Abstract
In a screen for suppressors of npr1-5–based salicylic acid (SA) insensitivity, we isolated a semidominant gain-of-function mutation, designated ssi4, that confers constitutive expression of several PR (pathogenesis-related) genes, induces SA accumulation, triggers programmed cell death, and enhances resistance to bacterial and oomycete pathogens. Through map-based cloning, ssi4 was identified and found to encode a putative protein belonging to the TIR-NBS-LRR (Toll Interleukin1 Receptor–Nucleotide Binding Site–Leu-Rich Repeat) class of R (resistance) proteins. Comparison between ssi4 and the corresponding wild-type sequence revealed a single amino acid substitution in the NBS. Epistasis analysis indicated that SA and EDS1 are required for ssi4-induced PR-1 expression and enhanced disease resistance; they also are required for the increased accumulation of SSI4 and EDS1 transcripts detected in the ssi4 mutant. Although high levels of ssi4 transcripts correlate with the appearance of the mutant phenotype, overexpression of the wild-type SSI4 gene failed to induce stunting, spontaneous lesion formation, or increased PR-1 expression associated with the ssi4 mutation. Thus, the ssi4 phenotype does not appear to be caused by overexpression of this R gene; rather, we propose that the NBS substitution generates a constitutively activated R protein. Furthermore, because SA treatment induced the expression of SSI4 and the closely related TIR-NBS-LRR genes RPP1 and RPS4 but had little effect on the expression of the coiled-coil NBS-LRR genes RPM1 and RPS2, we suggest that SA not only functions as a critical signal for downstream resistance events but also upregulates the expression of certain R genes.
INTRODUCTION
For eons, battles have raged between plants and pathogenic microbes intent on using them as a nutritional resource. To ward off infection, plants activate a variety of defense responses. In the infected leaf, these defenses include strengthening of cell walls, activation and/or synthesis of antimicrobial compounds, and expression of many defense-associated proteins, including the pathogenesis-related (PR) proteins (Durner et al., 1997; Dempsey and Klessig, 1999). A hypersensitive response, characterized by the formation of necrotic lesions and the restriction of the pathogen to the cells within and immediately surrounding these lesions, also frequently develops. After these local responses, the uninoculated portions of the plant usually exhibit increased PR gene expression and the appearance of an enduring resistance to a broad spectrum of pathogens known as systemic acquired resistance (SAR). For some plant–pathogen interactions, the signal transduction cascade leading to disease resistance is activated by the direct or indirect interaction between the products of a plant R (resistance) gene and a pathogen Avr (avirulence) gene (Flor, 1971; Keen, 1990). Based on this gene-for-gene model, if the cognate gene from either the plant or the pathogen is missing, plant defense responses either fail to be activated or are induced too weakly and/or too late to prevent pathogen colonization.
A variety of studies have demonstrated that salicylic acid (SA) plays a critical role in the defense signaling pathway. In many plant species, SA levels increase in conjunction with the activation of PR gene expression and disease resistance (Malamy et al., 1990; Métraux et al., 1990; Uknes et al., 1993). Furthermore, plants unable to accumulate SA as a result of the expression of the bacterial nahG gene fail to develop SAR and exhibit increased susceptibility to pathogen infection (Delaney et al., 1994; Vernooij et al., 1994). Analysis of various genetic mutants, including the cpr (Bowling et al., 1994, 1997; Clarke et al., 1998; Silva et al., 1999; Yoshioka et al., 2001), cim (Ryals et al., 1996), acd (Greenberg et al., 1994; Rate et al., 1999), ssi (Shah et al., 1999, 2001; Kachroo et al., 2001), and lsd (Dietrich et al., 1994; Weymann et al., 1995) mutants, also has revealed a correlation between increased SA levels, constitutive PR expression, and SAR. By contrast, mutations in the NPR1 gene cause a SA-insensitive phenotype, which is characterized by heightened disease susceptibility and the inability to develop SAR after SA treatment (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997).
During the past few years, many R genes have been cloned. Based on their predicted protein structures, R proteins can be divided into several groups. The largest group contains a nucleotide binding site (NBS) and a Leu-rich repeat (LRR) region (Ellis et al., 2000a). NBS domains have been identified in many prokaryotic and eukaryotic proteins, in which their ability to bind ATP or GTP is essential for their biological activity (Saraste et al., 1990). The ability of plant R proteins to bind nucleotides has yet to be demonstrated; however, they contain several conserved motifs, such as the P loop, which are known to be important for this function. LRR domains are known to mediate protein–protein interactions in diverse proteins (Jones and Jones, 1996). Based on the high degree of divergence between the LRRs of R protein homologs, they are thought to play a role in pathogen recognition (Bent, 1996; Jones, 1996; Ellis et al., 2000b; Nimchuk et al., 2001). The NBS-LRR class of R proteins can be subdivided further into two classes based on the secondary structure of the N terminus. The first class contains a putative coiled-coil region at the N terminus (CC-NBS-LRR). The second class contains an N-terminal TIR region, which shares homology with the Toll protein of Drosophila and the Interleukin1 receptor (IL-1R) of mammals (TIR-NBS-LRR) (Ellis et al., 2000a). Because the Toll protein and IL-1R play roles in activating innate immunity, the TIR domain has been proposed to transduce a signal that activates plant defense responses (Staskawicz et al., 1995; Baker et al., 1997).
Recent evidence indicates that the signals from the TIR- and CC-containing classes of R proteins are transduced through separate pathways (Aarts et al., 1998). The TIR-NBS-LRR proteins appear to induce disease resistance via a pathway that requires the EDS1 gene (Parker et al., 1996). By contrast, the CC-NBS-LRR proteins frequently use a pathway that requires the NDR1 gene (Century et al., 1995). However, a few CC-NBS-LRR proteins have been identified that require neither EDS1 nor NDR1 (McDowell et al., 2000; Bittner-Eddy and Benyon, 2001). Thus, a third defense signaling pathway(s) that is independent of these proteins also appears to exist.
The ability of different R proteins to activate a specific defense pathway suggests that diverse signal transduction strategies are used. Currently, the mechanisms by which a pathogen is perceived and the resistance signal is initiated and transduced are poorly understood. However, several recent studies have demonstrated that overexpression of R genes can result in enhanced disease resistance. For example, overexpression of the Pto gene in tomato induces constitutive PR gene expression, increased levels of SA, spontaneous lesion formation, and enhanced disease resistance (Tang et al., 1999). Broad-spectrum disease resistance also is induced by overexpression of Prf, a CC-NBS-LRR R gene whose product works in conjunction with Pto (Oldroyd and Staskawicz, 1998). In addition, overexpression of the Arabidopsis TIR-NBS-LRR R gene At4g16890 results in enhanced disease resistance (Stokes et al., 2002). It has been hypothesized that R proteins are found normally in a complex with a guard protein; while sequestered in this complex, they are unable to activate defense responses (Dangl and Jones, 2001). After interaction with the appropriate Avr protein, a change in the R protein would allow the complex to dissociate and a signal transduction event would be initiated. Thus, if R proteins are present normally in rate-limiting amounts, their overexpression might be sufficient to activate the defense signaling pathway.
To further investigate the SA signaling pathway leading to plant disease resistance, we used a genetic screen to isolate suppressors of the npr1-5 mutation. One mutant, designated ssi4, was identified based on its ability to constitutively express several PR genes. Analysis of the cloned ssi4 gene revealed that the predicted open reading frame (ORF) encodes a TIR-NBS-LRR protein with a single amino acid substitution in the NBS domain. Consistent with the presence of an altered TIR-NBS-LRR R gene, ssi4 mutants exhibited enhanced resistance to Pseudomonas syringae pv maculicola ES4326 and Peronospora parasitica biotype Emco5. Furthermore, both SA and EDS1 were required for ssi4-induced constitutive PR-1 expression and enhanced disease resistance. All plants exhibiting the ssi4 mutant phenotype contained increased levels of ssi4 transcripts. However, because SSI4-overexpressing plants failed to express PR-1 or to display enhanced disease resistance, the NBS mutation, rather than overexpression of this TIR-NBS-LRR gene, appears to be responsible for signaling defense responses. Interestingly, SA treatment induced the expression of SSI4 and several other TIR-NBS-LRR R genes. This finding suggests that SA is involved in a positive feedback loop that regulates the expression of a variety of defense-associated genes, including certain R genes.
RESULTS
The ssi4 Mutation Confers Constitutive PR Gene Expression
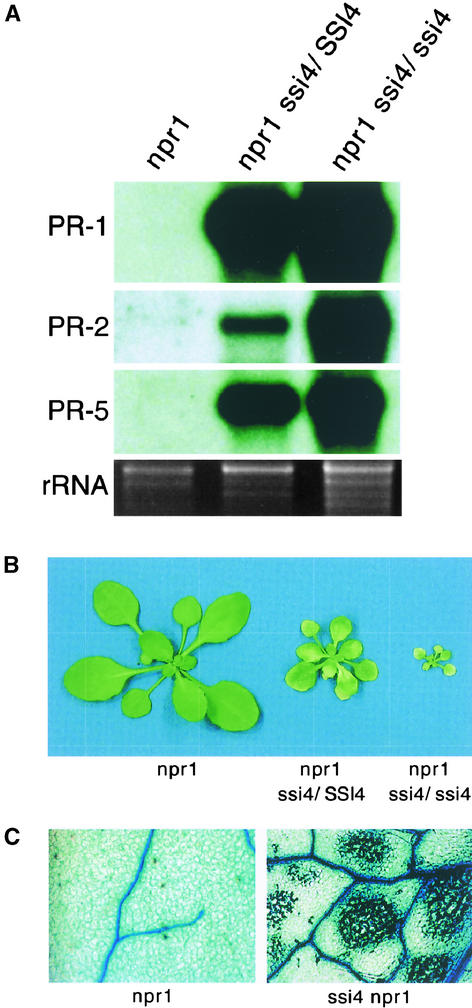
To identify signaling components of the SA defense pathway, 3- to 4-week-old M2 progeny of ethyl methanesulfonate–mutagenized npr1-5 seeds (ecotype Nö) were screened for individuals that exhibited constitutive PR gene expression. One individual, designated ssi4 (suppressor of salicylic acid insensitivity of npr1-5), was found to express the PR-1, PR-2, and PR-5 genes (Figure 1A). By contrast, the jasmonic acid– and ethylene-inducible PDF1.2 gene was not expressed (data not shown). The ssi4 mutant also exhibited several morphological abnormalities, including stunted growth, severe chlorosis, and the development of spontaneous lesions (Figure 1B). Because these lesions contained large areas of dead cells (Figure 1C) and were associated with the presence of autofluorescent material (data not shown), the cell death induced by ssi4 is similar to that seen during the hypersensitive response.
Figure 1.
Phenotypic Effects of the ssi4 Mutation.
(A) Expression of the PR-1, PR-2, and PR-5 genes in npr1-5, npr1-5 ssi4/SSI4, or npr1-5 ssi4/ssi4 plants. RNA gel blot analysis was performed on 10 μg of total RNA extracted from 3-week-old soil-grown plants. Ethidium bromide staining of rRNA (bottom gel) was used as a loading control.
(B) Comparison of the morphological phenotypes displayed by plants heterozygous or homozygous for the ssi4 mutant allele in the npr1-5 background. Plants were grown on soil and photographed when 3 weeks old.
(C) Microscopy of trypan blue–stained leaves from npr1-5 and ssi4 npr1-5 plants. Leaves from ssi4 npr1-5 plants contain intensely stained areas of dead cells.
ssi4 Is a Monogenic Gain-of-Function Mutation
To investigate the nature of the ssi4 mutation, we analyzed the progeny of a backcross between the ssi4 npr1-5 double mutant and the SSI4 npr1-5 parent. All of the F1 progeny exhibited constitutive PR gene expression and the stunted morphology associated with the ssi4 mutation. However, all of the ssi4-associated phenotypes were less pronounced in these heterozygous plants, suggesting that ssi4 is a semidominant mutation (data not shown). Analysis of F2 progeny confirmed this finding (Figures 1A and 1B).
To determine whether the ssi4 mutation suppresses the npr1-5 phenotype by functioning as an intragenic suppressor of the npr1-5 allele, ssi4 npr1-5 and wild-type (ecotype Nö) plants were crossed. ssi4 homozygous F2 progeny that also were homozygous for NPR1 were identified using a codominant cleaved amplified polymorphic sequence marker for npr1-5 (Shah et al., 1999). The ssi4 NPR1 plants showed the same phenotypes as ssi4 npr1-5 plants (data not shown). Thus, the ssi4 mutation is not a simple revertant of npr1-5, and the effects of this mutation are independent of the npr1-5 mutation.
We next determined whether the ssi4 mutation is attributable to a dominant gain of function or to haploinsufficiency by crossing ssi4 homozygous plants with Columbia tetraploid plants (CS3432). RNA gel blot analysis of the resulting triploid F1 progeny revealed that they constitutively expressed PR-1, and these plants also displayed extensive leaf chlorosis and formed lesions spontaneously (data not shown). This result suggests that the ssi4 phenotype is caused by a gain-of-function mutation rather than by haploinsufficiency.
Positional Cloning of ssi4
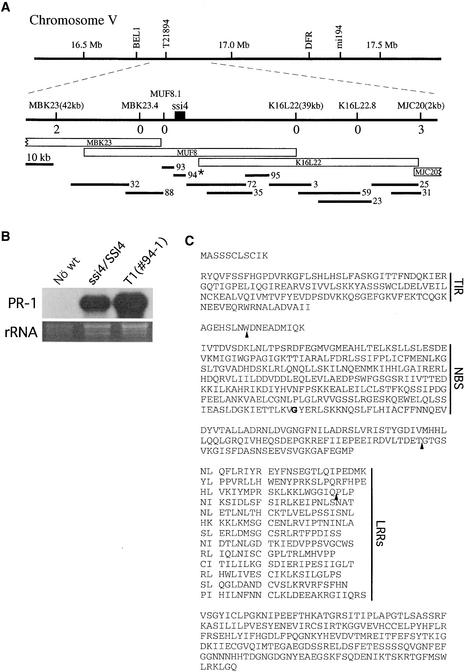
To determine the map position of the ssi4 locus, we crossed ssi4 plants with wild-type plants from ecotype Landsberg. As expected, the F2 progeny segregated in a 3:1 ratio (PR-1+:PR-1−) when scored for constitutive PR-1 gene expression. Because of the semi-dominant nature of the ssi4 mutation, we determined map position by analyzing F2 plants exhibiting the wild-type (PR-1−) phenotype. A total of 1179 wild-type–like F2 plants were analyzed; SSI4 was mapped to a 132-kb region of chromosome V. This region corresponds to 37 kb at the 3′ of the P1 clone MBK23, the entire region covered by overlapping clones MUF8 and K16L22, and 2 kb of MJC20 beyond the region of overlap between K16L22 and MJC20, as defined by the Arabidopsis sequencing project (Figure 2A).
Figure 2.
Isolation and Identification of the ssi4 Mutation.
(A) Genetic and physical map of the ssi4 region of chromosome V. The top line represents chromosome V, with the broken lines showing the points of recombination. The middle line represents a larger scale of the region within the recombination breakpoints, with markers indicated above it, the number of recombination events for each marker designated below it, and the MUF8.2 ORF (ssi4) denoted as a black box. P1 and TAC clones spanning this region are represented as labeled boxes (MBK23, MUF8, K16L22, and MJC20). Directly beneath these boxes are the numbered TAC-based clones representing the contig of this region generated from ssi4 genomic DNA. Clone 94, marked with an asterisk, contains the ssi4 mutation and maps to the MUF8.2 ORF.
(B) PR-1 expression in wild-type (Nö wt), ssi4/SSI4, and T1 (#94-1) plants. Ten micrograms of total RNA was used for RNA gel blot analysis. rRNA stained with ethidium bromide served as a loading control.
(C) Amino acid sequence of SSI4. Basic Local Alignment Search Tool (BLAST) analysis of the predicted SSI4 protein sequence revealed three conserved regions identified by the vertical lines: TIR, NBS, and LRR. The ssi4 mutation (G422R) is indicated in boldface type. The arrowheads indicate the intron positions in the corresponding genomic SSI4 sequence. The aligned spaces in the LRR region indicate the putative β-turn/β-strand motif as aligned with the porcine ribonuclease inhibitor structure.
Because ssi4 is a gain-of-function mutation, complementation analysis could be performed only using the ssi4 gene. Thus, a transformation-competent artificial chromosome (TAC) library containing ssi4 genomic DNA was constructed with an average insert size of 10 to 20 kb. After screening 100,000 colonies, those clones spanning the ssi4-containing region were isolated and organized into a contig (Figure 2A). Each clone was introduced into wild-type (ecotype Nö) plants by vacuum infiltration. Hygromycin-resistant T1 plants then were screened for the ssi4 phenotype. Only plants containing clone 94, which encompasses the MUF8.2 reading frame, exhibited stunted growth and extensive leaf chlorosis (data not shown). These plants also displayed constitutive PR-1 gene expression (Figure 2B), confirming that this clone contains the ssi4 mutation.
Sequence analysis of the MUF8.2 ORF from TAC clone 94, as well as reverse transcriptase (RT)–mediated PCR of the ssi4 mutant and SSI4 wild-type mRNAs, identified the ssi4 mutation as a G-to-A transition at codon 422 of MUF8.2, resulting in a Gly-to-Arg amino acid substitution. Sequencing of PCR-generated fragments using genomic DNA or cDNA from mutant or wild-type plants as the template produced the same result. Because the ssi4 mutation does not alter any restriction enzyme site, a derived cleaved amplified polymorphic sequence marker was created to monitor the segregation of mutant genes in transgenic plants containing clone 94. T2 analysis of these plants further confirmed that the ssi4 mutation is responsible for the mutant phenotype; the ssi4 transgene cosegregated with constitutive PR-1 gene expression and the stunted, chlorotic morphology exhibited by ssi4 plants.
Analysis of TAC clone 94 revealed that it contains a single ORF of 3434 nucleotides. Comparison of RT-PCR–generated SSI4 or ssi4 cDNA with genomic DNA indicated that this gene contains three introns that are spliced out to produce a transcript with a predicted protein of 1055 amino acids. A search of the GenBank database indicated that SSI4 is highly similar to several genes that encode functional R proteins, including N (Whitham et al., 1994), L6 (Lawrence et al., 1995), RPS4 (Gassmann et al., 1999), members of the RPP5 gene family (Parker et al., 1997; van der Biezen et al., 2002), and RPP1-WsA (Botella et al., 1998). Like the R proteins encoded by these genes, SSI4 contains a TIR-NBS-LRR structure (Figure 2C). Downstream of the TIR domain, SSI4 contains seven conserved motifs that are associated with the NBS domains of all other TIR-NBS-LRR R proteins (Meyers et al., 1999). The Gly-to-Arg substitution in ssi4 is located in a nonconserved region of the NBS just upstream of the resistance nucleotide binding site D TIR motif. The C-terminal portion of SSI4 contains an LRR domain consisting of 13 imperfect LRRs that range in length from 20 to 25 amino acids.
ssi4 Encodes an Activated TIR-NBS-LRR Protein
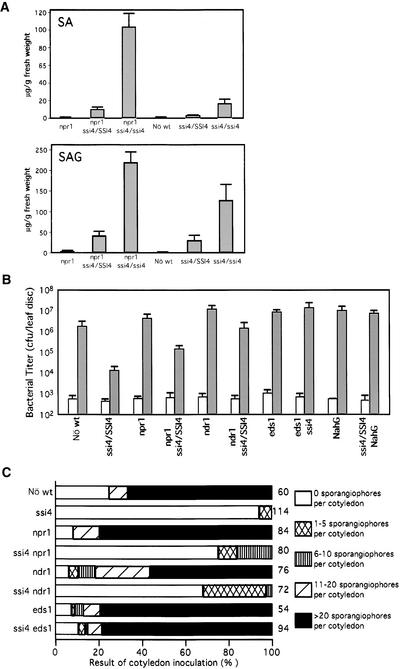
The discovery that SSI4 encodes a TIR-NBS-LRR protein raised the possibility that other defense responses, in addition to PR gene expression, might be activated by the NBS mutation. To assess this possibility, we assayed ssi4 mutants for enhanced disease resistance and increased levels of SA. SA levels in ssi4 npr1 homozygous plants were 300-fold higher than those detected in wild-type plants or in the parental npr1-5 mutant, and SA glucoside (SAG) levels were >200-fold greater (Figure 3A). Plants heterozygous for ssi4 in the npr1-5 background exhibited intermediate levels of SA and SAG. By contrast, ssi4 homozygous and heterozygous mutants carrying the NPR1 allele accumulated substantially lower levels of SA and SAG than the corresponding double mutants.
Figure 3.
The ssi4 Protein Stimulates SA Accumulation and Enhanced Disease Resistance.
(A) SA and SAG levels in the leaves of 3-week-old, soil-grown npr1 single mutants, npr1 mutants heterozygous or homozygous for ssi4, wild-type (Nö wt) plants, and ssi4 heterozygous and homozygous single mutants. The values presented are averages of four replicates.
(B) Growth of P. syringae pv maculicola ES4326. Leaves of different ssi4 plant genotypes were infiltrated with P. syringae pv maculicola ES4326 (OD600 = 0.002). Four leaf discs were collected immediately after inoculation (white bars), and six leaf discs were collected at 3 days after inoculation (gray bars). Colony-forming units (cfu) per leaf disc are expressed ±sd and represent averages of four or six samples.
(C) Growth of P. parasitica ecotype Emco5. Cotyledons of 7-day-old seedlings from the various plant genotypes listed at left were inoculated by applying a drop of conidiospore suspension (105 spores/mL). There was no difference in the size of cotyledons between wild-type and mutant plants at the time of infection. Pathogen growth was assayed by counting the number of sporangiophores per cotyledon at 7 days after inoculation. The shade of each box indicates the severity of infection, based on the number of sporangiophores per cotyledon (see key at right). Numbers to the right of the sample boxes indicate the number of cotyledons assayed.
To determine whether ssi4 plants exhibit enhanced disease resistance, inoculations were performed using the bacterial pathogen P. syringae pv maculicola ES4326 and the oomycete pathogen P. parasitica biotype Emco5. As a result of the extreme stunting of ssi4 homozygous plants, inoculations with P. syringae pv maculicola ES4326 were performed only on ssi4 heterozygous plants. By 3 days after inoculation with P. syringae pv maculicola ES4326, ssi4/SSI4 npr1 plants contained ∼15- or 20-fold less bacteria than either wild-type or npr1-5 parental plants, respectively (Figure 3B). ssi4-induced resistance was even higher in the NPR1 background; these plants contained 150-fold less bacteria than wild-type plants. Similar results were observed after inoculation with P. parasitica biotype Emco5. At 7 days after inoculation, the level of sporulation on the cotyledons of ssi4/ssi4 npr1 or ssi4/ssi4 NPR1 plants was reduced greatly compared with that detected on wild-type or npr1-5 single-mutant plants (Figure 3C).
ssi4 Induces Most Defense Responses via an SA- and EDS1-Dependent Pathway
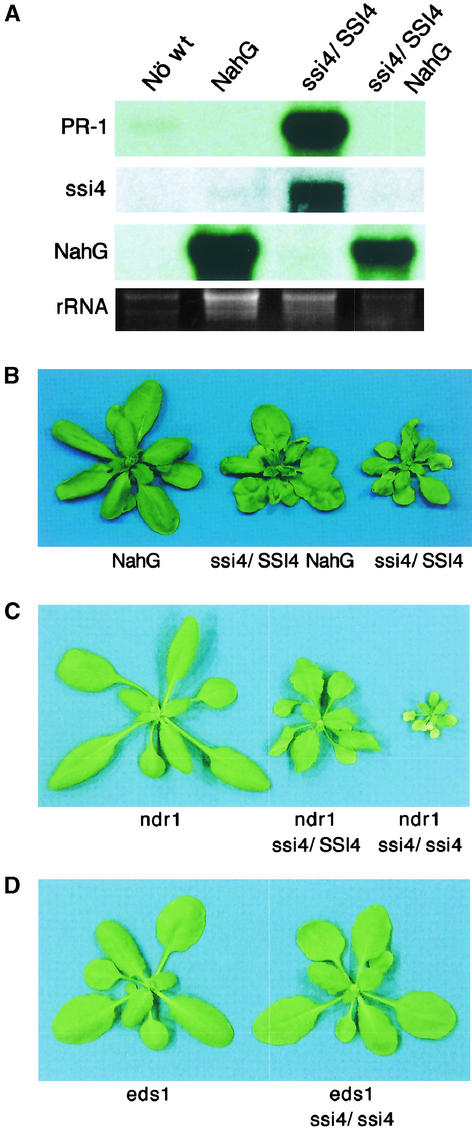
After activation, R proteins stimulate many defense responses via an SA-dependent pathway(s). To determine whether SA is required for ssi4-induced PR gene expression and disease resistance, a cross between ssi4 and nahG plants (ecotype Nö) was performed. None of the resulting F1 progeny constitutively expressed the PR-1 gene (Figure 4A), became chlorotic (Figure 4B), or exhibited enhanced resistance to P. syringae pv maculicola ES4326 (Figure 3B). Thus, SA is required for these ssi4-mediated phenomena. However, nahG-expressing ssi4 heterozygous plants exhibited some stunting, and their leaves were curled (Figure 4B). At the microscopic level, trypan blue staining revealed a significant number of dead cells surrounding the leaf vascular area (data not shown). Thus, ssi4 appears to induce stunting and spontaneous lesion formation via a pathway that is at least partly independent of SA. Although the residual level of SA in ssi4 nahG plants was not determined, it is likely to be similar to that in wild-type plants, because PR-1 gene expression was no longer constitutive and previous studies of ssi1 nahG and ssi2 nahG plants indicated that nahG in the ecotype Nö background was able to suppress SA accumulation to basal levels (Shah et al., 1999, 2001).
Figure 4.
Most ssi4 Phenotypes Are Activated via an SA- and EDS1-Dependent Pathway.
(A) Expression of the PR-1, nahG, and ssi4 genes in wild-type (Nö wt), nahG, ssi4/SSI4, and ssi4/SSI4 nahG plants. Ten micrograms of total RNA was used for RNA gel blot analysis, and ethidium bromide staining of rRNA was used as a loading control.
(B) The phenotypes of 3-week-old soil-grown nahG, ssi4/SSI4 nahG, and ssi4/SSI4 plants.
(C) Phenotypes of ndr1 plants and ndr1 plants heterozygous or homozygous for the ssi4 mutation. Plants were grown in soil and photographed when 3 weeks old.
(D) Phenotypes of eds1 and ssi4 eds1 plants. Plants were grown in soil and photographed when 3 weeks old.
Previous studies have demonstrated that several members of the TIR-NBS-LRR class of R genes activate disease resistance via an EDS1-dependent pathway, whereas most members of the CC-NBS-LRR class use NDR1 (Aarts et al., 1998). To determine whether ssi4 induces defense responses via either of these signal transducers, ssi4 plants were crossed with eds1-1 and ndr1-1 plants. Analysis of the resulting ssi4 ndr1 double mutants revealed that they were nearly as stunted and chlorotic as the ssi4 single mutant in the npr1-5 background (Figure 4C). In addition to these abnormalities, ssi4 ndr1 double mutants developed spontaneous lesions and constitutively expressed PR-1 (Figure 5A and data not shown). Furthermore, after inoculation with P. syringae pv maculicola ES4326 or P. parasitica biotype Emco5, ssi4 ndr1 plants exhibited greater resistance than ndr1 single mutants (Figures 3B and 3C). However, they were much less resistant than ssi4 single mutants; this result suggests that NDR1 either plays some role in the enhanced resistance conferred by the ssi4 mutation or has an additive effect on resistance. By contrast, ssi4 eds1 double mutants exhibited wild-type morphology, even in the ssi4 homozygous condition (Figure 4D). Furthermore, these plants did not develop spontaneous lesions, failed to constitutively express PR-1 (Figure 5A and data not shown), and exhibited heightened susceptibility to infection by P. syringae pv maculicola ES4326 and P. parasitica biotype Emco5 (Figures 3B and 3C). Based on these results, ssi4, like other TIR-NBS-LRR genes, requires a functional EDS1 gene to signal defense responses and disease resistance.
Figure 5.
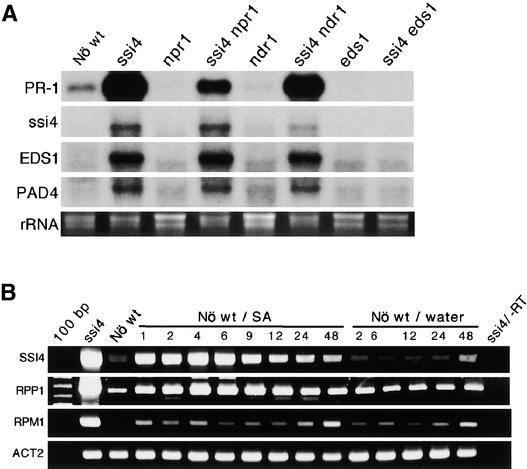
ssi4 Expression Requires EDS1 and Is SA Inducible.
(A) Expression of the PR-1, SSI4, EDS1, and PAD4 genes in wild-type (Nö wt) plants, ssi4, npr1, ndr1, and eds1 single mutants, and ssi4 npr1, ssi4 ndr1, and ssi4 eds1 double mutants (all of the mutant genes were present in the homozygous state). RNA gel blot analysis was performed using 10 μg of total RNA, and ethidium bromide staining of rRNA was used as a loading control.
(B) RT-PCR analysis of wild-type plants treated with 500 μM SA or sterile water. Plants were sprayed with SA or water, and leaves were harvested at the times indicated after treatment. RT-PCR was performed using total RNA and gene-specific primers; the products were visualized on an ethidium bromide–stained agarose gel. The level of Actin2 (ACT2) was used as an internal control to normalize the amount of cDNA template. A control with RNA from ssi4 processed without the addition of reverse transcriptase (−RT) is shown at right.
ssi4 Induces Increased ssi4 and EDS1 Gene Expression
It was demonstrated previously that overexpression of the R genes Pto, Prf, and At4g16890 induces constitutive PR gene expression, spontaneous cell death, increased SA levels, and/or enhanced disease resistance (Oldroyd and Staskawicz, 1998; Tang et al., 1999; Stokes et al., 2002). Because the ssi4 mutant exhibits a similar phenotype, we tried to determine whether the presence of the NBS substitution alters the expression of this gene. RNA gel blot analysis revealed that transcripts for the ssi4 gene accumulate to moderate levels in ssi4 mutant plants (Figure 5A). By contrast, the level of SSI4 transcripts in wild-type plants was extremely low and could be detected only by RT-PCR (Figures 5A and 5B). This increase in ssi4 expression does not appear to be caused by a promoter mutation, because analysis of 981 base pairs 5′ of the translational start site revealed no difference between the ssi4 and wild-type sequences. ssi4 transcripts also were readily detectable in ssi4 npr1 and ssi4 ndr1 mutant plants, but not in ssi4 eds1 (Figure 5A) and ssi4 nahG mutant plants (Figure 4A). These results suggest that increased expression of ssi4 requires a functional EDS1 gene and is SA dependent.
Because ssi4-induced defense responses appear to be activated via an EDS1-dependent pathway, we tested whether ssi4 plants also constitutively express EDS1. Correlating with ssi4 expression, transcripts for EDS1 were detected readily in ssi4 and ssi4 npr1 plants as well as in the ssi4 ndr1 double mutant (Figure 5A). By contrast, few or no transcripts were detected in wild-type plants or in the ssi4 eds1 double mutant. Because eds1-1 contains a point mutation resulting in a nonfunctional protein (Falk et al., 1999), the lack of EDS1 transcripts in ssi4 eds1 plants suggests that a functional EDS1 protein is required for the upregulation of both ssi4 and EDS1 expression. Analysis of PAD4, which encodes a protein that forms a complex with EDS1 (Feys et al., 2001), revealed that it, like ssi4 and EDS1, is expressed at readily detectable levels only in ssi4, ssi4 npr1, and ssi4 ndr1 plants (Figure 5A). Strikingly, transcripts for RPP1, a closely related TIR-NBS-LRR gene, also accumulate to increased levels in ssi4 plants (Figure 5B). Thus, the ssi4 mutation causes increased expression of a wide variety of genes involved in plant defense.
It is possible that the high levels of SA found in the ssi4 mutant induce the expression of ssi4, EDS1, and PAD4. Consistent with this possibility, EDS1 was shown to be induced by SA treatment (Falk et al., 1999). To determine whether SA also induces SSI4 expression, we treated wild-type (ecotype Nö) plants with SA. As expected, SA treatment strongly induced PR-1 expression by 6 h after treatment, and a very weak band corresponding to SSI4 also was detected at this time (data not shown). Thus, SA treatment induces SSI4 expression, although to a much lower level than that detected in ssi4 mutant plants. We then tested whether other TIR-NBS-LRR genes could be induced by SA treatment. Because R genes frequently are expressed at very low levels, this analysis was performed using RT-PCR. Consistent with our RNA gel blot analysis, transcripts for SSI4 increased until 6 h after SA treatment and then decreased gradually during the remainder of the time course (Figure 5B). The same trend was seen with RPP1, another TIR-NBS-LRR gene (Figure 5B). By contrast, the CC-NBS-LRR gene RPM1 exhibited little or no induction by SA treatment. In addition, RPS4, a TIR-NBS-LRR gene, was activated by SA, whereas RPS2, another CC-NBS-LRR gene, was not (data not shown).
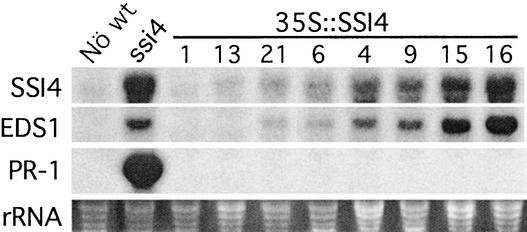
Overexpression of SSI4 Does Not Constitutively Activate Defense Responses
The correlation between increased ssi4 expression and the constitutive activation of defense responses suggested that overexpression of this TIR-NBS-LRR gene may be responsible for these effects. To assess this possibility, we transformed ecotype Nö plants with an SSI4 cDNA fused to the 35S promoter of Cauliflower mosaic virus in a pBI121 binary vector. Kanamycin-resistant T1 plants were isolated, and their level of SSI4 expression was determined by RNA gel blot analysis for eight independent T1 plants (Figure 6). None of the T1 plants, including those with highly increased levels of SSI4 transcripts, developed the severe ssi4 phenotype or constitutively expressed PR-1 (Figure 6 and data not shown). This was confirmed further by analyzing ∼30 to 40 T2 plants of T1 lines 9, 15, 16, and 21. Interestingly, overexpression of SSI4 resulted in a corresponding increase in EDS1 transcripts, suggesting that the increased levels of EDS1 mRNAs in ssi4 plants are attributable in part to overexpression of the ssi4 gene. However, increased levels of SSI4 and EDS1 transcripts in 35S::SSI4 T2 plants did not confer enhanced resistance to the oomycete pathogen P. parasitica biotype Emco5; ∼200 to 300 T2 plants of T1 lines 9, 15, and 16 were tested and found to be as susceptible as nontransformed control plants (data not shown). These results suggest that the constitutively activated defense responses exhibited by ssi4 mutant plants are not simply the result of overexpression of the ssi4 gene; rather, they are caused by the amino acid substitution in the NBS.
Figure 6.
Defense Gene Expression in Transgenic Plants Carrying the SSI4 cDNA Fused to the 35S Promoter of Cauliflower mosaic virus.
Ten micrograms of total RNA extracted from 4-week-old T1 plants was subjected to RNA gel blot analysis. The blot was probed sequentially for the SSI4, EDS1, and PR-1 gene transcripts. The numbers at top correspond to the different T1 plants. Ethidium bromide staining of rRNA served as a loading control.
DISCUSSION
To elucidate the components involved in defense signaling, a screen for suppressor mutations of npr1-5 was performed. One semidominant gain-of-function mutant, designated ssi4, was identified based on its ability to constitutively express several PR genes. In addition, ssi4 plants exhibited a stunted, chlorotic morphology and spontaneously developed lesions. Map-based cloning of the ssi4 gene revealed that it encodes a TIR-NBS-LRR protein. Thus, ssi4 exhibits structural similarity to a large class of plant R genes; nearly 100 members have been identified in the Arabidopsis genome (Dangl and Jones, 2001). Comparison with the wild-type SSI4 sequence revealed that the ssi4 gene contains a point mutation leading to a single amino acid substitution in the NBS domain. Although the crystal structures of a TIR domain and several LRR domains, all from other organisms, were solved recently, structural information on the NBS domain remains elusive (Xu et al., 2000; Kobe and Kajava, 2001). Recent sequence analysis of many R protein NBS domains has identified several conserved TIR-NBS-LRR–specific motifs that are dispersed between highly divergent nonconserved regions (Meyers et al., 1999). The Gly-to-Arg substitution caused by the ssi4 mutation was detected in a nonconserved region just upstream of the resistance NBS D motif. Thus, it is unclear whether this mutation affects the putative NBS of ssi4 or some other activity performed by this domain.
The combined discoveries that SSI4 encodes a TIR-NBS-LRR–type R protein and that the Gly-to-Arg substitution in its NBS confers constitutive PR gene expression led us to suspect that ssi4 is a constitutively activated R protein. Consistent with this hypothesis, ssi4 plants accumulated high levels of SA and SAG and exhibited enhanced resistance to both P. syringae pv maculicola ES4326 and P. parasitica biotype Emco5. Analysis of ssi4 nahG plants revealed that most of the ssi4-induced phenotypes, including constitutive PR gene expression, enhanced disease resistance, and chlorosis, require increased levels of SA. Because the ssi4 mutation was isolated in the npr1-5 background, all of these SA-dependent defenses must be activated via an NPR1-independent pathway. Analyses of several other Arabidopsis mutants also have revealed an SA-dependent, NPR1-independent pathway(s) that mediates PR gene expression and/or disease resistance (Bowling et al., 1997; Clarke et al., 1998; Li et al., 1999, 2001; Rate et al., 1999; Shah et al., 1999, 2001; Kachroo et al., 2001; Yoshioka et al., 2001). Because SA alone does not induce PR expression or enhanced resistance in npr1 plants, the activation of the SA-dependent, NPR1-independent pathway(s) is thought to require a second signal that works in conjunction with SA. As has been proposed by others, this second signal could be generated by pathogen infection or by various mutations, such as ssi4.
It is interesting that although NPR1 is not required for the ssi4-mediated activation of defense responses, it appears to play an important role in regulating SA accumulation. SA and SAG levels were substantially greater in ssi4 npr1-5 double mutants than in wild-type plants. SA and SAG levels in the ssi4 single mutant also were increased with respect to wild-type plants, although they were lower than those in the double mutant. A similar phenomenon was observed in snc1, cpr5, cpr6, and ssi2 single mutants, which accumulate less SA than snc1 npr1, cpr5 npr1, cpr6 npr1, and ssi2 npr1 double mutants (Clarke et al., 2000; Li et al., 2001; Shah et al., 2001). Together, these results suggest that NPR1 not only transduces the SA signal but also plays a role in downregulating SA levels, as was proposed previously by Clarke et al. (2000).
Most R proteins activate disease resistance via either an EDS1- or an NDR1-dependent pathway, with the structure of the R protein determining which pathway is used (Aarts et al., 1998). Like other TIR-NBS-LRR proteins, ssi4 requires EDS1 to activate resistance to P. syringae pv maculicola ES4326 and P. parasitica biotype Emco5. Indeed, ssi4 eds1 double mutants were indistinguishable from wild-type plants, demonstrating that EDS1 is required for all of the ssi4-induced phenotypes. In addition to a strong requirement for EDS1, NDR1 also appears to play a minimal role in signaling the ssi4 phenotype. ssi4 ndr1 double mutants exhibited less pronounced levels of PR gene expression, stunting, and resistance to P. syringae pv maculicola ES4326 compared with ssi4 single mutants. Similarly, the presence of an ndr1 allele reduced snc1- and RPS4-mediated resistance to strains of P. syringae and decreased RPP-mediated resistance to various P. parasitica isolates (Aarts et al., 1998; Li et al., 2001). Thus, these EDS1-dependent R genes also may incrementally activate defense responses via an NDR1-dependent pathway.
There have been several reports that R gene overexpression results in enhanced disease resistance (Oldroyd and Staskawicz, 1998; Tang et al., 1999; Stokes et al., 2002). Consistent with this possibility, increased levels of ssi4 transcripts correlated with constitutive SA accumulation, PR gene expression, and enhanced disease resistance. However, because overexpression of the wild-type SSI4 gene did not lead to stunting, constitutive PR-1 expression, or enhanced resistance to P. parasitica biotype Emco5, the Gly-to-Arg substitution in the NBS, rather than or in addition to overexpression of this R gene, appears to be required to activate defense responses. There are several possible explanations for how the NBS substitution in ssi4 can lead to constitutive defense response activation. For example, the Gly-to-Arg mutation might generate an aberrant protein that stresses the cell, thereby activating a nonspecific pathway for defense responses. Consistent with this possibility, previous studies have demonstrated that expression of various foreign or endogenous transgenes can induce PR gene expression and SAR (Durner et al., 1997). However, because SSI4 is a TIR-NBS-LRR–type R protein and ssi4 requires EDS1 but not NDR1 for defense response activation, it seems unlikely that this protein activates defenses via a nonspecific pathway. An alternative possibility is that the NBS mutation abolishes ssi4's ability to bind a guard protein that sequesters (and thereby silences) SSI4 in the absence of the appropriate Avr protein. Arguing against this possibility is the finding that overexpression of the SSI4 gene, which should titrate out the guard protein (as may have happened with overexpressed Pto, Prf, and At4g16890), did not induce defense responses. A more likely explanation is that the NBS mutation produces a constitutively activated R protein that constantly stimulates the defense signaling pathway, causing increased levels of SA, constitutive PR gene expression, and enhanced disease resistance. The observation that ssi4 is a semidominant gain-of-function mutation is consistent with this model. Note that the latter two models are not mutually exclusive.
In addition to stimulating downstream defense responses, the increased SA levels in the ssi4 mutant appear to enhance the expression of the ssi4 gene. Supporting this conclusion, ssi4 plants that express the nahG gene failed to accumulate increased levels of ssi4 transcripts. Analysis of the wild-type SSI4 gene and the closely related TIR-NBS-LRR R genes RPP1 and RPS4 suggests that their expression also is upregulated by SA. All of these genes were induced by SA treatment. Additionally, transcripts for RPP1 accumulated to higher levels in the ssi4 mutant than in wild-type plants. Another TIR-NBS-LRR R gene, 162J11T7, was shown recently to be highly induced by treatment with the SA analog BTH and to a lesser extent by infection with bacterial or oomycete pathogens (Maleck et al., 2000). Thus, increased levels of SA, such as those generated by pathogen infection, may upregulate the expression of at least some TIR-NBS-LRR genes. By contrast, SA treatment did not induce the CC-NBS-LRR genes RPM1 or RPS2, which suggests that upregulation of their expression either does not occur or involves a different mechanism.
Together, our results suggest that a positive feedback loop links SA accumulation and the expression of several TIR-NBS-LRR R genes, including SSI4. In this model, pathogen infection would lead to activation of the SSI4 protein; the activated protein then would transduce the resistance signal primarily via an EDS1-dependent signaling pathway. Increased SA levels generated during this response would not only activate downstream defenses directly but also would upregulate the expression of some R genes, thereby amplifying the defense response. Because defense signaling would not be initiated until the SSI4 protein was activated (presumably by a direct or indirect interaction with the pathogen Avr protein), plants that overexpress the SSI4 gene would not accumulate SA or display constitutive defense responses. By contrast, plants that express a constitutively activated R protein, such as ssi4, would accumulate increased levels of SA, which would stimulate additional expression of the ssi4 gene and activate the resistance signaling pathway.
METHODS
Isolation of the ssi4 Mutant and Genetic Analysis
Mutagenesis and screening for the ssi4 mutant were conducted as described previously (Shah et al., 1999). Briefly, 5000 seeds from Arabidopsis thaliana plants homozygous for the npr1-5 allele were mutagenized with 0.3% ethyl methanesulfonate (Sigma), and the M2 seeds from these plants were harvested in pools of ∼10 ethyl methanesulfonate–mutagenized M1 plants. Pooled M2 plants were screened for putative PR-1 overexpressors. Backcrosses were performed by pollinating flowers of the npr1-5 parental line (SSI4 npr1-5) with pollen from the ssi4 npr1-5 mutant. To isolate ssi4 mutants homozygous for the NPR1 wild-type allele, ssi4 npr1-5 mutant plants were crossed with wild-type ecotype Nö plants. Cleaved amplified polymorphic sequence (CAPS) marker analysis of 36 F2 plants then was used to identify plants that contained only the wild-type NPR1 allele. The genotype at the ssi4 locus in these F2 plants was determined by monitoring the segregation of PR gene expression in the F3 population using RNA gel blot analysis. To isolate ssi4 ndr1 and ssi4 eds1 double mutants, ndr1-1 (ecotype Columbia) or eds1-1 (ecotype Wassilewskija) plants were crossed with ssi4 homozygous plants. PCR markers for the ndr1-1, eds1-1, and ssi4 (see below) alleles then were used to identify the genotypes of the F2 progeny.
Map-Based Cloning of ssi4
ssi4 homozygous plants were crossed with a wild-type Landsberg plant, and F2 progeny lacking constitutive PR gene expression (homozygous for the SSI4 allele) were identified by RNA gel blot analysis. CAPS and simple sequence length polymorphism marker analysis performed on these F2 plants localized the ssi4 mutation to chromosome V, between PHYC and DFR. Sequence information (www.kazusa.or.jp/kaos/) then was used to generate novel CAPS and simple sequence length polymorphism markers in this region. Of 1179 F2 plants examined using these markers, two recombination events were identified using MBK23.42kb and three recombination events were detected using MJC20.2kb. No recombination events were identified using additional markers within this 132-kb region. Thus, ssi4 is located within a region spanning the latter half of P1 clone MBK23, the entire region of P1 clone MUF8, and the entire region of transformation-competent artificial chromosome (TAC) clone K16L22, as defined previously (www.kazusa.or.jp/kaos/).
An ssi4 TAC library was created as described by Liu et al. (1999). Briefly, genomic DNA from ssi4 homozygous mutant plants was partially digested with HindIII, and 10- to 20-kb fragments were purified by agarose gel electrophoresis and cloned into pYLTAC7 (Liu et al., 1999). The library was screened using the K16L22 TAC clone and PCR fragments from P1 clones MBK23 and MUF8 as probes. A contig was constructed based on the results of end-sequencing positive clones with vector-based primers. Two gaps in the contig were identified and filled by PCR-generated clones (Advantage-HF2; Clontech, Palo Alto, CA). Clones representing the entire contig then were transformed into wild-type ecotype Nö plants using the vacuum infiltration method (Bechtold and Pelletier, 1998). Transformants were selected on Murashige and Skoog (1962) medium containing 15 μg/mL hygromycin, and the presence of the entire T-DNA region was confirmed by PCR analysis using primers for the sacB and hygromycin resistance genes, indicating the left and right borders of the vector, respectively (Liu et al., 1999).
Segregation analysis of the ssi4 mutation among F2 and T2 plants was conducted by creating a derived CAPS marker. An ∼100-bp fragment was amplified using primers MUF8.2BglIIF (5′-GGTTCATCTTTGCGTGGGGAGAGCAAGCAA-3′) and MUF8.2BglIIR (5′-GTGAAGGAATAGAGATTGATTTTTCTTCGACAATCTTTCAGATC-3′) and digested with BglII. This generated either a 140-bp undigested fragment in the amplified product from the wild-type plants or 100- and 40-bp fragments in the amplified product from the ssi4 plants.
RNA Extraction and RNA Gel Blot Analysis
Total RNA was extracted from 3- to 4-week-old leaves using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA). RNA was separated on a 1.2% denaturing agarose gel, transferred onto a Hybond-NX membrane (Amersham Pharmacia), and hybridized with radiolabeled probes that were generated as described previously (Shah et al., 1997). PCR-amplified products of SSI4 (SSI4F, 5′-CTC-AAGAGAGTATGCTTCTCTTTCCATAACCC-3′; SSI4R, 5′-CTGGTT-TGGTCTTCATGAGACTCCATGAG-3′), EDS1 (EDS3F, 5′-GGATAG-AAGATGAATACAAGCC-3′; EDS1R, 5′-ACCTAAGGTTCAGGTATC-TGT-3′), and PAD4 (PAD4F, 5′-ATGGACGATTGTCGATTCGAG-3′; PAD4R, 5′-CTAAGTCTCCATTGCGTCACT-3′) were used as probes to detect these transcripts by RNA gel blot analysis.
Trypan Blue Staining
Leaf samples were taken from 2-week-old plants grown on soil. Trypan blue staining was performed as described previously (Bowling et al., 1994).
Salicylic Acid and Salicylic Acid Glucoside Measurement
Salicylic acid (SA) and SA glucoside were extracted and measured from 0.25 g (fresh weight) of leaf tissue as described previously (Bowling et al., 1994).
Pathogen Infection
Infection with Pseudomonas syringae pv maculicola ES4326 was performed on 4-week-old soil-grown plants as described previously (Shah et al., 1997). Leaves were infiltrated with a bacterial suspension (OD600 = 0.002) in 10 mM MgCl2. For each genotype, four leaf discs (4 mm in diameter) were harvested at day 0 and six discs were harvested at day 3; the discs were homogenized in 10 mM MgCl2, and colony-forming units were counted. Infection with Peronospora parasitica biotype Emco5 was performed by spraying or applying a single drop of asexual inoculum suspension (105 conidiosporangia/mL) per cotyledon of 7-day-old seedlings. The seedlings were grown at 19°C and >90% RH with an 8-h photoperiod. P. parasitica biotype Emco5 growth was assayed visually by counting the number of sporangiophores per cotyledon at 7 days after inoculation.
SA Treatment of Plants
Three-week-old plants were sprayed and irrigated for 10 min with a solution of SA (500 μM) or water. Leaves were harvested at the times indicated (in Figure 5B), and RNA was extracted as described above.
Reverse Transcriptase–Mediated PCR
cDNA for reverse transcriptase (RT)–mediated PCR was generated using SuperScript Reverse Transcriptase (Invitrogen Life Technologies) according to the manufacturer's instructions. PCR was performed using Advantage 2 polymerase (Clontech). The gene-specific primers used for RT-PCR analysis were as follows: for SSI4, SSI4F (5′-CTCAAGAGAGTATGCTTCTCTTTCCATAACCC-3′) and SSI4R (5′-CTGGTTTGGTCTTCATGAGACTCCATGAG-3′); for RPP1, RPP1F (5′-GTGGAGCTCCCCGCTATCGAGAATGCGAC-3′) and RPP1R (5′-GCAAGGGAATCTGGAAGTTGGGGGAGTGATACC-3′); for RPM1, RPM1F (5′-GCATACATGGGACCTAGGTTGCGTTTTGCACAAGG-3′) and RPM1R, (5′-GCCTTGGCCGCCTAAGATGAGAGGCTCAC-3′); and for Actin2, ACT2F (5′-CTAAGCTCTCAAGATCAAAGGCTT-AAAAAGCTGGGG-3′) and ACT2R (5′-CTTATACAATACTTATATTAACATTGCAAAGAGTTTCAAGGT-3′).
Construction of SSI4-Overexpressing Plants
An SSI4 cDNA of 3226 bp was amplified from Arabidopsis wild-type ecotype Nö RNA by RT-PCR using primers MUF8.2F (5′-CTT-TCTTTCAAGCATTTGTGATCTCTCATGGCTTCT-3′) and MUF8.2R (5′-CTGGTTTGGTCTTCATGAGACTCCATGAG-3′) and cloned into the pCR2.1-TOPO vector (Invitrogen Life Technologies). The cDNA clone was sequence verified, and the SSI4 coding sequence then was subcloned into the XbaI and SacI sites of pBI121 (Clontech), which placed the SSI4 gene under the control of the 35S promoter of Cauliflower mosaic virus. This construct then was transformed into wild-type ecotype Nö plants by vacuum infiltration (Bechtold and Pelletier, 1998). Transformants were selected on Murashige and Skoog (1962) medium containing 50 μg/mL kanamycin for 2 weeks, transferred to soil, and grown for 2 additional weeks before analysis of SSI4 expression.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for the SSI4 genomic sequence is AY179750.
Acknowledgments
We thank Daisuke Shibata for kindly providing the pYLTAC7 vector used in this study as well as S. Tabata for the P1 and TAC clones. We also thank J.E. Parker for eds1-1 seeds and P. Repetti and B.J. Staskawicz for ndr1-1 seeds and marker information. We are grateful to Shashi Sharma for conducting HPLC analysis for SA and SAG quantitation and to Keiko Yoshioka and Fasong Zhou for helpful advice. We also gratefully acknowledge D'Maris Dempsey for critical reading of the manuscript. This work was supported by Grant MCB-9723952/0110404 from the National Science Foundation to D.F.K.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005348.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (1996). Plant disease resistance genes: Function meets structure. Plant Cell 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., and Benyon, J.L. (2001). The Arabidopsis downy mildew resistance gene Rpp13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant-Microbe Interact. 14, 416–421. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Benyon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Holub, E.B., and Staskawicz, B.J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledfort, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance responses. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000. a). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3, 279–284. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000. b). The generation of plant disease resistance gene specificities. Trends Plant Sci. 5, 373–379. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D.G., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.-A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H. (1971). Current status of gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T., Guo, A., Klessig, D.F., and Ausubel, F.M. (1994). Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77, 551–563. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., and Jones, J.D.G. (1996). The roles of leucine rich repeats in plant defences. Adv. Bot. Res. Adv. Plant Pathol. 24, 90–167. [Google Scholar]
- Jones, J.D.G. (1996). Please disease resistance genes: Structure, function and evolution. Curr. Opin. Biotechnol. 7, 155–160. [Google Scholar]
- Kachroo, P., Shanklin, J., Shah, J., Whittle, E.J., and Klessig, D.F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. USA 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen, N.T. (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24, 447–463. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Kajava, A.V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Clarke, J.D., Zhang, Y., and Dong, X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant-Microbe Interact. 14, 1131–1139. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Shirano, Y., Fukaki, H., Yanai, Y., Tasaka, M., Tabata, S., and Shibata, D. (1999). Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome vector accelerates positional cloning. Proc. Natl. Acad. Sci. USA 96, 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J., Carr, J.P., Klessig, D.F., and Raskin, I. (1990). Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Cuzick, A., Can, C., Benyon, J.L., Dangl, J.L., and Holub, E.B. (2000). Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J. 22, 523–529. [DOI] [PubMed] [Google Scholar]
- Métraux, J.-P., Signer, H., Ryals, J., Ward, E., Wyss-Benz, M., Gaudin, J., Raschdorf, K., Schmid, E., Blum, W., and Inverardi, B. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Dickerman, A.W., Michelmore, R.W., Sivarsmakrishnan, S., Sobral, B.W., and Young, N.D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nimchuk, Z., Rohmer, L., Chang, J.H., and Dangl, J.L. (2001). Knowing the dancer from the dance: R-gene products and their interactions with other proteins from host and pathogen. Curr. Opin. Plant Biol. 4, 288–294. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E.D., and Staskawicz, B.J. (1998). Genetically engineered broad-spectrum disease resistance in tomato. Proc. Natl. Acad. Sci. USA 95, 10300–10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabò, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and Interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, M., Sibbald, P.R., and Wittinghofer, A. (1990). The P-loop, a common motif in ATP- and GTP-binding proteins. Trends Biotechnol. 15, 430–435. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., Nandi, A., and Klessig, D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. [DOI] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Silva, H., Yoshioka, K., Dooner, H.K., and Klessig, D.F. (1999). Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol. Plant-Microbe Interact. 12, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J., Ausubel, F.M., Baker, B.J., Ellis, J.G., and Jones, J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268, 661–667. [DOI] [PubMed] [Google Scholar]
- Stokes, T.L., Kunkel, B.N., and Richards, E.J. (2002). Epigenetic variation in Arabidopsis disease resistance. Genes Dev. 16, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X., Xie, M., Kim, Y.J., Zhou, J., Klessig, D.F., and Martin, G.B. (1999). Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes, S., Winter, A.M., Delaney, T., Vernooij, B., Morse, A., Friedrich, L., Nye, G., Potter, S., Ward, E., and Ryals, J. (1993). Biological induction of systemic acquired resistance in Arabidopsis. Mol. Plant-Microbe Interact. 6, 692–698. [Google Scholar]
- van der Biezen, E.A., Freddie, C.T., Kahn, K., Parker, J.E., and Jones, J.D.G. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NBS-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Vernooij, B., Friedrich, L., Morse, A., Reist, R., Kolditz-Jawhar, R., Ward, E., Uknes, S., Kessmann, H., and Ryals, J. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann, K., Hunt, M., Uknes, S., Neuenschwander, U., Lawton, K., Steiner, H.-Y., and Ryals, J. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7, 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Tao, X., Shen, B., Horng, T., Medzhitov, R., Manley, J.L., and Tong, L. (2000). Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408, 111–115. [DOI] [PubMed] [Google Scholar]
- Yoshioka, K., Kachroo, P., Tsui, F., Sharma, S.B., Shah, J., and Klessig, D.F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]