/DPa (De Veylder et al., 2002), supporting a role for AtE2Fc as gene repressor. The repression of cell cycle genes should be important in maintaining a differentiated state and for progress through the cell cycle, because a temporally concerted upregulation and downregulation of certain genes is required. Together, these data indicate that AtE2Fc likely functions as a transcriptional repressor, although we cannot fully exclude the possibility that it might act as an activator, depending on the cellular/environmental context.
Recently, new data regarding Arabidopsis E2F function were obtained by analysis of transgenic plants. Overexpression of AtE2Fa, synergistically with AtDPa, induces ectopic cell division and DNA endoreplication and produces a severe effect on plant development, suggesting that an excess of AtE2Fa/AtDPa affects cell division and differentiation during plant development (De Veylder et al., 2002). These authors conclude that AtE2Fa/AtDPa function as a positive regulator of the cell cycle. By contrast, morphological analysis showed that overexpression of a stable form of AtE2Fc also seems to affect cell division, because the cells appear to be larger and more irregular in shape and many of them lack the division plane, as if the number of cell divisions would have been reduced. Likewise, the negative effect of AtE2Fc on cell division could explain the delayed development of the first true leaves, in which the rate of cell division is important for organ growth (Cockcroft et al., 2000). Together, our data support a negative role of AtE2Fc in cell division. However, further experiments will be needed to determine whether the truncated ΔE2Fc functions as a wild-type protein or a negative- or positive-dominant protein. Analysis of loss-of-function mutants will help to clarify this point; unfortunately, to date, we have been unable to find an AtE2Fc mutant in different collections.
A variety of reports have implicated the ubiquitin-proteasome pathway in the control of eukaryotic cell cycle progression (Krek, 1998; Bartek and Lukas, 2001; Vodermaier, 2001), multiple environmental responses, and developmental processes (Deshaies, 1999; Callis and Vierstra, 2000). Interestingly, we found that the AtE2Fc protein is degraded rapidly in a ubiquitin-dependent manner in response to light, suggesting a possible role of this protein during skotomorphogenesis, likely as a transcriptional repressor. We can speculate that AtE2Fc is required to maintain skotomorphogenesis, during which cell division is prevented. Upon light stimulation, when cell proliferation is reactivated, AtE2Fc (or most of it) is degraded. However, some AtE2Fc activity, consistent with its higher expression in early S-phase, could participate in modulating (likely repressing) the expression of genes that need to be shut off for further progression through the cell cycle. The positive photomorphogenesis regulator HY5 offers the best example of ubiquitin-mediated proteolysis controlled by light (Osterlund et al., 2000). In this case, the COP-signalosome complex plays a crucial role in controlling the stability of HY5 (Osterlund et al., 2000). These data indicate that the ubiquitin-signalosome pathway is important in the regulation of the transition from skotomorphogenesis to photomorphogenesis. Furthermore, recent findings have revealed that the signalosome is involved in removing RUB1 from AtCUL1, a necessary process for efficient AXR2 degradation (Schwechheimer et al., 2001). Interestingly, human CSN1 (a component of the signalosome complex) interacts with SKP2 (Lyapina et al., 2001), suggesting that the activity of SKP2 is related to signalosome activity, which also has been implicated in cell cycle control (Tomoda et al., 2002). In this context, it is noteworthy that the promoter of one of the subunits of the signalosome, Arabidopsis AtCOP9/FUS7, contains an E2F motif, suggesting that this gene might be regulated by E2F activities (E. Ramirez-Parra, C. Fründt, and C. Gutierrez, unpublished data). Therefore, it would be interesting to analyze the role of COP9/FUS7 in cell proliferation and its regulation by Arabidopsis E2F family members.
In the majority of cases, the F-box proteins interact exclusively with the phosphorylated forms of their target proteins (Deshaies, 1999). For example, the SCFcdc4 complex binds exclusively to phosphorylated Sic1 (Feldman et al., 1997; Skowyra et al., 1997). Human E2F1 is phosphorylated by the CDK2/CYCA complex (Yang et al., 1999) and is targeted for ubiquitin-mediated proteolysis by the SCFSKP2 complex (Marti et al., 1999). Nevertheless, whether the phosphorylation of human E2F1 is needed for SCFSKP2 recruitment remains a matter of controversy (Marti et al., 1999), because it has been reported that human E2F1 can be ubiquitylated in a phosphorylation-independent manner (Ohta and Xiong, 2001). We have found that AtE2Fc is phosphorylated by the AtCDKA;1/CYCA2;2 and AtCDKA;1/CYCD2;1 complexes. These in vitro data, together with the fact that AtE2Fc interacts with AtCDKA;1 present in plant extracts, suggest that AtE2Fc activity is regulated by CDK-dependent phosphory-lation in vivo. In fact, AtSKP2 recruits CDKA-dependent phosphorylated AtE2Fc but not the nonphosphorylated form, suggesting that this phosphorylation is necessary to target AtE2Fc for proteolysis. Consistent with this finding, roscovitine, a CDK activity inhibitor, seems to reduce ubiquitin-dependent AtE2Fc proteolysis. The weak reduction in AtE2Fc degradation observed might be explained by a low inhibitory efficiency, because in vitro assays showed that roscovitine inhibited CDKA activity slightly (data not shown). Alternatively, AtE2Fc might be phosphorylated, and targeted subsequently for proteolysis, by other types of kinases, or it might be degraded in a phosphorylation-independent manner, as has been reported for human E2F1 (Ohta and Xiong, 2001). Together, our data support the conclusion that the phosphorylation of AtE2Fc is likely a requirement for its ubiquitin-mediated degradation in vivo.
In plants, one of the best ubiquitin-dependent responses known is auxin signaling. Molecular analyses of the auxin response mutants axr1 and tir1 support a model in which auxin signaling is mediated by an intricate ubiquitin-RUB-SCF pathway (Gray and Estelle, 2000; Leyser, 2001; Rogg and Bartel, 2001; Hellmann and Estelle, 2002). We found that AtE2Fc is stabilized in the axr1-12 mutant. The fact that AtE2Fc is more stable in the axr1-12 mutant and that AtE2Fc interacts preferentially with the modified AtCUL1 isoform strongly suggests that the modification of AtCUL1 with AtRUB1 is important for the efficient SCF-dependent degradation of AtE2Fc. Similarly, the ubiquitin-mediated proteolysis of the auxin response transcription factor AtAXR2/IAA7, which is dependent on the F-box protein AtTIR1, is less efficient in the axr1 mutant background, suggesting that the modification of AtCUL1 with AtRUB1 somehow is necessary for its efficient degradation (Gray et al., 2001). Modification of AtCUL1 with AtRUB1 is a process dependent on the function of the AtAXR1 gene (del Pozo et al., 1998, 2002; del Pozo and Estelle, 1999). This modification of AtCUL1 has been shown to be important for a normal response to the hormone auxin (Gray and Estelle, 2000; del Pozo et al., 2002). Although the mechanism is not clear, the modification of cullin with RUB seems to be involved in regulating the activity and/or the subcellular localization of the SCF complexes (Lammer et al., 1998; Freed et al., 1999; Osaka et al., 2000). Recently, it was suggested that RUB modification is important to recruit the ubiquitin-charged E2 enzyme to the SCF complex and to transfer the ubiquitin moiety to the target proteins efficiently (Kawakami et al., 2001; Zheng et al., 2002). Therefore, it is likely that the reduction of RUB-modified AtCUL1 limits the degradation of AtE2Fc, among other proteins.
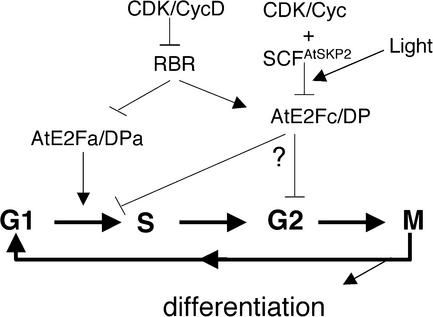
We propose that AtE2Fc, alone and/or in cooperation with RBR, functions as a negative regulator of cell division, repressing the expression of cell division genes. Because cyclin D2 and A function likely is important for G1- and S-phase progression as well as for cell cycle reentry, it is tempting to speculate that the ubiquitin-mediated destruction of AtE2Fc repressor might be necessary for these processes (Figure 8). Likewise, the destruction of AtE2Fc during the transition from dark- to light-grown conditions is necessary to activate the gene expression required for photomorphogenesis (Figure 8).
Figure 8.
Model Showing the Function of AtE2Fc in Relation to Cell Division and Development.
Information for AtE2Fa/DPa is adapted from De Veylder et al. (2002).
METHODS
Plant Material
Arabidopsis thaliana cultured cells were subcultured 1:10 (v/v) in MSC medium (1 × Murashige and Skoog [1962] [MS] salts, 1 × MS vitamins, pH 5.8, 3% Suc, 4.5 μM 2,4-D, and 0.45 μM kinetin). For synchronization experiments, 2 days after subculturing, aphidicolin was added (8 μg/mL) over 24 h. To eliminate the drug, cells were washed twice with 10 volumes of fresh medium. Where indicated, MG132 (Affiniti, Mamhead, UK) was used at 50 μM, and as a control, the same volume of DMSO was added. Arabidopsis seedlings (ecotype Columbia-0) were grown in MSS medium (0.5 × MS salts, pH 5.8, 1 × MS vitamins, 1% Suc, and 1% agar). For MG132 treatment on MS medium plates, the proteasome inhibitor was added fresh when the agar was almost solidified, and the plates were used immediately.
Transgenic Plants and Calli Lines
To generate PNE2Fc:N-GUS, a 1.8-kb genomic region containing the promoter region of AtE2Fc and the first 100 amino acids of the AtE2Fc protein was fused in frame to the GUS marker, using pBI101.2 vector. To generate ΔE2Fc-overexpressing lines, the cDNA coding for amino acids 95 to 457 was cloned in frame with the hemagglutinin epitope using pPily vector (Ferrando et al., 2000). This construction was cloned subsequently into pBin19 binary plasmid. PNE2Fc:N-GUS and ΔE2Fc were introduced into Agrobacterium tumefaciens, which was used to transform Arabidopsis plants (Bechtold and Pelletier, 1998). T3 homozygous lines were selected and used for this work. We further analyzed five independent PNE2Fc:N-GUS lines, which showed a similar GUS staining pattern and similar NE2Fc-GUS stability behavior. As a control, we generated transgenic lines harboring pBin19 plus two copies of the 35S promoter. The PNE2Fc:N-GUS transgene also was introduced into the axr1-12 mutant by genetic crossing, and homozygous plants for the transgene and the mutation were selected (axr1-12/PNE2Fc:N-GUS).
To generate calli lines, PNE2Fc:N-GUS or 35S:GUS seedlings were germinated in the dark in B5 medium (B5 salts plus vitamins [Sigma], pH 5.8, 2% Glc, 4.5 μM 2,4-D, and 0.45 μM kinetin) over 3 weeks. Afterward, the seedlings were sliced into small pieces, which were transferred to fresh B5 medium and grown in the dark for another 3 weeks. Small pieces of the formed calli were transferred to fresh B5 medium every 2 weeks.
DNA and RNA Manipulation
All standard DNA manipulations were performed as described (Sambrook et al., 1989). Total RNA was isolated from cultured cells or Arabidopsis seedlings using Trizol (Gibco BRL). Approximately 25 μg of total RNA was loaded per lane and fractionated on 1% agarose gels containing 2.2 M formaldehyde. The RNA was transferred to nylon membranes by capillary blotting. The blots were prehybridized and hybridized using perfectHyb buffer (Sigma). The blots were hybridized with AtE2Fc histone H4 (Reichheld et al., 1998) and cyclin B1;1 (Hemerly et al., 1992) probes. To normalize the amount of RNA in the blots, AtRP probe (Kim et al., 1990) was used as a loading control. For reverse transcriptase–mediated PCR analysis, 1 μg of total RNA from the different organs and growing conditions tested was used for cDNA synthesis using the Invitrogen kit (Carlsbad, CA). The amount of cDNA synthesized was normalized in both samples using the constitutive expressed gene AtDHR1 (5′-AAGAGGAGCAGA-TATCGTGGTTG-3′ and 5′-TTGTCTCCATGTATAGCAGCAGC-3′) and then used for PCR amplification with the specific primers for AtCDC6 (5′-ACCTACAATGCCTGCAAATCC-3′ and 5′-GCCTTCTTCGCT-CCATACAAC-3′) or AtE2Fc (5′-CAGGTTGGATGATCTTATAAG-GGAACGAC-3′ and 5′-CACATAATTGCCACATCCCTTGAATGACC-3′). The PCR program was as follows: 1 cycle at 94°C for 1 min; 20 cycles at 94°C for 20 s, 55°C for 45 s, and 72°C for 60 s; and 1 cycle at 72°C for 10 min. The amplified DNA was fractionated on a 1.2% agarose gel and transferred to nylon membranes (Sambrook et al., 1989). In the case of semiquantitative reverse transcriptase–mediated PCR to analyze AtCDC6 expression in the transgenic lines, the blots were prehybridized and hybridized using perfectHyb buffer (Sigma) and radiolabeled AtCDC6 or AtDHR1 probe (Okanami et al., 1998; Castellano et al., 2001).
Antibodies and Protein Gel Blot Analysis
AtDPb (At5g03410) was expressed in bacteria as a GST-DPb fusion protein using pGEX-2T vector (Amersham). AtE2Fc (At1g47870) and the N-terminal deleted protein, which lacks the first 100 amino acids, were expressed as GST fusion or His fusion proteins (GST-E2Fc, GST-ΔE2Fc, His-E2Fc, and His-ΔE2Fc) using pGEX-2T or QE (Qiagen, Valencia, CA) vector. AtSKP2 cDNA was cloned into pMal vector, and the protein was expressed as MBP-SKP2 recombinant protein (New England Biolabs, Beverly, MA). All recombinant proteins were expressed in bacteria at 30°C and used to generate immune serum in rats or rabbits. The antiserum against ZmRb was generated in rabbits using the recombinant GST-ZmRBR1A/B pocket (Boniotti and Gutierrez, 2001). Anti-AtE2Fc, anti-AtSKP2, and anti-ZmRb IgGs were affinity purified against the recombinant proteins using a published procedure (Pringle et al., 1989). Protein gel blot analysis was performed in standard conditions using the anti-AtE2Fc IgGs (1:250), anti-AtSKP2 (1:1000), and anti-ZmRb (1:500). To detect HisAtCYCA2;2, HisAtCYCD2;1, or AtCDKA;1, monoclonal anti-His antibodies (1:3000; Sigma) and rabbit polyclonal anti-PSTAIRE antibodies (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA) were used.
For protein gel blot analysis, total proteins of Arabidopsis seedlings or cultures were extracted in cold buffer A (50 mM Tris-HCl, pH 7.0, 50 mM NaCl, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 × protease inhibitor cocktail [Boehringer Mannheim]) and supplemented, where indicated, with 50 μM MG132.
AtCDKA and CYC Expression in Insect Cells and Kinase Assays
The AtCYCA2;2 or AtCYCD2;1 coding region was inserted in frame with a His tail into transfer vector pFastBacHta (HisAtCYCA2;2), whereas the AtCDKA;1 coding region was cloned into vector pFastBac1 (Gibco BRL). Both plasmids were transformed into Escherichia coli strain DH10BAC for transposition into the bacmid. The recombinant bacmids were transfected into HIGH5 insect cells in the presence of LipofectAMINE PLUS reagent (Gibco BRL). Active AtCDKA;1/HisCYCA2;2 complexes were produced in HIGH5 cells that had been coinfected with recombinant baculoviruses. Whole cell lysates were prepared by sonication of infected cells in kinase buffer (20 mM Tris-HCl, pH 8.0, 10 mM MgCl2, and 1 mM EGTA) containing 0.1% Nonidet P-40, 1 mM PMSF, 10 μg/mL aprotinin, 5 μg/mL pepstatin, 10 μg/mL leupeptin, 1 mM NaF, 200 μM sodium orthovanadate, and 2.5 mM sodium pyrophosphate. The extracts were cleared by centrifugation, and 10% glycerol was added.
For the kinase assays, the substrates (1 μg of either purified GST or GST-AtE2Fc) was incubated with AtCDKA;1/HisCYCA2;2 or AtCDKA/HisCYCD2;1 insect cell extracts (2 μL) in a final volume of 15 μL of kinase buffer in the presence of 5 μCi of γ-32P-ATP, 25 μM unlabeled ATP, and 1 mM DTT. Kinase reactions were performed at 30°C for 30 min. Afterward, 0.5 mL of PBS, 0.1% Tween 20, and 25 μL of glutathione beads were added and incubated for 1 h at 4°C. The beads were washed three times for 10 min each with 1 mL of cold PBS and 0.1% Tween 20.
Pulldown and Immunoprecipitation Assays
Total protein of Arabidopsis seedlings was extracted in buffer A plus 50 μM MG132, and after 15 min on ice, the extracts were cleared by centrifugation. To use these extracts for pulldown or immunoprecipitation experiments, we added 100 mM NaCl, and after 10 min on ice, the extracts were cleared again. We incubated 2 mg of total protein with GST or GST-AtE2Fc (1 μg bound to glutathione beads) or with MBP or MBP-AtSKP2 (1 μg bound to amylose beads). To study the interaction between AtCDKA;1, AtCYCA2;2, or AtCYCD2;1 and AtE2Fc, different volumes of insect cell extracts containing equivalent quantities of the expressed proteins (AtCDKA;1 or AtCYCA2;2) in Tris-buffered saline (TBS) and 0.5% Nonidet P-40 were incubated with GST, GST-AtE2Fc, MBP, or MBP-AtSKP2 (1 μg bound to glutathione or amylose beads).
To study the interaction between recombinant AtE2Fc and AtSKP2 proteins, we incubated recombinant GST-AtE2Fc with MBP or MBP-AtSKP2 (1 μg bound to amylose beads) at 4°C over 2 h in TBS and 0.5% Nonidet P-40. GST-AtE2Fc–purified protein was phosphorylated by the AtCDKA;1/CYCA2;2 or AtCDKA;1/CYCD2;1 complex and then was incubated with 1 μg of MBP or MBP-AtSKP2 bound to amylose in 0.5 mL of TBS and 0.5% Nonidet P-40 for 2 h at 4°C.
Total protein extracts from PNE2Fc:N-GUS and 35S:GUS calli were prepared in TBS and 0.5% Nonidet P-40. Approximately 1 × 105 units of GUS activity was incubated with MBP or MBP-AtSKP2 (1 μg bound to amylose beads) for 2 h at 4°C. After the washing steps, the beads were resuspended in the GUS buffer, and GUS activity was determined (see below).
In all cases, after the incubation period, the beads were washed twice for 10 min with the same buffer and twice in the same buffer supplemented with 150 mM NaCl for 5 min. Proteins were released by boiling in SDS loading buffer and fractionated by SDS-PAGE.
GUS Assays
For whole-mount staining, transgenic PNE2Fc:N-GUS and axr1-12/PNE2Fc:N-GUS seedlings were grown for 3 days in the dark or for 3 days in the dark and then transferred to fresh medium containing DMSO or 50 μM MG132 for 7 h in the light. GUS staining was performed as described (del Pozo et al., 2002). Total protein from these transgenic lines was extracted in GUS buffer (50 mM phosphate buffer, pH 7.0, 50 mM NaCl, 0.1% Triton X-100, 10 mM β-mercaptoethanol, 1 mM PMSF, 1 × protease inhibitor cocktail, and 50 μM MG132). Ten microliters of these extracts was mixed with 10 μL of 1 mM 4-methylumbelliferyl-d-glucuronide and incubated for 10 min at 30°C. The reactions were stopped by the addition of 2 mL of 0.2 M Na2CO3, and fluorescence was quantified in a TKO100 fluorometer (Hoefer, Muskegon, MI) that had been adjusted previously according to the manufacturer's instructions. GUS activity was represented as arbitrary units per microgram of protein, and the values and standard errors were calculated as the average of three independent experiments. In the case of NE2Fc-GUS or GUS interaction with MBP or MBP-AtSKP2, after the pulldown, the beads were incubated in GUS buffer containing 1 mM 4-methylumbelliferyl-d-glucuronide (GUS substrate; Sigma) for 10 min at 30°C. The reactions were stopped by the addition of 2 mL of 0.2 M Na2CO3, and fluorescence was quantified.
Degradation Assays
Total protein from PNE2Fc:N-GUS and 35S:GUS calli was extracted in buffer A. One hundred micrograms of protein was incubated at 30°C in a degradation assay containing 5 mM ATP, 0.2 mM DTT, 1 μM ubiquitin, 0.01 unit/μL creatine kinase, and 5 mM phosphocreatine in buffer A and with 50 μM MG132 or DMSO in the control reaction. After the incubation period, 20 μL was removed from the sample, and GUS activity was quantified as described above. To analyze the stability of the recombinant AtE2Fc protein, protein extracts were prepared from exponential Arabidopsis cultured cells in buffer A without MG132. The degradation reaction contained 25 μg of cell-free protein extract, 5 mM ATP, 0.2 mM DTT, 1 μM ubiquitin, 0.01 unit/μL creatine kinase, and 5 mM phosphocreatine in buffer A and incubated at 30°C. In these degradation assays, we used purified recombinant His-AtE2Fc or phosphorylated His-AtE2Fc. In some cases, the cell-free protein extracts were preincubated with roscovitine and/or MG132 at a final concentration of 50 μM each for 5 min before mixing with the recombinant AtE2Fc protein.
Microscopic Analysis
Control transgenic (harboring pBin19) or ΔE2Fc-overexpressing seeds were germinated in the dark for 3 days in MSS medium. Afterward, the dark-grown seedlings or the first rosette leaves were incubated in 95% ethanol at 90°C for 5 min and in lactophenol (33% [w/v] phenol, 33% lactic acid, and 33% glycerol) overnight at room temperature. The seedlings were mounted on slides and analyzed by Nomarski microscopy using an Axiophot microscope (Zeiss, Jena, Germany), and the images were captured with a digital Coolsnap FX camera (Roper Scientific, Trenton, NJ). Area measurements were performed using NIH Image software (version 1.61) that was calibrated to calculate the area in square micrometers using the correlation between the pixels of the image and the micrometers calculated by the camera. The values represent the mean area of at least 150 cells taken from three different dark-grown cotyledons.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession number for AtE2Fc is AF242581. Accession numbers for other genes mentioned in this article are Z31402 (AtCYCA2;2), X83370 (AtCYCD2;1), and X57839 (AtCDKA;1).
Acknowledgments
We thank J. Sanchez-Serrano for kindly providing the Arabidopsis culture cells, M. Estelle for the axr1-12 seeds and anti-AtCUL1 serum, D. Inzé for the AtCYCA2;2 clone, and C. Koncz for the Arabidopsis cDNA library. J.C.d.P. was supported by a Spanish “Ramon y Cajal” contract from the Ministerio de Ciencia y Tecnologia. This work was supported by Grant BMC2000-1004 from the Ministerio de Ciencia y Tecnologia, Grant 07G/0033/00 from the Comunidad de Madrid, and an institutional grant from Fundación Ramón Areces.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006791.
References
- Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C., Helin, K., and Cella, R. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem. 275, 19258–19267. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408, 816–820. [DOI] [PubMed] [Google Scholar]
- Bartek, J., and Lukas, J. (2001). Cell cycle: Order from destruction. Science 294, 66–67. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Boniotti, M.B., and Gutierrez, C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDK/cyclin D complex. Plant J. 28, 341–350. [DOI] [PubMed] [Google Scholar]
- Callis, J., and Vierstra, R.D. (2000). Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- Castellano, M.M., del Pozo, J.C., Ramirez-Parra, E., Brown, S., and Gutierrez, C. (2001). Expression and stability of Arabidopsis CDC6 are associated with endoreplication. Plant Cell 13, 2671–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G.W., Healy, J.M., and Murray, J.A.H. (2000). Cyclin D control of growth rate in plants. Nature 405, 575–579. [DOI] [PubMed] [Google Scholar]
- Cooper, S., and Shayman, J.A. (2001). Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell. Mol. Life Sci. 58, 580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager, S.M., Menges, M., Bauer, U.M., and Murray, J.A.H. (2001). Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtDCD6 and is a member of a multigene family with differential activities. Plant Mol. Biol. 47, 555–568. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1-ECR1–dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Timpte, C., Tan, S., Callis, J., and Estelle, M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., de Almeida-Engler, J., Ormenese, S., Maes, S., Naudts, M., Van Der Schueren, E., Jacqmard, A., Engler, G., and Inze, D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelkrout, E.M., Robertson, D., and Hanley-Bowdoin, L. (2001). Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, R.M., Correll, C.C., Kaplan, K.B., and Deshaies, R.J. (1997). A complex of Cdc4p, Skp1p and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91, 221–230. [DOI] [PubMed] [Google Scholar]
- Ferrando, A., Farras, R., Jasik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope, a tool for facile detection and purification of proteins expressed in Agrobacterium transformed plant cells. Plant J. 22, 553–560. [DOI] [PubMed] [Google Scholar]
- Freed, E., Lacey, K.R., Huie, P., Lyapina, S.A., Deshaies, R.J., Stearns, T., and Jackson, P.K. (1999). Components of the SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication. Genes Dev. 13, 2242–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel, M., Jimenez, B., and Herrera-Estrella, L. (1996). The first intron of the Arabidopsis thaliana gene coding for elongation factor 1 beta contains an enhancer-like element. Gene 170, 201–206. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (1998). The retinoblastoma pathway in plant cell cycle and development. Curr. Opin. Plant Biol. 1, 492–497. [DOI] [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000). The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 14, 2393–2409. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Hemerly, A., Bergounioux, C., Van Montagu, M., Inze, D., and Ferreira, P. (1992). Genes regulating the plant cell cycle: Isolation of a mitotic-like cyclin from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 89, 3295–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., Zhang, H., and Scholl, R.L. (1990). Two evolutionarily divergent genes encode a cytoplasmic ribosomal protein of Arabidopsis thaliana. Gene 93, 177–182. [DOI] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. a). Interaction of Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol. 128, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi, S., and Ohashi, Y. (2002. b). E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. Plant J. 29, 45–59. [DOI] [PubMed] [Google Scholar]
- Krek, W. (1998). Proteolysis and the G1-S transition: The SCF connection. Curr. Opin. Genet. Dev. 8, 36–42. [DOI] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2001). Auxin signalling: The beginning, the middle and the end. Curr. Opin. Plant Biol. 4, 382–386. [DOI] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. (2002). The E2F family of transcription factors from Arabidopsis thaliana: Novel and conserved components of the pRB/E2F pathway in plants. J. Biol. Chem. 277, 9911–9919. [DOI] [PubMed] [Google Scholar]
- Marti, A., Wirbelauer, C., Scheffner, M., and Krek, W. (1999). Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1, 14–19. [DOI] [PubMed] [Google Scholar]
- Meijer, M., and Murray, J.A.H. (2000). The role and regulation of D-type cyclins in the plant cell cycle. Plant Mol. Biol. 43, 621–633. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A.H. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30, 203–212. [DOI] [PubMed] [Google Scholar]
- Morello, L., Bardini, M., Sala, F., and Breviario, D. (2002). A long leader intron of the Ostub16 rice beta-tubulin gene is required for high-level gene expression and can autonomously promote transcription both in vivo and in vitro. Plant J. 29, 33–44. [DOI] [PubMed] [Google Scholar]
- Müller, H., Bracken, A.P., Vernell, R., Moroni, M.C., Christians, F., Grassilli, E., Prosperini, E., Vigo, E., Oliner, J.D., and Helin, K. (2001). E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15, 267–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, H., and Helin, K. (2000). The E2F transcription factors, key regulators of cell proliferation. Biochim. Biophys. Acta 1470, M1–12. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Ohta, T., and Xiong, Y. (2001). Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res. 61, 1347–1353. [PubMed] [Google Scholar]
- Okanami, M., Meshi, T., and Iwabuchi, M. (1998). Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana. Nucleic Acids Res. 26, 2638–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka, F., Saeki, M., Katayama, S., Aida, N., Toh-E, A., Kominami, K., Toda, T., Suzuki, T., Chiba, T., Tanaka, K., and Kato, S. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., Preston, R.A., Adams, A.E., Stearns, T., Drubin, D.G., Haarer, B.K., and Jones, E.W. (1989). Fluorescence microscopy methods for yeast. Methods Cell Biol. 31, 357–435. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Xie, Q., Boniotti, M.B., and Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G(1)/S regulators. Nucleic Acids Res. 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld, J.P., Gigot, C., and Chaubet-Gigot, N. (1998). Multilevel regulation of histone gene expression during the cell cycle in tobacco cells. Nucleic Acids Res. 26, 3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg, L.E., and Bartel, B. (2001). Auxin signaling, derepression through regulated proteolysis. Dev. Cell 1, 595–604. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H., and Shinmyo, A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Tomoda, K., Kubota, Y., Arata, Y., Mori, S., Maeda, M., Tanaka, T., Yoshida, M., Yoneda-Kato, N., and Kato, J.Y. (2002). The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J. Biol. Chem. 277, 2302–2310. [DOI] [PubMed] [Google Scholar]
- Trimarchi, J.M., Fairchild, B., Wen, J., and Lees, J.A. (2001). The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98, 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S., and Inze, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodermaier, H.C. (2001). Cell cycle: Waiters serving the destruction machinery. Curr. Biol. 11, R834–R837. [DOI] [PubMed] [Google Scholar]
- Yang, R., Muller, C., Huynh, V., Fung, Y.K., Yee, A.S., and Koeffler, H.P. (1999). Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Mol. Cell. Biol. 19, 2400–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F box-Skp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]