Abstract
Brassinosteroids (BRs) are plant steroid hormones that are essential for normal plant development. To gain better understanding of the conservation of BR signaling, the partially BR-insensitive tomato mutant altered brassinolide sensitivity1 (abs1) was identified and found to be a weak allele at the curl3 (cu3) locus. BR content is increased in both of these mutants and is associated with increased expression of Dwarf. The tomato homolog of the Arabidopsis Brassinosteroid Insensitive1 Leu-rich repeat (LRR) receptor-like kinase, named tBri1, was isolated using degenerate primers. Sequence analysis of tBRI1 in the mutants cu3 and abs1 revealed that cu3 is a nonsense mutant and that abs1 is a missense mutant. A comparison of BRI1 homolog sequences highlights conserved features of BRI1 sequences, with the LRRs in close proximity to the island domain showing more conservation than N-terminal LRRs. The most homologous sequences were found in the kinase and transmembrane regions. tBRI1 (SR160) also has been isolated as the putative receptor for systemin, a plant peptide hormone. This finding suggests a possible dual role for tBRI1 in steroid hormone and peptide hormone signaling.
INTRODUCTION
Brassinosteroids (BRs) are steroid hormones that affect plant growth and development, including the promotion of stem elongation, xylogenesis, and leaf bending, as reviewed by Mandava (1988) and Clouse and Sasse (1998). Mutants in the biosynthesis of the most bioactive BR, brassinolide (BL), are dark green dwarfs that are deetiolated when grown in the dark, which indicates that they lack the normal dark-grown developmental program (skotomorphogenesis). These dramatic phenotypes highlight the importance of steroid hormones in plants, and many reviews have discussed how recent advances in our understanding of BR biosynthesis and signaling are based on the analysis of such mutants (Altmann, 1999; Schumacher and Chory, 2000; Bishop and Yokota, 2001; Friedrichsen and Chory, 2001; Mussig and Altmann, 2001; Bishop and Koncz, 2002).
The initial genetic approach adopted to determine both how steroids in plants are perceived and how the subsequent signaling processes work has been to isolate BR-insensitive mutants. Initially, genetic screens were performed based on the fact that BRs inhibit normal root elongation (Clouse et al., 1996). Subsequent screens have involved selecting mutants that are phenotypically similar to the BR biosynthesis mutants and testing these for BR responsiveness (Kauschmann et al., 1996; Li and Chory, 1997). Initial results from such screens in Arabidopsis identified numerous recessive mutants that were found to be alleles at the Brassinosteroid Insensitive1 (BRI1) locus (Li and Chory, 1997), indicating that this is the major nonredundant component of BR signaling. Phenotypically, bri1 mutants are similar to the BR biosynthesis mutants in that they are dark green dwarfs that are deetiolated when grown in the dark. Other characteristics of bri1 mutants include increased transcript accumulation of BR biosynthesis genes and increased BR content (Noguchi et al., 1999).
Bri1 encodes a putative Leu-rich repeat receptor-like kinase (LRR-RLK) (Li and Chory, 1997) that is similar to RLKs involved in meristem proliferation (CLAVATA1 [Clark et al., 1997]), pathogen defense (Xa21 [Song et al., 1995] and FLS2 [Gomez-Gomez and Boller, 2000]), abscission (HAESA HAESA [Jinn et al., 2000]), and plant stature (ERECTA [Torii et al., 1996]). LRR-RLKs are involved in the signaling of the plant peptide hormones phytosulfokine (Matsubayashi et al., 2002) and systemin (Scheer and Ryan, 2002) and also are required to establish symbiotic relationships with microbes (Endre et al., 2002; Stracke et al., 2002). The topological domain structure of BRI1 includes an N-terminal signal peptide, a Leu zipper motif, and 25 imperfect LRRs that are interrupted by an island domain (Li and Chory, 1997). Paired Cys residues are found at either end of the extracellular LRR region, and this external region is followed by a transmembrane domain and the C-terminal kinase. Like many plant RLKs, BRI1 has Ser/Thr kinase activity but lacks Tyr kinase activity (Friedrichsen et al., 2000; Oh et al., 2000). Detailed characterization of sequences surrounding phosphorylated residues highlights a consensus sequence that is a substrate for BRI1 phosphorylation (Oh et al., 2000).
Further compelling evidence indicates that BRI1 is the BL receptor. A chimeric receptor was made with the extracellular LRR domains and the transmembrane region of BRI1 fused to the kinase domain of Xa21 (He et al., 2000). When transformed into rice cells with BL applied, this receptor was found to elicit downstream responses similar to those seen when the Xa21 LRR-RLK was stimulated with its elicitor (He et al., 2000). This finding indicates that the extracellular domain of BRI1 is sufficient to sense BL. More recent evidence that BRI1 is the BL receptor was gained through the use of radiolabeled BL and a transgenic line harboring a BRI1 receptor with a C-terminal green fluorescent protein fusion, BRI-GFP (Wang et al., 2001). The key experiment using these materials showed that BRI1 coimmunoprecipitates with BL and that the addition of BL promoted autophosphorylation (Wang et al., 2001). This finding is highly suggestive of BL binding to BRI1. In addition, the BRI-GFP fusion has been shown to localize to the plasma membrane and is expressed ubiquitously, although at higher levels, in meristematic tissues, which indicates that the receptor is both abundant and in an appropriate location for the perception of an extracellular ligand (Friedrichsen et al., 2000).
BRI1-Associated Receptor Kinase1 (BAK1) has been isolated via activation tagging and suppression of the weak bri1-5 phenotype (Li et al., 2002) and by BAK1's ability to interact with BRI1 in the yeast two-hybrid system (Nam and Li, 2002). Analysis of BAK1 indicates that it interacts with BRI11 and most likely forms a heterodimer that activates downstream phosphorylation, although exactly how BR interacts with this receptor complex remains to be determined.
In Arabidopsis, further genetic screens have identified additional components involved in BR signaling: bri1 Suppressor1 (BRS1) (Li et al., 2001a), Brassinosteroid-Insensitive2 (BIN2) (Li et al., 2001b; Li and Nam, 2002), BIN3 and BIN5 (Yin et al., 2002a), bri1-EMS-Suppressor1 (BES1) (Yin et al., 2002b), and Brassinazole-Resistant1 (BZR1) (Wang et al., 2002). brs1 is an activation-tagged suppressor mutant of the weak bri1-5 allele that results in the overexpression of a Ser carboxypeptidase (Li et al., 2001a). BIN2 is a homolog of glycogen synthase kinase/shaggy kinases; a dominant mutation of this kinase generates a BR-insensitive mutant similar to bri1 (Li and Nam, 2002). BIN2 acts as a kinase that negatively alters the accumulation of BES1 (Yin et al., 2002b) and BZR1 (He et al., 2002) in the nucleus. BZR1 and BES1 are novel nucleus-localized proteins that, when mutated, suppress BR-associated dwarfism. However, their exact roles in the nucleus remain to be defined. Genetic analysis of these mutants suggests that BRS1 acts upstream of BRI1, whereas BIN2, BZR1, and BES1 act downstream. However, the connections between BRI1 and these signaling components have yet to be determined. BIN5 is one of the SPO11 homologs in Arabidopsis, and BIN3 is a putative homolog of archeabacterial topoisomerase VI B (Yin et al., 2002a). It is unlikely that BIN5 and BIN3 play a direct role in BR signaling, but they may modulate the expression of BR-regulated gene expression by altering chromatin structure (Yin et al., 2002a).
Conservation of BL signaling in plants has been shown through the identification of BR-insensitive mutants in species other than Arabidopsis. The discovery of rice dwarf mutants that are defective in OsBRI1, the rice BRI1 homolog, highlights the conservation function of BRI1 between monocotyledonous plants (rice) and dicotyledonous plants (Arabidopsis) (Yamamuro et al., 2000). These rice dwarf mutants are only partially insensitive to BRs and do not have a dramatic dwarfing phenotype. This fact suggests that these are not null but weak alleles. Interestingly, OsBRI1 lacks three of the extracellular LRRs, suggesting evolutionary diversification of this sequence (Yamamuro et al., 2000). In addition to Arabidopsis and rice, BR-insensitive mutants have been identified in tomato (Koka et al., 2000) and pea (Nomura et al., 1997). These mutants are candidates to be defective in a BRI1 homolog, and their characterization is important to clarify the conservation of BRI1's role in plant development. In addition to the identification of conserved functions of BRI1, other BRI1 mutants may identify novel roles for BRI1 that are not revealed in one species. Such a parallel study of BR mutants has proved invaluable in the identification of the enzyme involved in the C-6 oxidation of BRs. The cloning of the tomato Dwarf gene (D) (Bishop et al., 1996) and characterization of the corresponding mutants identified DWARF as a C-6 oxidase of BRs (Bishop et al., 1999) for which no equivalent Arabidopsis mutant has been recovered. The lack of an Arabidopsis mutant equivalent to dwarf is most likely the consequence of genetic redundancy. It appears that there are two homologs of D in the Arabidopsis genome sequence (Arabidopsis Genome Initiative, 2000), one of which has been shown to have C-6 oxidase activity (Shimada et al., 2001).
A recent survey comparing tomato EST and Arabidopsis sequences highlights the fact that tomato, with a larger genome size, may have less genetic redundancy than Arabidopsis (Van der Hoeven et al., 2002). This finding suggests that by using tomato, it may be possible to recover novel mutations in BR signaling that may not be isolated in other species. To identify such novel mutants, we have characterized two tomato mutations that have BR insensitivity and that have been found to be defective in a BRI1 homolog (tBRI1). This has confirmed the important role of BRI1 homologs in plant steroid signaling.
It has been observed that the putative systemin receptor, SR160, has sequence homology with BRI1 (Scheer and Ryan, 2002). Sequence analysis between tBRI1 and SR160 has shown that these sequences are identical apart from single nucleotide polymorphisms, highlighting both the possible dual role of tBRI1/SR160 LRR-RLK in systemin and BR signaling and also the utility of using tomato to recover novel features of plant hormone signaling.
RESULTS
abs1, a New Allele at the cu3 Locus
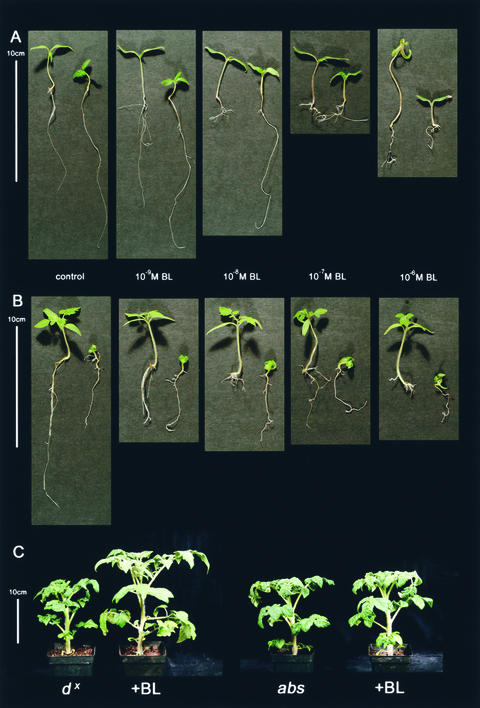
A tomato dwarf mutant was provided by C. Rick from a collection of many uncharacterized mutations maintained at the Rick Stock Center (University of California, Davis) for BR sensitivity tests. This choice was based on phenotypic similarity to the BR-deficient mutants extreme dwarf (dx) (Bishop et al., 1999) and dumpy (Koka et al., 2000)—that is, being a curly-leafed dark green dwarf. When grown in medium containing BL, the roots of this dwarf line were found to be partially sensitive to BL, but the aerial portion of the plant lacked any observable change in phenotype (Figure 1A). This mutant was named altered brassinolide sensitivity1 (abs1), because it appeared to be different from the previously reported curl3 (cu3) (Yu, 1982) mutant that has been shown to be insensitive to BRs (Koka et al., 2000). To directly compare the altered response of the roots to different concentrations of BL, cu3 mutants were grown under the same conditions as abs1 mutants. The cu3 mutants had similar root growth on medium containing no BL and at high concentrations of BL (Figure 1B). To verify that the aerial portions of abs1 plants were insensitive to BL and that this finding was not the consequence of poor BL transport from the medium, the abs1 and BR-responsive dx mutants were sprayed with BL. The abs1 mutant did not respond to a thrice weekly spray regime, whereas dx did (Figure 1C), indicating that aerial portions of the abs1 mutant were insensitive to BL. The cu3 mutant also did not respond to this spray regime (data not shown). An additional feature of the abs1 mutants is that they are fertile compared with the sterile cu3 mutants and have longer roots than wild-type plants (Figure 1A).
Figure 1.
Phenotypes of cu3 and abs Mutants in Response to BL.
(A) and (B) Photographs of seedlings at 18 days after sowing on medium containing various concentrations of BL.
(A) Pairs of seedlings: wild type (left) and abs1 (right).
(B) Pairs of seedlings: wild type (left) and cu3 (right).
(C) Phenotypes of 54-day-old plants (dx and abs1) sprayed with water control and 10−6 M BL (+BL).
Initially, it was thought that the abs1 mutant represented a new mutation in BL signaling as a result of the different response to BL. Allelism tests performed with cu3 and abs1, however, indicated that they were allelic. This was observed first in the progeny of the cross between a cu3 heterozygote and an abs1 line in which ∼50% were dwarf (abs1 like) and ∼50% were wild type (Table 1). Self progeny of the tall individuals all segregated ∼3:1 wild type:abs1 (Table 1). Self progeny of the dwarf individuals segregated ∼3:1 abs1:cu3 (Table 1). These data indicate that abs1 is a weak (intermediate) recessive allele at the cu3 locus; therefore, the abs allele of cu3 is now referred to as cu3-abs or the abs1 allele.
Table 1.
Segregation of abs and cu3 Visual Phenotypes
| Population | Observed Phenotypes | Number | χ2a |
|---|---|---|---|
| +/cu3 × abs/abs | Tall Short abs-like Short cu3-like |
8 14 0 |
1.64 |
| Tall selfedb | Tall | 29 | 0.59 |
| Short abs-like | 7 | ||
| Short cu3-like | 0 | ||
| Tall selfedb | Tall | 10 | 0.03 |
| Short abs-like | 3 | ||
| Short cu3-like | 0 | ||
| Short selfedc | Tall | 0 | 2.57 |
| Short abs-like | 36 | ||
| Short cu3-like | 6 | ||
| Short selfedc | Tall | 0 | 0.44 |
| Short abs-like | 9 | ||
| Short cu3-like | 3 | ||
| +/abs × abs/abs | Tall | 5 | 0 |
| Short abs-like | 5 | ||
| +/abs selfed | Tall | 24 | 0.35 |
| Short abs-like | 10 |
The ratio tested assumes that mutations are allelic.
Tall individual from cross +/cu3 × abs/abs.
Short individual from cross +/cu3 × abs/abs.
Endogenous BR Content
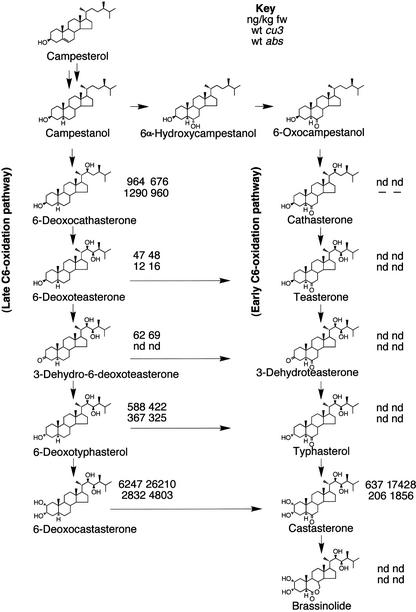
It has been shown that in the BR-insensitive mutants of Arabidopsis (Noguchi et al., 1999), rice (Yamamuro et al., 2000), and pea (Nomura et al., 1997), there are increased concentrations of BR intermediates. To determine whether the tomato mutants also have increased BR levels, the BR contents of cu3 and cu3-abs mutants were measured using deuterated internal standards and gas chromatography–mass spectrometry. Plant material for the cu3 line was harvested at 44 days after sowing. Wild-type plants averaged 812 ± 20 mm tall (n = 35), and cu3 plants averaged 41 ± 1 mm tall (n = 47). All of the cu3 mutant aerial tissue was harvested for analysis (68.7 g), whereas only tissue from the fifth leaf to the apex of the wild-type plants was taken (191.6 g). Plant material for the cu3-abs BR measurements was 93 days old. Wild-type plants were 918 ± 10 mm tall (n = 32), and abs mutants were 507 ± 5 mm tall (n = 24). Equivalent tissue was harvested from the eighth leaf to the apex. The BR content values shown in Figure 2 indicate that cu3 mutants have an ∼27-fold increase in castasterone (CS) compared with the wild-type equivalent, whereas cu3-abs mutants have an ∼9-fold increase, which is consistent with these mutants being defective in BR homeostasis. One striking feature from these data is the lack of the early C-6 oxidation pathway in tomato and the apparent deficiency of BL in the insensitive mutants. This finding highlights the potential role of CS as a bioactive BR and is consistent with the previous observation that tomato lacks an early C-6 oxidation pathway (Yokota et al., 1997; Bishop et al., 1999; Nomura et al., 2001).
Figure 2.
BR Biosynthesis Pathway and BR Contents of Tomato Mutants.
BR biosynthesis pathway adapted from Bishop and Yokota (2001), indicating the relative concentrations (ng/kg fresh weight [fw]) of BRs from wild-type (wt), cu-3, and abs1 plants. nd, not detected; -, not analyzed.
Transcript Analysis
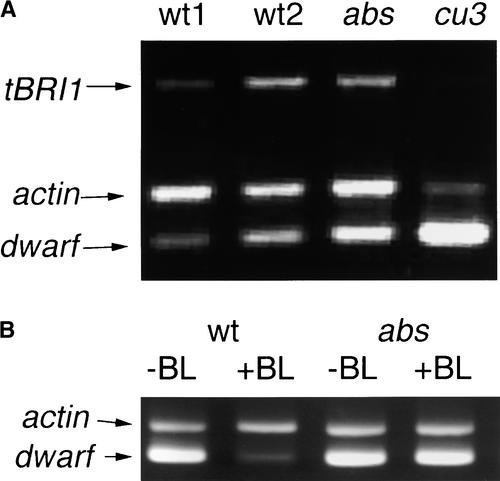
The increased endogenous BR concentrations in the cu3 and cu3-abs mutants suggested that the expression of BR biosynthesis genes would be increased in these lines. To determine whether these mutants had increased transcript levels of a BR biosynthesis gene, semiquantitative reverse transcriptase–PCR was performed using RNA from leaf tissue from the mutants and their wild-type equivalents. The relative transcript abundance of the tomato BR C-6 oxidase (Dwarf) gene was compared with the level of actin (Figure 3A). Both mutants showed increased levels of Dwarf transcript, indicating a lack of downregulation of this gene.
Figure 3.
Transcript Analysis.
(A) An ethidium bromide–stained agarose gel harboring products from reverse transcriptase–PCR of two wild-type (wt1, L. esculentum control for abs1; wt2, L. pimpinellifolium control for cu3) and two mutant (cu3 and abs1) RNAs. Primers for actin, tBRI1, and Dwarf genes were used in the same PCR. The gel shows the PCR products after 29 cycles of PCR.
(B) An ethidium bromide–stained agarose gel harboring products from reverse transcriptase–PCR of wild-type (wt; L. esculentum) and mutant (abs) RNAs from plants exposed to 10−6 M BL (+BL) or control (−BL). Primers for actin and Dwarf genes were used in the same PCR. The gel shows the PCR products after 33 cycles of PCR.
Recently, it was shown that the transcript accumulation of the Arabidopsis homologs of Dwarf is downregulated by BL (Bancos et al., 2002). In tomato, the regulation of Dwarf transcript levels by BL was confirmed by growing wild-type and cu3-abs mutant plants on medium for 19 days and subsequently transferring them to media with or without 10−6 M BL. After 4 h, the relative levels of Dwarf transcript were assayed as described above. Wild-type seedlings had decreased levels of Dwarf transcript, whereas there was no change in the cu3-abs mutants (Figure 3B), confirming that the cu3-abs mutant is defective in BL signaling.
Isolation of tBRI1 Homologs
These results were highly suggestive that cu3 and cu3-abs were defective in a homolog of the Arabidopsis BRI1 LRR-RLK. Concurrent with the BR sensitivity characterization of cu3-abs, a PCR-based approach was used to isolate the tomato homolog of the Arabidopsis BRI1 LRR-RLK. The method adopted used degenerate primers based on conserved amino acid residues within the kinase domains of BRI1 and the most homologous sequence known at the time of the experiment, a rice EST, and a confidential preview of the rice BRI1 sequence (Yamamuro et al., 2000). Degenerate primers were made based on the conserved amino acid sequences AEMETIGKIKH and EARVSDFGMA that were homologous between the expected BRI1 sequences but different from other RLK kinase domains. These primers were used on genomic tomato DNA and, assuming that this region would lack introns (i.e., it would be similar to Arabidopsis BRI1), a PCR product of ∼318 bp was expected. A PCR band of the appropriate size was obtained (data not shown) and cloned. The cloned band yielded two sequences with close homology with kinases, and gene-specific primers were designed to isolate larger portions of each sequence from a cDNA library (data not shown). One of these two kinases had higher homology with the Arabidopsis BRI1 sequence, and the complete sequence of this gene was obtained using inverse PCR. The other LRR-RLK was not characterized further. However, more recently, it was found to be 100% homologous with the LRR-RLK sequenced in the region of the lateral suppressor gene of tomato (Rossberg et al., 2001).
Because all of the phenotypic characterization experiments suggested that cu3 and cu3-abs were likely to be defective in the tBRI1 homolog, this gene was amplified from these mutants and their corresponding wild-type cultivars. Sequence comparison of the resulting PCR products revealed that cu3 contained a nonsense mutation, G749Z (Figure 4), which generates a TspRI restriction endonuclease polymorphism. The cu3-abs mutant has a missense mutation in the kinase domain, H1012Y (Figure 4). These mutations were consistent with the respective severity of the phenotypes, suggesting that cu3 is a null allele and that cu3-abs may have tBRI1 activity and may give rise to a weak allele.
Figure 4.
Alignment of BRI1 Protein Sequences from Tomato, Arabidopsis, Pea, and Rice.
Output from the alignment of rice, pea (T. Nomura, G.J. Bishop, and T. Yokota, unpublished data), and tomato (tom) sequences with the Arabidopsis (ara) BRI1 sequence using the program PileUp (Genetics Computer Group). The output was formatted using GeneDoc. Black boxes indicate identical amino acid residues. Spaces were introduced to separate domain regions as described in the text, with the LRR1 region containing 15 LRRs, the LRR2 region containing 6 LRRs, and the LRR3 region containing 4 LRRs. Currently known mutations resulting from a single base change in Arabidopsis (1-101, 1-103, 1-105, 1-113, and 1-115 [Li and Chory, 1997]; 1-1, 1-102, 1-114, and 1-117 [Friedrichsen et al., 2000]; and 1-5, 1-6, 1-8, and 1-9 [Noguchi et al., 1999]), in rice (d61-1 and d61-2 [Yamamuro et al., 2000]) and in tomato (cu3 and abs) are highlighted to indicate the change of amino acid residue.
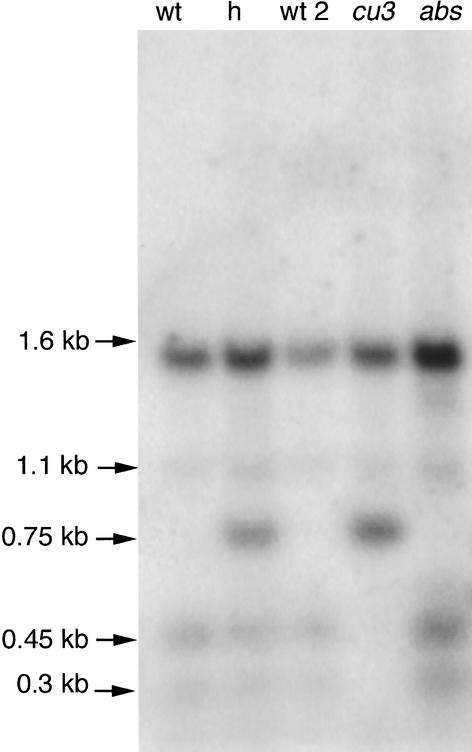
To determine the copy number of tBRI1 sequences and to confirm the predicted TspRI restriction site polymorphism present in the cu3 allele, DNA gel blot analysis was performed. TspRI-cut genomic DNA was hybridized with tBRI1 LRR regions, and after washing and exposure, the resulting image indicated that the gene was a single-copy gene (Figure 5). The image also indicates that there are no major deletions in the mutants and confirmed the predicted loss of a TspRI restriction site in the cu3 mutant.
Figure 5.
DNA Gel Blot Analysis.
Autoradiograph of a DNA gel blot hybridized with ∼2.9 kb of 32P-labeled tBRI1 sequence. Genomic DNA from the wild-type equivalent to abs (wt), the cu3 heterozygote (h), the wild-type equivalent of cu3 (wt 2), the cu3 mutant (cu3), and the abs mutant (abs) was digested with TspR1 restriction endonuclease and separated on a 0.8% agarose gel. Note the polymorphism in cu3, lacking bands of ∼0.45 and ∼0.3 kb and the presence of a 0.75-kb band.
Cosegregation Analysis
Sequence analysis indicated that not only does the cu3 allele lack a TspRI restriction site but also that the abs allele harbors the loss of a HphI restriction site. To further confirm that the sequence changes observed were linked to the mutant phenotype, coamplified polymorphism analysis of DNA isolated from individuals in the segregating populations described in Table 1 was performed. Photographs of representative gels highlighting the cosegregation of mutant phenotypes with sequence alteration can be seen in Figures 6A and 6B. Results from the genotypic analysis shown in Table 2 indicate that none of the plants tested showed a recombinant genotype from the observed phenotype and that cu3 and abs1 behave as allelic mutations.
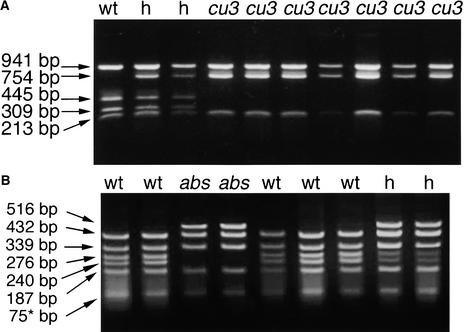
Figure 6.
Coamplified Polymorphism Analysis of the cu3 and abs1 Mutations.
Ethidium bromide–stained agarose gels highlighting the polymorphisms observed between wild-type and mutant DNAs.
(A) TspRI digestion of PCR-amplified DNA, using primers TBR27 and 5-1, from genomic DNA isolated from the wild type (wt), cu3 heterozygotes (h), and cu3 tomato mutants (cu3). Note that cu3 mutants lack a TspRI site yielding DNA fragments of 445 and 309 bp.
(B) HphI digestion of PCR-amplified DNA using primers TBR29 and 5-1 from genomic DNA isolated from the wild type (wt), abs1 heterozygotes (h), and abs1 tomato mutants (abs1). The 75-bp band indicated with the asterisk represents a doublet.
Table 2.
Genotypes of Segregating Individuals
| ABS/CU3 Genotypes Identified by PCR Restriction Fragment Length Polymorphism |
|||||
|---|---|---|---|---|---|
| Populationa | Observed Phenotype | ABS | CU3 | Number | χ2b |
| +/cu3 × abs/abs | Tall | +/abs | +/+ | 8 | 1.64 |
| Short abs-like | +/abs | +/cu3 | 14 | ||
| Tall selfedc | Tall | +/+ | +/+ | 16 | 1.65 |
| +/abs | +/+ | 23 | |||
| Short abs-like | abs/abs | +/+ | 10 | ||
| Short selfedd | Short abs-like | abs/abs | +/+ | 14 | 1.26 |
| +/abs | +/cu3 | 30 | |||
| Short cu3-like | +/+ | cu3/cu3 | 10 | ||
| +/abs × abs/abs | Tall | +/abs | nse | 5 | 0 |
| Short abs-like | abs/abs | ns | 5 | ||
| +/abs selfed | Tall | +/+ | ns | 8 | 0.35 |
| +/abs | ns | 16 | |||
| Short abs-like | abs/abs | ns | 10 | ||
Tall selfed and short selfed populations consist of combined populations from Table 1.
χ2 analysis based on allelic mutations.
Tall individual from cross +/cu3 × abs/abs.
Short individual from cross +/cu3 × abs/abs.
ns, not segregating in this population and not tested.
DISCUSSION
cu3 and cu3-abs Mutants Indicate Similar Roles of BRI1 Homologs
The identification of cu3 and cu3-abs mutants as defective in BR sensitivity aids our understanding of the central role that BRI1 plays in plant steroid signaling. The dramatic dwarfism associated with these tomato BR-insensitive mutants is similar to that of the BRI1 mutants of Arabidopsis. However, these tomato mutants have revealed some unique features in BR biology. First, it is apparent that in the insensitive line, there is a lack of early C-6 oxidation pathway intermediates and BL but an accumulation of CS (Figure 2). This feature reinforces the previous suggestions that the late C-6 oxidation pathway predominates in tomato and that CS may be a bioactive BR (Bishop et al., 1999). However, tomato seedlings respond to BL in a dose-dependent manner, as they do to CS (Bishop et al., 1999), indicating that although it may be absent in tomato, BL also can be used to screen for BR-insensitive mutants. The apparent lack of BL may be attributable to the fact that it remains below the level of detection in the tissue used for analysis (leaves and stems). This warrants further investigation using tissues in which BL is expected to be present at higher concentrations (e.g., pollen and seeds). A second novel feature of tomato BR signaling mutants is that the cu3-abs allele has a unique phenotype in that it has longer roots that have the ability to partially respond to BL. It is a mystery why the root and shoot tissues respond in different ways, but this may be associated with the recently observed differences in the levels of BR intermediates between such tissues (Bancos et al., 2002).
The cu3-abs allele is a weak allele, with the mutant plants being ∼50% of the height of wild-type plants. By contrast, the cu3 mutants are a mere ∼5% of wild-type height, and the presence of a stop codon suggests that cu3 is a null allele. The less dramatic phenotype of the cu3-abs mutants suggests that the mutated tBRI1 may have kinase activity, and it is possible that the altered root growth in these mutants may be associated with a novel activity of a mutated kinase domain. The H1012Y conversion in this allele is at a His in subdomain VIb (HRDMKSSN) of the kinase, which is highly conserved in Ser/The kinases, including LRR-RLKs. However, Tyr is found at this location in cyclic nucleotide–dependent and protein kinase C subfamilies (Hanks and Quinn, 1991). Subdomain VIb plays a key role in the kinase active site, with the Asp residue (D) being implicated as the catalytic base and the Lys residue (K) being in contact with the γ-phosphate of ATP (Schenk and Snaar-Jagalska, 1999). Because the His-to-Tyr conversion is within this catalytic loop, it is conceivable that the mutated tBRI1 in cu3-abs may not only have lower kinase activity for components of BR signaling but also may have altered substrate specificity and therefore subtly affect other signaling pathways. If such altered kinase activity exists, it may provide a link to the altered root growth in the abs1 mutants. This possibility is worthy of further investigation.
The cosegregation analysis of cu3 and cu3-abs visual phenotypes with the observed sequence alterations in tBRI1 provides convincing evidence that these mutants are defective in a BRI1 homolog. It is worth noting, however, that because of the different species from which these mutations were recovered (cu3-abs from Lycopersicon esculentum and cu3 from L. pimpinellifolium), the progeny from the crosses between the mutants produced a range of phenotypes, and partial suppression of the weak cu3-abs phenotype was observed. However, in all cases tested, the visual phenotype scored was consistent with the genotype expected.
Sequence Conservation
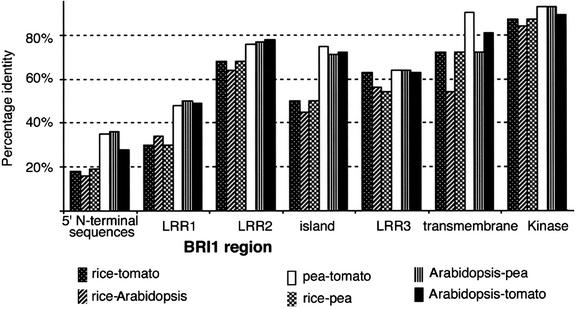
BRI1 is unique among LRR-RLKs because mutants have been identified with a similar phenotypic morphology in several plant species. Therefore, sequence analysis of these homologs provides an exceptional opportunity to reveal key regions in this LRR-RLK that are required for normal function. Initial inspection of the sequence alignment (Figure 4) indicates that the highest similarity is in the kinase domain. This finding is not unexpected, considering the enzymatic role this domain performs, and reinforces the utility of the use of a degenerate oligonucleotide approach to obtain tBRI1. To dissect the conservation of BRI1, homolog sequences were compared by subdividing BRI1 into the following regions: the amino signal peptide and Leu zipper motif (5′ N-terminal region), 15 N-terminal LRRs (LRR1), the 6 LRRs N-terminally adjacent to the island domain (LRR2), the island domain, the 4 LRRs C-terminally adjacent to the island domain (LRR3), the juxtamembrane region, the transmembrane region, the second juxtamembrane region, the kinase domain, and the carboxyl tail (Figure 4).
Results from pairwise sequence identity comparison in these selected regions (Figure 7) indicate several novel features. First, although key importance has been placed on the island domain because of the disproportionate isolation of Arabidopsis bri1 mutations in this region, it is not as conserved as the transmembrane and kinase domains, especially when comparing rice BRI1 with its pea, tomato, and Arabidopsis homologs. Although there is reduced conservation of sequences, all of the mutations found in this region were at Gly residues conserved between all species (Figure 4). In addition, the LRR regions on both sides of the island domain (LRR2 and LRR3) (Figures 4 and 7) are more conserved than the N-terminal LRRs (LRR1), where OsBRI1 lacks three LRRs. This observation suggests that the island and LRRs near this region, especially toward the transmembrane region, are important for BRI1 function and may interact either with the ligand or with other components in BRI1 signaling (i.e., BAK1) to form a receptor complex and promote downstream signaling events.
Figure 7.
Sequence Comparison of Different Regions of BRI1 Homologs.
Pairwise sequence identity comparison of different regions of BRI1 homologs using the output from the PileUp analysis of the four BRI1 homologs identified in Figure 5.
Tomato Cf-4 and Cf-9 are highly homologous LRR-containing genes that are involved in pathogen resistance and that share >91% amino acid identity (Thomas et al., 1997). These proteins have an extracellular interrupted LRR/island feature similar to that of BRI1 and are involved in the signaling of the two distinct avirulence proteins Avr 4 (Thomas et al., 1997) and Avr 9 (Jones et al., 1994). Interestingly, the sequence differences between these predicted proteins are confined to the N-terminal LRRs (Thomas et al., 1997) equivalent to the LRR1 region in Figure 4, indicating that the recognition/signaling determinants that differentiate these proteins lie within this region. More recently, the RLKs involved in symbiotic relationships with microbes were isolated. These RLKs have only a few extracellular LRRs that are adjacent to the membrane, with a relatively large N-terminal domain (Endre et al., 2002; Stracke et al., 2002). The LRR region of these kinases would be equivalent to region LRR3 in Figure 4, and this observation supports the idea that the LRRs adjacent to the membrane may perform a structural role rather than a role in the recognition of a ligand. Therefore, it is surprising that the N-terminal region, which is a candidate ligand interaction region, is one with the least homology in BRI1 sequences that have a similar BR ligand. It is exciting to speculate that this may be the consequence of BL interacting with a BL binding protein, which then binds BRI1 to activate BRI1 signaling, as discussed in a recent review (Bishop and Koncz, 2002). Therefore, the variation in the LRR1 region would reflect the recognition of different BL binding proteins. An alternative possibility is that the transmembrane domain plays a key role in BL recognition, because this is an environment in which steroids may be present, and it also is a highly conserved region.
The isolation of other BRI1 homologs will prove informative in developing a rational approach to test key regions for their effect on BRI1 function. This will be aided by the fact that in Arabidopsis, three closely related homologs have been identified, two of which have been reported to bind BL (Yin et al., 2002c). There also are homologous sequences in tomato and other species that may provide additional important information in determining the key regions necessary for BRI1 function. Additionally, it may now be possible to rapidly isolate other BRI1 homologs from numerous species using the degenerate oligonucleotide PCR approach described above. This approach will be useful not only for evolutionary aspects of BRI1 function but also will enable the isolation of BRI1 homologs from important crops and provide the tools for the manipulation of plant architecture.
Is tBRI1 a Dual Ligand Receptor?
Highlighted in the introduction was the recent and intriguing finding that the peptide hormone systemin binds to a plasma membrane–localized protein, SR160 (Scheer and Ryan, 1999), which is homologous with BRI1 (Scheer and Ryan, 2002). Sequence comparison of this putative systemin receptor and tBRI indicates that the sequences are identical apart from single nucleotide polymorphisms derived from the different species from which these sequences were isolated (tBRI1 from L. esculentum and SR160 from L. peruvianum) (data not shown). Systemin is a member of an increasing family of peptide hormones that are being discovered in plants (Ryan et al., 2002). The observation that systemin interacts with a LRR-RLK is not unusual, because the LRR motif is implicated in protein–protein interactions (Kobe and Deisenhofer, 1994, 1995) and the proposed ligand for CLV1 is the polypeptide CLV3 (Fletcher et al., 1999). More recently, it was observed that a phytosulfokine, which is another plant peptide hormone, interacts with a LRR-RLK (Matsubayashi et al., 2002). Systemin is an 18–amino acid peptide involved in defense responses to insect attack and wounding. Signaling induced by systemin causes an increase in jasmonate production that leads to defense gene expression.
The binding of systemin to tBRI1/SR160 is a novel and exciting result that suggests an overlapping role between BL and systemin signaling. As discussed by Yin et al. (2002c), the identification of a mutant in tBRI1/SR160 will be invaluable in confirming whether this LRR-RLK plays a dual role in systemin and BR signaling. Such confirmation can be sought through the predicted lack of labeled systemin or BL binding to membranes extracted from cu3 plants. Further supportive evidence also could be obtained to determine whether the tBRI1 mutants are defective in systemin signaling responses (e.g., jasmonate production or defense gene expression). Once confirmed, this will open a new and exciting area of research, because tBRI1 is a plant receptor with two quite different ligands.
METHODS
Plant Growth
Tomato (Lycopersicon esculentum) plants were grown either in environmentally controlled cabinets (16-h days at 25°C) or in a glasshouse with supplementary heating and lighting (16-h days).
Brassinosteroid Analysis
Brassinosteroid (BR) contents of leaf and shoot material after the removal of fruits and flowers were determined using deuterated BR internal standards and extraction procedures described previously (Yokota et al., 1997; Bishop et al., 1999; Nomura et al., 2001).
tBRI1 Cloning Strategy
Exact details of cloning are available upon request. The strategy adopted used degenerate primers dBR2 and dBR5 (Table 3) on genomic DNA from tomato. Both vector and gene-specific primers based on cloned fragments were used on a cDNA library made from 5-day-old light-grown seedlings, and 3′ sequences were determined. Full-length tBRI1 was obtained by inverse PCR as described by Thomas et al. (1994) using self-ligated tomato genomic DNA that had been digested previously with XbaI enzyme. The resulting ∼4-kb product was sequenced, but it lacked 5′ sequences of tBRI1. To obtain the 5′ sequences, inverse PCR was repeated using self-ligated DNA after PvuI digestion. A PCR product of ∼900 bp was cloned, sequenced, and confirmed to be homologous with Arabidopsis BRI1; it also contained the ATG start codon and part of the promotor. To ensure that there were no PCR errors, primers were used to amplify the coding sequences, and PCR bands of the expected size were sequenced.
Table 3.
Primer Sequences Used
| Primer Name |
5′ to 3′ Sequence |
|---|---|
| dBR2 | GCNGARATGGARACNATHGGNAARATHAARCA |
| dBR5 | GCCATNCCRAARTCNSWNACNCKNGCMTC |
| DW52 | TTCTTTTGAAATTTTGAGGTGCATC |
| DW53 | CTCCCATATCTGGCTCTTTG |
| Oligo-(dT) | GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTT |
| TACT1 | ATTGCTCTTGACTATGAACAGG |
| TACT2 | CTTGCTCATCCTATCAGCAATACC |
| 4-9 | CAAGCTGAATTGGCCTGCAAG |
| 4-10 | CAAGACATTACTTGATTTCATGTCC |
| 5-1 | TGGATGGGAACTAGTGGTACATAC |
| TBR 20 | GTCAATCTCTCCAAGCAATGTAAGACTG |
| TBR 27 | CTGCTGAATTGGGTAATTGCCAGAG |
| TBR 29 | TCCTCAACAACTTGGAGGCTTGAAG |
| TBR 31 | TTGGAGACTTTGGATATGAGTTC |
| TBR 32 | GGATCTTTGCAAATCCCAGATGG |
| TBR 35 | AGATCTATGGAGTCTAAAGTAACC |
| TBR 41 | CATCAAGAGCTCAAGCTATAGATTCAAG |
| TBR 42 | CCCCAGAGATTAGAGTGTGTTCTC |
Sequence Analysis of abs1 and cu3
PCR products using primer pairs TBR42 plus TBR32, TBR31 plus TBR20, TBR29 plus 4-10, and 4-9 plus 5-1 (Table 3) from DNA of mutants and their respective wild-type equivalents were sequenced directly using gene-specific primers. All ambiguous regions were reamplified and resequenced. Sequence analysis was performed using the Genetics Computer Group (Madison, WI) software program PileUp and Staden (Cambridge, UK) sequencing software programs PreGap4 and Gap4, using the default settings where appropriate. Output from the PileUp program was edited using GeneDoc software.
DNA Gel Blot Analysis
Genomic DNA (10 μg) was digested using 30 units of TspRI. Products were run on a 0.8% agarose gel and transferred onto Hybond N+ membranes (Amersham) according to the manufacturer's instructions. Membranes were prehybridized for 2 h in hybridization solution (Church and Gilbert, 1984). The radiolabeled DNA probe of 2.9-kb tBRI1 sequences lacking the kinase region was generated by random priming using the High Prime Labeling Kit (Roche, Indianapolis, IN) according to the manufacturer's instructions. The denatured probe then was added to the prehybridization solution, and membranes were hybridized overnight at 65°C. Two washes in 2 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at room temperature and two washes in 0.1 × SSC and 0.1% SDS at 65°C were performed to remove nonspecific hybridization. Membranes then were exposed to PhosphorImager screens (Molecular Dynamics, Sunnyvale, CA) and scanned using a Thyphoon 8600 scanner (Amersham, Little Chalfont, UK).
Reverse Transcriptase–PCR
Total RNA from leaves was isolated using Total RNA Isolation Reagent from ABgene (Epsom, UK). Single-strand DNA was synthesized using 1.4 μg of total RNA, reverse transcriptase from Moloney murine leukemia virus (Life Technologies, Paisley, UK), and oligo(dT) anchor primer according to the manufacturer's instructions. Three picomoles of oligo(dT) anchor primer and 10 nmol of deoxynucleotide triphosphate in a total volume of 15 μL were denatured by heating at 65°C for 5 min and stored on ice for 5 min. Five microliters of 5 × buffer (250 mM Tris-HCI [pH 8.3], 375 mM KCI, 15 mM MgCl2), 2.5 μL of 0.1 M DTT, 50 units of RNase inhibitor, and 250 units of reverse transcriptase from Moloney murine leukemia virus were added, and the reaction was incubated at 37°C for 1 h. The reaction was stopped by incubating at 70°C for 15 min. Single-stranded DNA was amplified by PCR using the following primers at the given concentrations: 0.4 μM TBR41 and TBR35 (to amplify tBRI1); 0.3 μM DW52 and DW53 (to amplify Dwarf); and 0.14 μM TACT1 and TACT2 (to amplify actin as a control). A total of 29 and 32 cycles were performed, and results from the 29 cycles were selected for the analysis. Gels stained with ethidium bromide were photographed using GeneSnap (Syngene, Cambridge, UK) and analyzed using GeneTools manual quantification (Syngene).
Coamplified Polymorphism Analysis
Polymorphisms between abs1 and the wild type were detected by amplifying genomic DNA with primers TBR29 plus 5-1 and digesting with HphI restriction enzyme. Polymorphisms between cu3 and the wild type were identified by amplifying genomic DNA with primers TBR27 plus 5-1 and digesting with TspRI restriction endonuclease. Digestion products were separated on 2% (w/v) agarose gels.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are CAC36401, AY179606 (tBRI1) tomato BRI1 sequence, AY112661 (putative systemin receptor SR160), D39976 (a rice EST), AP003453.3 (rice BRI1 sequence), and AAC49810 (Arabidopsis BRI1 sequence).
Acknowledgments
We thank members of the Botany Gardens for plant growth and care. We also thank Anthony Pugh and Gwen Jenkins for photography and help and advice in the preparation of the figures. We thank M. Matsuoka for providing a copy of the rice BRI1 protein sequence before publication and Y. Kamiya for his support. We also are grateful to both S. Clouse and C. Rick of the Rick Stock Center for tomato seeds. We thank J. Scheer and C. Ryan for providing SR160 sequence prior to publication. We appreciate J. Chory's helpful comments on the manuscript and for providing preprint articles. We thank D. Jones for help in defining the LRRs of tBRI1. G.J.B. and T.N. thank the Royal Society for support. This work contributes to the Human Frontier Research Program (Grant 2000-162). G.J.B. also acknowledges support from the Biotechnology and Biosciences Research Council (Grant P13908) and the British Council. T.N. acknowledges the support of a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006379.
References
- Altmann, T. (1999). Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208, 1–11. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome se-quence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bancos, S., Nomura, T., Sato, T., Molnár, G., Bishop, G.J., Koncz, C., Yokota, T., Nagy, F., and Szekeres, M. (2002). Regulation of transcript levels of the Arabidopsis cytochrome P450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D.G. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging en-codes the first member of a new cytochrome P450 family. Plant Cell 8, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., and Koncz, C. (2002). Brassinosteroids and plant steroid hormone signaling. Plant Cell 14 (suppl.), S97.–S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D.G., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96, 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, G.J., and Yokota, T. (2001). Plants steroid hormones, brassinosteroids: Current highlights of molecular aspects of their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 42, 114–120. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse, S.D., and Sasse, J.M. (1998). Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966. [DOI] [PubMed] [Google Scholar]
- Fletcher, L.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D., and Chory, J. (2001). Steroid signaling in plants: From the cell surface to the nucleus. Bioessays 23, 1028–1036. [DOI] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Joazeiro, C.A.P., Li, J.M., Hunter, T., and Chory, J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., and Quinn, A.M. (1991). Protein-kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200, 38–62. [DOI] [PubMed] [Google Scholar]
- He, J.-X., Gendron, J.M., Yang, Y., Li, J., and Wang, Z.-Y. (2002). The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z.H., Wang, Z.Y., Li, J.M., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., and Walker, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14, 108–117. [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D.G. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Kauschmann, A., Jessop, A., Koncz, C., Sekeres, M., Willmitzer, L., and Altmann, T. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9, 701–713. [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 19, 415–421. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Koka, C.V., Cerny, R.E., Gardner, R.G., Noguchi, T., Fujioka, S., Takatsuto, S., Yoshida, S., and Clouse, S.D. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122, 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Lease, K.A., Tax, F.E., and Walker, J.C. (2001. a). BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wen, J., Lease, K.A., Doke, J.T., Tax, F.E., and Walker, J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Li, J.M., and Nam, K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li, J.M., Nam, K.H., Vafeados, D., and Chory, J. (2001. b). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava, B.N. (1988). Plant growth promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 23–52. [Google Scholar]
- Matsubayashi, Y., Ogawa, M., Morita, A., and Sakagami, Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472. [DOI] [PubMed] [Google Scholar]
- Mussig, C., and Altmann, T. (2001). Brassinosteroid signaling in plants. Trends Endocrinol. Metab. 12, 398–402. [DOI] [PubMed] [Google Scholar]
- Nam, K.H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., Feldmann, K.A., and Tax, F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Nakayama, M., Reid, J.B., Takeuchi, Y., and Yokota, T. (1997). Blockage of brassinosteroid biosynthesis and sensitivity causes dwarfism in garden pea. Plant Physiol. 113, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T., Sato, T., Bishop, G.J., Kamiya, Y., Takatsuto, S., and Yokota, T. (2001). Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry 57, 171–178. [DOI] [PubMed] [Google Scholar]
- Oh, M.H., Ray, W.K., Huber, S.C., Asara, J.M., Gage, D.A., and Clouse, S.D. (2000). Recombinant brassinosteroid insensitive 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 124, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossberg, M., Theres, K., Acarkan, A., Herrero, R., Schmitt, T., Schumacher, K., Schmitz, G., and Schmidt, R. (2001). Comparative sequence analysis reveals extensive microcolinearity in the Lateral suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, C.A., Pearce, G., Scheer, J., and Moura, D.S. (2002). Polypeptide hormones. Plant Cell 14 (suppl.), S251.–S264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, J., and Ryan, C.A. (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, J.M., and Ryan, C.A. (1999). A 160-kD systemin receptor on the surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell 11, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.W., and Snaar-Jagalska, B.E. (1999). Signal perception and transduction: The role of protein kinases. Biochim. Biophys. Acta 1449, 1–24. [DOI] [PubMed] [Google Scholar]
- Schumacher, K., and Chory, J. (2000). Brassinosteroid signal transduction: Still casting the actors. Curr. Opin. Plant Biol. 3, 79–84. [DOI] [PubMed] [Google Scholar]
- Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., Yokota, T., Kamiya, Y., Bishop, G.J., and Yoshida, S. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126, 770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, XA21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczglowski, K., and Parniske, M. (2002). A plant receptor like-kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M., Jones, D.A., English, J.J., Carroll, B.J., Bennetzen, J.L., Harrison, K., Burbidge, A., Bishop, G.J., and Jones, J.D.G. (1994). Analysis of the chromosomal distribution of transposon-carrying T-DNAs in tomato using the inverse polymerase chain-reaction. Mol. Gen. Genet. 242, 573–585. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M., Jones, D.A., Parniske, M., Harrison, K., Balint-Kurti, P.J., Hatzixanthis, K., and Jones, J.D.G. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., Nakano, T., Gendron, J., He, J.X., Chen, M., Vafeados, D., Yang, Y.L., Fujioka, S., Yoshida, S., Asami, T., and Chory, J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. [DOI] [PubMed] [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Cheong, H., Friedrichsen, D., Zhao, Y., Hu, J., Mora-Garcia, S., and Chory, J. (2002. a). A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc. Natl. Acad. Sci. USA 99, 10191–10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y.H., Wang, Z.Y., Mora-Garcia, S., Li, J.M., Yoshida, S., Asami, T., and Chory, J. (2002. b). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yin, Y.H., Wu, D., and Chory, J. (2002. c). Plant receptor kinases: Systemin receptor identified. Proc. Natl. Acad. Sci. USA 99, 9090–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, T., Nomura, T., and Nakayama, M. (1997). Identification of brassinosteroids that appear to be derived from campesterol and cholesterol in tomato shoots. Plant Cell Physiol. 38, 1291–1294. [Google Scholar]
- Yu, M.H. (1982). The dwarf curly leaf tomato mutant. J. Hered. 73, 470–472. [Google Scholar]