Abstract
The rice basic domain/Leu zipper factor TRAB1 binds to abscisic acid (ABA) response elements and mediates ABA signals to activate transcription. We show that TRAB1 is phosphorylated rapidly in an in vivo labeling experiment and by phosphatase-sensitive mobility shifts on SDS–polyacrylamide gels. We had shown previously that a chimeric promoter containing GAL4 binding sites became ABA inducible when a GAL4 binding domain–TRAB1 fusion protein was present. This expression system allowed us to assay the ABA response function of TRAB1. Using this system, we show that Ser-102 of TRAB1 is critical for this function. Because no ABA-induced mobility shift was observed when Ser-102 was replaced by Ala, we suggest that this Ser residue is phosphorylated in response to ABA. Cell fractionation experiments, as well as fluorescence microscopy observations of transiently expressed green fluorescent protein–TRAB1 fusion protein, indicated that TRAB1 was localized in the nucleus independently of ABA. Our results suggest that the terminal or nearly terminal event of the primary ABA signal transduction pathway is the phosphorylation in the nucleus of preexisting TRAB1.
INTRODUCTION
The plant hormone abscisic acid (ABA) mediates plant responses to environmental stress, such as high salinity, drought, low temperature, and mechanical wounding (Giraudat et al., 1994). ABA also plays important roles in the accumulation of storage proteins and other reserve materials, the acquisition of desiccation tolerance, the arrest of embryonic development, and the achievement of dormancy during seed maturation (McCarty, 1995).
ABA activates various genes associated with these processes (Giraudat et al., 1994; Busk and Pages, 1998). Analyses of the ABA-regulated promoters in a search for cis elements required for ABA induction have revealed ABA-responsive elements (ABREs) (Marcotte et al., 1989; Guiltinan et al., 1990; Mundy et al., 1990; Skriver et al., 1991; for review, see Busk and Pages, 1998). Because multimerized ABREs can confer ABA responsiveness to a heterologous, minimal promoter (Skriver et al., 1991; Vasil et al., 1995), this sequence intrinsically possesses the capacity to mediate ABA signals. However, in a natural promoter context, an ABRE requires a second sequence element called a coupling element. The two elements together, or with other additional elements, constitute an ABA-responsive complex and synergistically activate transcription in response to ABA (Shen and Ho, 1995; Shen et al., 1996). Although two distinct coupling elements, namely CE1 (Shen and Ho, 1995) and CE3 (Shen et al., 1996), were reported originally, the latter has been shown to be a variant of ABRE (Hobo et al., 1999a). Therefore, two or more copies of ABREs also appear to constitute an ABA-responsive complex (Hobo et al., 1999a).
Basic domain/Leu zipper (bZIP) factors such as the wheat EmBP1 (Guiltinan et al., 1990), the tobacco TAF-1 (Oeda et al., 1991), and the rice OSBZ8 (Nakagawa et al., 1996) and osZIP-1a (Nantel and Quatrano, 1996) have been reported as candidates for ABRE binding factors. However, none of these proteins has been shown conclusively to be involved in ABA-responsive gene expression. By contrast, a distinct subfamily of bZIP proteins has been shown to be responsible for ABA-induced transcription in recent studies (Hobo et al., 1999b; Choi et al., 2000; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Uno et al., 2000; Kang et al., 2002). TRAB1, one of these factors, has been cloned by yeast two-hybrid screening in a search for proteins that interact with VP1/ABI3 (Hobo et al., 1999b), a transcription factor that is required for ABA-regulated gene expression during seed development (McCarty et al., 1991; Giraudat et al., 1992; Hattori et al., 1992, 1994). Although VP1/ABI3 binds to Sph/RY elements via its B3 domain (Suzuki et al., 1997), it also acts through ABRE (Hattori et al., 1995; Vasil et al., 1995). However, VP1/ABI3 does not bind directly to ABRE; instead, it functions via an interaction with ABRE binding factors such as TRAB1 (Hobo et al., 1999b) and ABI5 (Nakamura et al., 2001). TRAB1 has been demonstrated to have the ability to mediate ABA signals, because transcription from a chimeric promoter containing GAL4 binding sites becomes ABA inducible in the presence of a GAL4 DNA binding domain–TRAB1 fusion protein (Hobo et al., 1999b).
ABI5 is an Arabidopsis bZIP factor homologous with TRAB1 (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). The abi5 mutant displays reduced sensitivity to ABA upon seed germination. The transcript levels for some LEA genes have been shown to be lower in abi5 mutant seeds (Finkelstein, 1994; Finkelstein and Lynch, 2000; Lopez- Molina and Chua, 2000). Physical interaction between ABI3 and ABI5 also has been demonstrated by yeast two-hybrid assays (Nakamura et al., 2001). In addition to its function in late embryogenesis, ABI5 is suggested to play a role in a checkpoint mechanism by which growth is arrested under stressed conditions during a limited period of postgerminative development (Lopez-Molina et al., 2001). In Arabidopsis, other TRAB1 homologs (AREB1/ABF2, AREB2/ABF4, AREB3, ABF1, and ABF3) also have been cloned (Choi et al., 2000; Uno et al., 2000). These factors were identified on the basis of their ability to bind ABREs by the yeast one-hybrid screen. Some of these factors have been shown to activate an ABA-responsive promoter and to enhance its ABA responsiveness (Uno et al., 2000).
Because ABA-induced transcription via ABREs basically does not require de novo protein synthesis (Mundy and Chua, 1988), TRAB1 and other ABRE binding factors represent the terminal factors of the primary ABA signaling pathway. In addition to these transcription factors, several components involved in ABA signal transduction have been identified. ABI1 and ABI2 each encodes a protein phosphatase 2C that is homologous with the other (Leung et al., 1994, 1997; Meyer et al., 1994), and both of them have been shown to negatively regulate ABA signals (Gosti et al., 1999; Merlot et al., 2001). Overexpression of calcium-dependent protein kinases (CDPKs) has been shown to activate ABA-regulated promoters (Sheen, 1996). AAPK, an ABA-activated protein kinase that is involved in ABA-induced stomatal closure, has been cloned from Vicia faba (Li et al., 2000). Second messenger molecules, such as cyclic ADP-Rib, phosphatidic acid, and phosphoinositides that regulate intracellular Ca2+ levels, and the enzymes that produce these molecules, are implicated in ABA signaling (Wu et al., 1997; Ritchie and Gilroy, 1998; Sanchez and Chua, 2001).
Although information about ABA signal transduction is increasing, how ABA signals are transmitted to the transcription machinery and lead to the activation of gene expression remains largely unknown. In the present study, we investigated the molecular changes in TRAB1 protein that are caused by ABA signals. We show here that TRAB1 is phosphorylated rapidly in response to ABA. We also demonstrate that this ABA-induced phosphorylation of TRAB1 occurs at a specific Ser residue that is essential to its capacity to mediate the ABA signal.
RESULTS
Conserved Sequence Blocks and Phosphorylation Signatures in TRAB1
To date, six different bZIP proteins from Arabidopsis have been reported, all of which have bZIP structures highly homologous with that of TRAB1 and all of which are implicated in ABA regulation, in addition to those reported previously for Helianthus (Kim et al., 1997). When the amino acid sequence of TRAB1 is compared with those of homologs from other plant species, three sequence blocks are found, designated regions I to III, that are highly conserved among these factors in the N-terminal region (Figure 1). The sequence comparison and examination of the genomic sequence corresponding to the TRAB1 gene obtained from the Syngenta draft sequence database (http://portal.tmri.org/rice/RiceAccess.html; Goff et al., 2002) revealed that the protein encoded by the previously reported cDNA clone (Hobo et al., 1999b) lacked the N-terminal 56 amino acids, which include half of the conserved region I. The conserved regions contain an interesting repeated motif, Thr-hy-ac-ac, where hy and ac represent hydrophobic and acidic amino acid residues, respectively. This motif includes a potential casein kinase II phosphorylation site predicted by PROSITE searching. The PROSITE search also revealed a potential protein kinase C phosphorylation site in region II. In addition, another motif, QGSLT, which also contains a potential phosphorylation site (Lopez-Molina et al., 2001), was found to be repeated twice in regions I and II.
Figure 1.
Conserved Sequence Blocks I to III of the TRAB1 Family Proteins.
The structure of TRAB1 is shown in the scheme at top, where regions I to III and the bZIP are indicated with black and hatched boxes, respectively. Amino acids identical, similar, and dissimilar to those of TRAB1 and gaps are indicated by dashes, uppercase letters, lowercase letters, and underlined letters, respectively. The repeated sequence motifs Thr-hy-ac-ac and QGSLT (see text) are indicated by solid and dotted underlines, respectively. The Ser residue in the protein kinase C phosphorylation signature is indicated by a dot. The arrow points to the Met residue formerly predicted to be the N terminus from the sequence of the TRAB1 cDNA clone. The amino acids of TRAB1 are numbered based on the putative full-length sequence, which includes the N-terminal 56 amino acids predicted from the genome sequence obtained from the Syngenta draft sequence database (see text).
Although the cDNA clone for TRAB1 turned out to be partial, we performed further analysis with this clone because it retains the ability to mediate ABA signal and to bind ABREs (Hobo et al., 1999b).
Role of the N-Terminal Conserved Sequence Blocks
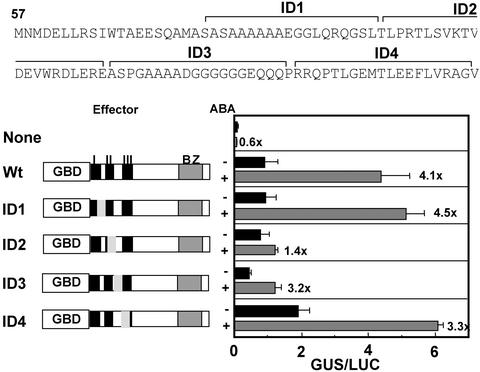
We examined the role of the conserved sequence blocks in the N-terminal region. Serial internal deletions of 20 amino acids were introduced in the N-terminal region of the TRAB1 part of GBD-TRAB1 (DNA binding domain–TRAB1 fusion protein), and each of these deletion constructs was cotransfected with the UAS-TATA-GUS reporter gene (a β-glucuronidase [GUS] reporter gene under the regulation of a chimeric promoter consisting of GAL4 binding sites and the 35S minimal promoter of Cauliflower mosaic virus) into rice protoplasts (Figure 2). The expression of the UAS-TATA-GUS reporter gene was induced by ABA when it was cotransfected with the wild-type GBD-TRAB1, as reported previously (Hobo et al., 1999b). This experimental system allowed us to assay the activating and/or ABA signaling function of TRAB1 independently of its DNA binding function. The deletions that removed conserved regions I and III (Figure 2, constructs ID1 and ID4) did not affect the ABA induction of the reporter gene expression. By contrast, the deletion in ID2, which removed most of region II, resulted in a significant decrease of ABA-induced transcription as well as basal transcription. Construct ID3, carrying a deletion between regions II and III, gave a decreased level of reporter gene expression in both the absence and presence of ABA; however, it retained the ability to respond to ABA. These results indicate that region II but not region III is essential for mediating ABA signal. Because we did not test a deletion of the N-terminal 20 amino acids of the truncated version of TRAB1, it is possible that this part of region I also might be essential. In addition, the possibility cannot be excluded that the N-terminal sequence missing in our TRAB1 clone might contribute to greater ABA responsiveness or activation.
Figure 2.
Effects of Internal Deletions in the N-Terminal Region of TRAB1 on the Ability to Mediate ABA-Responsive Transcription.
Ten micrograms each of the effector plasmid for wild-type (Wt) GBD-TRAB1 or its derivatives with an internal deletion of 20 amino acids (ID1 to ID4) as indicated in the sequence at top and the UAS-TATA-GUS reporter gene containing GAL4 binding sites was cotransfected into protoplasts of rice suspension-cultured cells by electroporation. Each transfection also included 5 μg of a ubiquitin promoter::luciferase (LUC) plasmid as an internal standard. GUS activities were normalized with luciferase activity. The deleted region is indicated by the shaded area in the scheme of each mutant construct. The value of induction by ABA is shown with the ABA + bars. BZ, bZIP.
TRAB1 Is Phosphorylated in Response to ABA
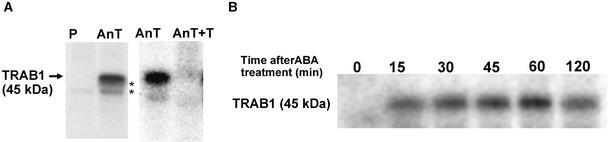
Phosphorylation is a frequently used mechanism to regulate the activity of transcription factors in lower and higher organisms. TRAB1 was tested for this by in vivo 32P labeling followed by immunoprecipitation experiments. Rice cells were cultured in the presence of 32Pi for 1 h and treated with 50 μM ABA for various time periods. Nuclear extracts were prepared from the labeled cells and subjected to immunoprecipitation with antibody raised against recombinant TRAB1 protein. As shown in Figure 3A, a 45-kD 32P-labeled polypeptide was detected in the immunoprecipitates from the ABA-treated cells. Neither control preimmune antibody nor anti-TRAB1 antibody preincubated with an excess amount of recombinant TRAB1 protein was able to precipitate the labeled polypeptide. This result indicates that TRAB1 is, in fact, phosphorylated. A time course experiment (Figure 3B) showed that TRAB1 was not phosphorylated in the absence of ABA but was phosphorylated within 15 min after ABA treatment. The level of radioactivity in the TRAB1 polypeptide remained relatively constant up to 120 min. Because the amount of TRAB1 in the nuclear extract did not increase greatly after ABA treatment (Figure 4A and data not shown), the incorporation of 32P label into TRAB1 protein after ABA treatment was not attributable to de novo synthesis of the protein. These results indicate that TRAB1 is phosphorylated in an ABA-dependent manner.
Figure 3.
TRAB1 Is Phosphorylated Rapidly in Response to ABA.
Rice suspension-cultured cells (Oc) were labeled with 32Pi for 1 h. Cells then were treated with 50 μM ABA. Nuclear extracts were prepared from the labeled cells and subjected to immunoprecipitation with anti-TRAB1 antibody followed by SDS-PAGE.
(A) After ABA treatment for 30 min, immunoprecipitation was performed with preimmune serum (P), anti-TRAB1 antibody (AnT), or anti-TRAB1 antibody preincubated with an excess amount of recombinant TRAB1 (AnT+T). The position of the 45-kD TRAB1 band is indicated by the arrow. The smaller minor bands indicated by asterisks are considered to be immunologically related to TRAB1. However, it is not known whether they are partially degraded forms of TRAB1 or the products of related genes.
(B) Immunoprecipitation with anti-TRAB1 antibody using nuclear extracts from the labeled cells treated with ABA for the indicated times.
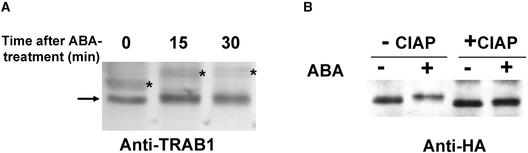
Figure 4.
Mobility Shift of TRAB1 on SDS-PAGE Caused by ABA-Induced Phosphorylation.
(A) Nuclear extracts of rice suspension-cultured cells (Oc cells) treated with ABA for the indicated times (0 indicates untreated cells) were analyzed by immunoblotting with anti-TRAB1 antibody. The arrow and asterisks indicate TRAB1-specific and nonspecific bands, respectively. Although the expression of the nonspecific bands was affected by ABA, the nature of these polypeptides is unknown.
(B) TRAB1-dHA/His expressed in transgenic cells treated without (−) or with (+) ABA for 30 min was recovered with nickel–nitrilotriacetic acid agarose resin, incubated without (−) or with (+) CIAP, and analyzed by immunoblotting with anti-HA antibody. The signals seen at the top of the +CIAP lanes are parts of bulky CIAP bands that reacted nonspecifically with the anti-HA antibody.
A retardation of mobility on SDS-PAGE often is observed when a polypeptide is phosphorylated (Wang et al., 1998; Karniol et al., 1999; Ishida et al., 2000; Lopez-Molina et al., 2001). Although not very clear, the electrophoretic mobility of TRAB1 appeared to decrease when cells were treated with ABA (Figure 4A). This observation was consistent with the labeling experiments. To characterize this mobility shift more clearly and to exclude the interference of cross-reacting polypeptides, TRAB1 protein tagged with double hemagglutinin and His6 epitopes (TRAB1-dHA/His) was expressed in transgenic rice callus and detected with anti-HA antibody. Again, a decrease in the electrophoretic mobility of TRAB1-dHA/His polypeptides by ABA treatment was observed (Figure 4B). To confirm that this mobility shift in response to ABA was caused by phosphorylation, TRAB1-dHA/His was purified partially from the extract of control or ABA-treated cells by nickel–nitrilotriacetic acid agarose resin and treated with calf intestine alkaline phosphatase (CIAP). The phosphatase treatment of TRAB1-dHA/His from both control and ABA-treated cells resulted in an increase in mobility that was indistinguishable between the two (Figure 4B). These results indicate that the ABA-induced mobility retardation of TRAB1 was the result of phosphorylation. In addition, the observation that the mobility of TRAB1-dHA/His polypeptide from the control cells also was increased by the phosphatase treatment indicates that TRAB1 is phosphorylated not only in response to ABA but also constitutively at a residue(s) different from the one that undergoes ABA-induced phosphorylation. TRAB1-dHA/His protein, from both control and ABA-treated cell extracts, sometimes split into doublet bands depending on slight changes in the electrophoretic conditions, suggesting that the constitutive phosphorylation occurs at multiple sites.
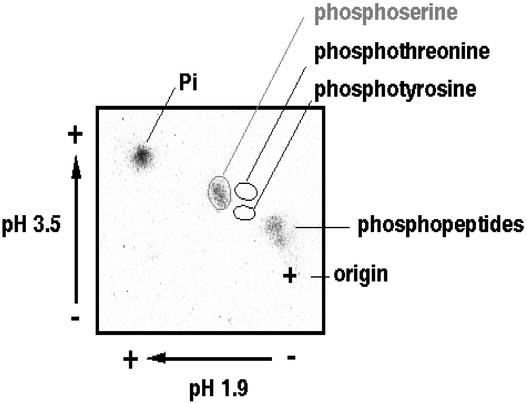
To determine the amino acid species that was phosphorylated in response to ABA, 32P-phosphorylated TRAB1 protein, detected as in Figure 3, was recovered from the polyacrylamide gel and subjected to acid hydrolysis followed by two-dimensional thin layer electrophoresis. As shown in Figure 5, 32P-radioactivity was found predominantly in the spot corresponding to phosphoserine but not in the phosphothreonine or phosphotyrosine spot. These results show that only a Ser residue in TRAB is phosphorylated in response to ABA.
Figure 5.
Determination of Which Amino Acid Species Is Phosphorylated in Response to ABA.
32P-labeled TRAB1 protein obtained as described for Figure 3 was recovered from the polyacrylamide gel and subjected to phosphoamino acid analysis. The autoradiogram made after two-dimensional thin layer electrophoresis is shown. The spots corresponding to phosphoserine, phosphotyrosine, incompletely digested phosphopeptide, and free phosphate are indicated.
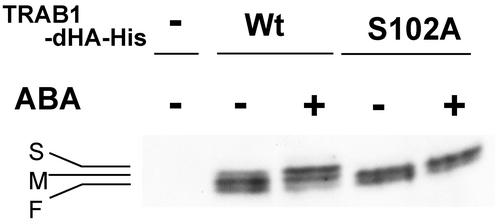
Ser-102 of TRAB1 Is Phosphorylated and Essential for ABA-Induced Transcription
Because the deletion of amino acid residues 97 to 116 of TRAB1 resulted in the loss of the capacity to mediate the ABA signal, we suspected that the Ser residue in this region might be phosphorylated in response to ABA. This region contains a single Ser residue (Ser-102), which the PROSITE search predicted to be a potential protein kinase C phosphorylation site. To test this possibility, a mutant TRAB-dHA/His (S102A), in which Ser-102 was replaced by Ala, was compared with wild-type TRAB-dHA/His expressed transiently in protoplasts prepared from cultured rice cells (Figure 6). By immunoblot analysis, wild-type TRAB1-dHA/His was detected as triplet bands, which are designated bands F (fast), M (middle), and S (slow) for their order of mobility (Figure 6). In contrast to wild-type TRAB1-dHA/His, the S102A mutant protein gave only two bands, F and M (Figure 6). Therefore, the S band of the wild-type protein depends on the presence of Ser-102. ABA treatment of the cells resulted in an increase in the intensity of band S of the wild-type protein and a decrease in the intensity of band F. This change in the relative intensities of the two bands corresponded to the phosphorylation-associated mobility shift observed in Figure 4. ABA treatment affected neither the mobility nor the intensity of the two bands of the mutant protein. These results, in conjunction with those from the experiments using the transgenic callus, strongly suggest that Ser-102 is phosphorylated in response to ABA.
Figure 6.
ABA-Dependent Phosphorylation of TRAB1-dHA/His Is Abolished by the S102A Mutation.
Protoplast cells transfected with the expression plasmid for wild-type TRAB1-dHA/His (Wt), its S102A mutant derivative (S102A), or the empty vector 35S-shΔ-stop (−) were treated without (−) or with (+) ABA for 1 h. Total cellular extracts were analyzed by immunoblotting with anti-HA antibody. The positions of the S, M, and F bands (see text) are indicated.
Because constitutive phosphorylation appears to occur at multiple sites, as suggested above, the F and M bands in the absence of ABA signal most likely result from different levels of constitutive phosphorylation. This notion was confirmed by CIAP treatment, which converted both the three bands of wild-type TRAB1-dHA/His and the two bands of the S102A mutant into single bands with the same mobility, which was greater than that of the F band (data not shown).
ABA-induced phosphorylation at Ser-102 probably converted the F band to M as well as M to F. Therefore, the M band would represent both Ser-102 phosphorylated and unphosphorylated forms with lower and higher levels, respectively, of constitutive phosphorylation. This could explain the ABA-induced increase and decrease in the intensities of the F and S bands, respectively, without change in the intensity of the M band. The weak S band of the wild-type protein, the Ser-102–phosphorylated form, found in the control cell extracts probably was produced by partial ABA activation or some other signal from the osmotic stress caused by the protoplast culture medium.
Ser-102 of TRAB1 Is Essential for ABA-Induced Transcription
Because ABA-induced phosphorylation of Ser-102 was suggested by the experiment described above, the functional importance of this residue was tested by introducing the S102A mutation into GBD-TRAB1 (Figure 7). Indeed, the S102A mutation greatly reduced the ability of the GBD-TRAB1 fusion protein to confer ABA responsiveness to the UAS-TATA-GUS reporter gene. A control mutation (S94A) did not affect ABA induction. These results indicate that Ser-102 is essential for the ABA-induced activation of transcription, which is likely to be mediated by the phosphorylation of this Ser residue. The level of reporter gene expression in the absence of ABA also was decreased by the mutation. In the transient expression system, the basal level of reporter gene expression might be attributable in part to slight phosphorylation of Ser-102, as seen in Figure 6. Therefore, the loss of this phosphorylated form by mutation may have resulted in a decrease in the basal level of reporter gene expression.
Figure 7.
Effects of Mutations at Ser-102 on the Ability of TRAB1 to Mediate ABA-Responsive Transcription.
Cotransfection assays were performed as described for Figure 2 using wild-type (Wt) GBD-TRAB1 or its derivatives that carry mutations at Ser-102. “None” indicates assays using the empty effector plasmid 35S-shΔ-stop. The mutations are illustrated in the sequence at top. The value of induction by ABA is shown with the ABA + bars.
(A) Effect of the S102A mutation.
(B) Effect of the S102D mutation.
To further confirm the significance of phosphorylation at Ser-102, this residue was changed to an acidic residue, which might mimic a phosphoamino acid residue. As shown in Figure 7B, the S102D mutation significantly increased the level of activation in the absence of ABA. These results strongly support the importance of phosphorylation at Ser-102. Unexpectedly, the S102D mutation still responded to ABA, resulting in a level of reporter expression greater than that obtained with wild-type GBD-TRAB1 in the presence of ABA. This finding suggests that another ABA-dependent step is involved in the ABA-responsive activation of TRAB1, although ABA-induced phosphorylation at Ser-102 is a prerequisite. As seen in Figure 6, a portion of the wild-type protein remained unphosphorylated at Ser-102 (the F band and probably a portion the M band) after ABA treatment. By contrast, expression of the S102D mutant mimicked complete phosphorylation at Ser-102. This difference could explain the higher levels of basal and ABA-induced reporter gene expression with S102D compared with those seen with the wild-type protein.
TRAB1 Is Localized in the Nucleus Independently of ABA
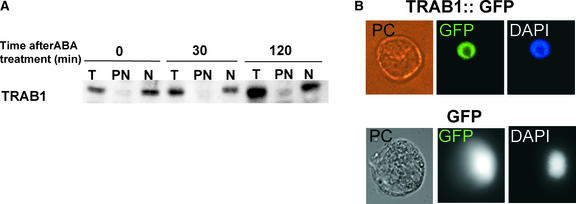
Previous studies have shown that transport from the cytoplasm to the nucleus of some bZIP proteins is regulated by signal-dependent phosphorylation (Harter et al., 1994; Droge-Laser et al., 1997; Kircher et al., 1999). To determine whether this is the case with TRAB1, subcellular fractionation was performed followed by immunoblot analysis. As shown in Figure 8, TRAB1 always was recovered in the nuclear fraction, irrespective of ABA treatment. The nuclear localization of TRAB1 was further examined in an experiment using a green fluorescent protein (GFP) fusion with TRAB1. Rice protoplast cells transformed with an expression plasmid for the GFP-TRAB1 fusion protein exhibited GFP fluorescence localized predominantly in the nucleus in the absence of ABA. By contrast, GFP fluorescence was observed in the entire region of the cell when normal GFP was expressed. These results confirmed the constant nuclear localization of TRAB1. Together with the results of experiments described above, our findings indicate that preexisting TRAB1 in the nucleus undergoes phosphorylation in response to ABA.
Figure 8.
TRAB1 Is Present in the Nucleus Independently of ABA.
(A) Rice suspension-cultured cells (Oc cells) were treated with 50 μM ABA for the indicated times (0 indicates untreated cells). Total cell (T), crude nuclear (N), and postnuclear (PN) fractions were prepared and subjected to immunoblot analysis with anti-TRAB1 antibody. Each lane was loaded with protein corresponding to an approximately equal volume of cells.
(B) Phase contrast and epifluorescence microscopy images deriving from GFP or 4′,6-diamidino-2-phenylindole (DAPI) of a rice protoplast transiently expressing GFP or TRAB1-GFP. Oc cell protoplasts were transformed with a GFP or TRAB1-GFP expression plasmid by electroporation and cultured for 12 h before microscopic observation. Protoplasts were not treated with ABA. DIC, differential interference contrast.
DISCUSSION
Phosphorylation/dephosphorylation is a frequently used mechanism to regulate the activity of a transcription factor in both eukaryotes and prokaryotes. In the present study, we demonstrate that TRAB1 is phosphorylated rapidly in vivo at a specific Ser residue in response to ABA. The S102A mutation led to the loss of an ABA-induced mobility shift of TRAB1 on SDS-PAGE. Furthermore, the mutation resulted in a great reduction of the capacity to mediate the ABA-induced activation of transcription. Therefore, Ser-102 is most likely phosphorylated in response to ABA, although the possibility that Ser-102 instead is critical to recognition by the responsible kinase cannot be excluded. In either case, the phosphorylation of TRAB1 is important to mediate the ABA-induced activation of transcription.
Conversion of Ser-102 to Asp in GBD-TRAB1 did not result in a simple constitutive active form; instead, it led to increases of both basal and induced levels of reporter gene expression. This result suggests that a more complex mechanism operates for the ABA-induced activation of TRAB1. Although ABA-induced phosphorylation at Ser-102, which can be mimicked by the S102D mutation, is essential, TRAB1 is believed to undergo additional ABA-dependent change(s) for full activation. One such change could be the covalent modification of TRAB1, such as phosphorylation at another residue, which would not cause a detectable mobility shift on SDS-PAGE. Alternatively, the activity of a protein that interacts with TRAB1 could be regulated by ABA.
Although Ser-102 is conserved in most TRAB1-related bZIP proteins, including those identified from the Arabidopsis genome sequence, this Ser residue is replaced by a Cys residue in ABI5 and AREB3. One reason for this may be that ABI5 and AREB3 have another phosphorylation site that functions in place of this Ser. In fact, phosphorylation of ABI5 has been reported by Lopez-Molina et al. (2001). However, their results were somewhat puzzling because 32P labeling resulted in the same rate of 32P incorporation into ABI5 irrespective of ABA treatment, whereas they observed a phosphatase-sensitive mobility shift that was induced by ABA. From their results, ABI5 appears to undergo both constitutive and ABA-induced phosphorylation at different sites, but only the ABA-induced phosphorylation causes the apparent mobility shift. They suggested a rearrangement of the phospho group within the molecule. They also reported a relatively high rate of ABI5 protein turnover in the absence of ABA. This rapid turnover of ABI5 may be responsible for the incorporation of the 32P label in the absence ABA, which was not observed for TRAB1 within a similar time course of labeling.
Although both TRAB1 and ABI5 are responsible for ABA-regulated transcription and interact with VP1/ABI3, their mechanisms for mediating ABA signals may be slightly different. The level of ABI5 itself is upregulated significantly by ABA through an increase in the transcript levels as well as the stability of the protein (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001). The expression of ABI5 also is under developmental regulation. For example, the level of ABI5 transcript is considerably higher at the very late stage than at the middle phase of embryogenesis (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), and the ABA-induced accumulation of ABI5 protein is observed within only a limited period of the postgerminative seedling stage (Lopez-Molina et al., 2001). Therefore, a major part of transcriptional regulation via ABI5 could be accounted for by the level of the protein. In this sense, ABI5 could mediate an ABA response even if it is constitutively active. By contrast, the level of TRAB1 is affected only moderately by ABA (Hobo et al., 1999b). Thus, TRAB1 must be regulated tightly by its activity. The lack in ABI5 of the Ser residue corresponding to Ser-102, which is critical for TRAB1 to mediate ABA signals, as well as the differences in the phosphorylation characteristics might account for the difference in the major mode of regulation between the two ABRE binding factors.
Ser-102 is found within a protein kinase C phosphorylation signature site predicted by PROSITE. However, a different type of protein kinase is thought to be responsible for the phosphorylation of TRAB1, because the presence of protein kinase C in plants is unlikely. Several protein kinases have been reported to be involved in ABA regulation. Constitutive overexpression of CDPK in maize mesophyll protoplasts has been reported to activate ABA-regulated promoters (Sheen, 1996). However, CDPK is proposed to function at the same level or upstream of ABI1/ABI2 protein phosphatase (Sheen, 1998), which is believed to operate at an early step of the signal transduction pathway. Thus, CDPK is not likely to be directly responsible for the ABA-regulated phosphorylation of TRAB1 or other equivalent transcription factors, which is the most downstream event in the primary ABA signaling pathway. Recently, Uno et al. (2000) reported an in vitro ABA-dependent protein kinase activity from Arabidopsis cultured cells using an N-terminal fragment of AREB1. The substrate fragment they used included the Ser residue corresponding to Ser-102 of TRAB1. Although the significance of this protein kinase activity in vivo has not been demonstrated, this or a similar kinase would be the most likely candidate for the ABA-dependent TRAB1 kinase. Another ABA-activated protein kinase, AAPK from guard cells of V. faba, has been characterized molecularly and physiologically and implicated in the regulation of the ABA activation of slow anion channels and thereby stomatal closure (Li et al., 2000). The position of AAPK in the ABA signaling pathway is not known. It might be close to the bottom step of the signaling pathway leading to stomatal closure, namely, activation of the anion channel. Although all ABA signaling steps that lead to stomatal closure are considered to be cytosolic events, ABA-dependent phosphorylation of TRAB1, or other ABRE binding factors of similar function, should occur in the nucleus, as discussed below. However, the kinase could function in both the cytosol and the nucleus. In addition, the Arabidopsis genome sequence predicts a family of AAPK homologs consisting of at least nine members. Therefore, one or some member(s) of the family could operate in the nucleus to activate TRAB1 and the related ABRE binding factors.
The regulated phosphorylation of several plant bZIP factors has been reported. These factors include OPAQUE2 (Ciceri et al., 1997), HY5 (Hardtke et al., 2000; Osterlund et al., 2000), G-box binding factors (Klimczak et al., 1992; Harter et al., 1994), H/GBF1 (Droge-Laser et al., 1997), and CPRFs (Kircher et al., 1999; Wellmer et al., 1999, 2001). The signal-induced phosphorylation of these factors has been demonstrated or suggested to alter their subcellular localization, stability, or affinity to the binding sequence. However, the consequence of ABA-induced TRAB1 phosphorylation is likely to be different. As shown previously and here, GBD-TRAB1 can activate transcription in response to ABA through a heterologous DNA binding domain and its target sequence (Hobo et al., 1999b). Thus, the major regulatory step for ABA-induced transcription through TRAB1 does not appear to lie at the level of DNA binding. The only possibility for GBD-TRAB1 to bind the GAL4 target sequence in an ABA-dependent manner is via a global conformational change in TRAB1, which would unmask the DNA binding surface covered by TRAB1 polypeptide in the inactive state. The constant nuclear localization, independent of ABA, observed in the present study precludes the possibility of regulated nuclear targeting. Thus, ABA-induced phosphorylation is more likely to regulate the activation function. We showed previously that TRAB1 interacts physically with VP1, which has a strong acidic transcriptional activation domain at its N terminus (McCarty et al., 1991; Hobo et al., 1999b). In addition, the possibility of ABA-regulated interaction between the two factors has been suggested (Hobo et al., 1999b). Although we have not mapped the domain in TRAB1 responsible for this interaction, the N-terminal region that includes Ser-102 would be a candidate for the interacting domain, which might have a higher affinity for VP1 when phosphorylated. Alternatively, phosphorylation may cause a conformational change that results in the exposure of an interacting surface that resides in a different region.
In addition to ABA-induced phosphorylation, we found that TRAB1 is phosphorylated constitutively at different sites. Because the constitutive phosphorylation is likely to occur at multiple sites, the Thr residues in the repeated conserved motif Thr-hy-ac-ac, which is within the context of the casein kinase II phosphorylation site signature, could serve as such sites. The functional significance of the constitutive phosphorylation is unknown. It might be important for other regulation, such as desensitization of ABA signals, or in cross-talk with other signals.
Because most of the ABA signaling components identified to date are common to ABA-induced stomatal closure and gene expression, which occur in different intracellular compartments, the signaling pathway must branch at some point. However, little is known about the branched pathway specific to each different output. The ABA-induced phosphorylation of TRAB1 identifies the terminal or nearly terminal event of the gene expression branch of the ABA signaling pathway. Because phosphorylation is suggested to be a nuclear event, TRAB1 kinase may be specific to the gene expression pathway. Alternatively, the same kinase may function in both the cytoplasm and the nucleus. Identification and cloning of the TRAB1 kinase will clarify this point and enable us to trace backward along the ABA signaling pathway, revealing the components of the gene expression branch.
METHODS
Plasmid Constructions
The internal deletion series of the GBD-TRAB1 expression plasmids were constructed from the wild-type p35S-ShΔ-GBD–TRAB1 plasmid using the ExSite PCR-based site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the following primer pairs: 5′-CTG-CCGCGCACTCTCAGCGTCAAG-3′ and 5′-CGCCATCGCTTGGCT-CTCCTCCGC-3′ for ID1; 5′-GCGTCGCCGGGGGCGGCTGCT-3′ and 5′-GGTGAGCGACCCCTGCCTCTGCAG-3′ for ID2; 5′-AGGCGG-CAGCCGACGCTCGGGGAG-3′ and 5′-CTCTCGCTCTAAGTCCCG-CCACAC-3′ for ID3; and 5′-GTCAGAGAGAATACCGCTGCGGCG-3′ and 5′-GGGCTGCTGCTGCTCCCCGCC-3′ for ID4. The presence of desired mutations and the absence of undesired mutations were confirmed by sequencing the entire TRAB1 coding region. Because the mutagenesis method included PCR amplification of the entire plasmid, an EcoRI-BamHI fragment of each mutated plasmid, which corresponds to the N-terminal half of the TRAB1 coding region, was excised and used to replace the corresponding fragment in the original wild-type p35S-ShΔ-GBD–TRAB1 plasmid to avoid undesired mutations outside of the TRAB1 coding region. The GBD-TRAB1 expression plasmids with the S102A and S102D mutations were constructed similarly using a common prime (5′-GTGCGCGGCAGGGTG-AGCGA-3′) and a specific primer (5′-TCTCGCCGTCAAGACGGT-GGACGA-3′ and 5′-TCTCGACGTCAAGACGGTGGACGA-3′ for S102A and S102D, respectively).
A double hemagglutinin (dHA)–His tag sequence was obtained by annealing the synthetic oligonucleotides 5′-TCGACCCTTACCCAT-ACGACGTTCCAGACTACGCTGGTTACCCATACGACGTTCCAGACT-ACGCTAGATCCGGTCACCACCACCACCACCACTAA-3′ and 5′-GAT-CTTAGTGGTGGTGGTGGTGGTGACCGGATCTAGCGTAGTCTGGAA-CGTCGTATGGGTAACCAGCGTAGTCTGGAACGTCGTATGGGTAAG-GG-3′ and ligated into SalI- and BglII-digested p35S-ShΔ-stop (Suzuki et al., 2001) to produce p35S-ShΔ-dHA/His. TRAB1 sequence was amplified from the pBluescript II SK− subclone by PCR using the M13 reverse primer and a synthetic oligonucleotide, 5′-TTATTA-TGTCGACCCAGGGACCTGTCAATG-3′, digested with EcoRI and SalI, and cloned into the corresponding site of p35S-ShΔ-dHA/His to obtain the TRAB1-dHA/His expression plasmid (p35S-ShΔ-TRAB1-dHA/His) for transient expression.
For Agrobacterium tumefaciens–mediated transformation, pIG121- Hm (Ohta et al., 1991) was modified by inserting a synthetic linker composed of 5′-AGCTTCTCGAGGCGGCCGCACTAGTGAGCT-3′ and 5′-CACTAGTGCGGCCGCCTCGAGA-3′ between the HindIII and SacI sites to create pIG121-Hm-HXS. A DNA fragment containing the 35S promoter of Cauliflower mosaic virus, truncated Sh1 intron, and TRAB1-dHA/His sequence was obtained by cutting p35S-ShΔ-TRAB1-dHA/His with SphI followed by partial digestion with BglII and blunt ended with T4 DNA polymerase. XhoI- and SacI-cut ends were added to this blunt-ended fragment at the 5′ end of the promoter and the 3′ end of dHA/His sequence, respectively, by subcloning into the EcoRV site of pBluescript II and ligated into XhoI- and SacI-digested pIG121-Hm-HXS. The resulting plasmid was transformed into Agrobacterium (EHA101).
pCaMV35S-sGFP (S65T)–NOS3′ (Chiu et al., 1996) was digested with NotI, blunt ended with the Klenow fragment, and then digested with SalI to obtain a green fluorescent protein (GFP) fragment. This GFP fragment was ligated to the SalI and BglII (blunt ended) sites of p35S-ShΔ-stop to create the straight GFP expression vector (p35S-ShΔ-GFP). The TRAB1 fragment was excised from p35S-ShΔ-TRAB1-dHA/His with EcoRI and SalI and inserted into the corresponding site of p35S-ShΔ-GFP to obtain the TRAB1-GFP fusion protein expression vector (p35S-ShΔ-TRAB1-GFP).
Transient Expression Assays
Transient expression assays by electroporation with the p35S-ShΔ-GBD–TRAB1 and UAS-TATA-GUS plasmids were performed as described previously (Hobo et al., 1999b). For expression of TRAB1-dHA/ His, the protoplasts were electroporated with 4 μg of the expression plasmid and cultured for 12 h. The culture was divided into two equal portions, each of which then was cultured further either in the absence or in the presence of abscisic acid (ABA; 50 μM) for 1 h.
Plant Transformation
Agrobacterium-mediated rice (Oryza sativa) transformation was performed as described by Hiei et al. (1994). Instead of regenerating plants, hygromycin-resistant transformed calli were maintained as suspension cultures in R2S liquid medium (Kyozuka and Shimamoto, 1991) containing 50 mg/L hygromycin and 250 mg/L carbenicillin.
Antibody Production
The entire TRAB1 cDNA fragment excised with EcoRI and SalI was cloned into pET32-a(+) (Novagen, Madison, WI), and the recombinant His-thioredoxin–tagged TRAB1 was produced in Escherichia coli BL21(DE3). Nickel–nitrilotriacetic acid agarose (Qiagen, Valencia, CA) was used to affinity purify the recombinant protein. Polyclonal anti-TRAB1 serum was obtained by immunizing rabbits with the recombinant protein.
Immunoblot Analysis
Proteins were separated on 10% SDS–polyacrylamide gels and transferred to a nitrocellulose membrane (Transblot Transfer Medium; Bio-Rad, Hercules, CA) by tank electroblotting using 10 mM cyclohexylaminopropane sulfonic acid buffer, pH 10.5, containing 10% methanol. The membrane was blocked at room temperature for 2 h in TBST buffer (10 mM Tris, pH 7.5, 100 mM NaCl, and 0.1% Tween 20) containing 1% BSA, 3% gelatin, and 5% skim milk and then incubated for 2 h in the same buffer with anti-TRAB1 or anti-HA antibodies (Covance, Richmond, CA). After incubation with horseradish peroxidase–conjugated secondary antibody, the antigen bands were detected using ECL (enhanced chemiluminescence) protein gel blot detection reagents (Amersham Pharmacia Biotech, Buckinghamshire, UK) and exposing the membrane to an x-ray film (Hyperfilm ECL; Amersham Pharmacia Biotech).
Labeling and Immunoprecipitation Experiments
Rice suspension-cultured cells (Oc cells) were precultured in R2S medium (Kyozuka and Shimamoto, 1991) depleted of Pi for 1.5 h. After the preculture, the cells were cultured in the same medium containing 0.2 mCi/mL 32P-orthophosphoric acid for 30 min and then treated with 50 μM ABA for various periods (0 to 120 min). After the ABA treatment, the cells were harvested and washed with the culture medium containing 10 mM potassium phosphate. Nuclear extracts were prepared from the recovered cells, diluted with ninth volume of TBST, and incubated with anti-TRAB1 antibody (1:200 dilution). The antigen-antibody complexes recovered on protein A–agarose (Pierce, Rockford, IL) were washed with TBST and separated on a 10% SDS–polyacrylamide gel and autoradiographed. Phosphoamino acid analysis was performed essentially as described by Kamps and Sefton (1989) using the radioactive TRAB1 polypeptide recovered from the gel.
Cell Fractionation and Preparation of Protein Extracts
Rice cells were disrupted using a Mini-BeadBeater-8 (BioSpec Products, Bartlesville, OK) in the presence of nuclei preparation buffer (0.5 M hexylene glycol, 20 mM KCl, 20 mM Pipes, pH 6.5, 0.5 mM EDTA, 0.4% Triton X-100, 0.05 mM spermine, 0.125 mM spermidine, 7 mM 2-mercaptoethanol, 5 mM NaF, 0.2 mM sodium vanadate, 2 mM p-nitrophenylphosphoric acid disodium salt, and 1 × COMPLETE [Roche Diagnostics, Mannheim, Germany]). Alternatively, the cells were ground into powder with a mortar and pestle in the presence of liquid N2 and extracted with nuclei preparation buffer. The postnuclear and crude nuclear fractions were separated by centrifugation at 5000g for 5 min. The crude nuclear pellets were washed once with nuclei wash buffer (40 mM Hepes-NaOH, pH 7.3, 10% [v/v] glycerol, and 0.1% Tween 20) and extracted with nuclei wash buffer containing 350 mM NaCl, 5 mM NaF, 0.2 mM sodium vanadate, 2 mM p-nitrophenylphosphoric acid disodium salt, 1 × COMPLETE, and 0.4% Triton X-100.
Phosphatase Treatments
TRAB1-dHA/His protein in the nuclear or protoplast extracts was recovered with nickel-nitrilotriacetic acid agarose resin (Qiagen). After changing the buffer to calf intestine alkaline phosphatase buffer (50 mM Tris-HCl, pH 7.6, and 0.1 mM EDTA) using a gel-filtration spin column, the protein solution was incubated with or without calf intestine alkaline phosphatase (Takara-Shuzo, Kyoto, Japan) at 37°C for 30 min. Protoplast extracts were treated similarly. After treatment, proteins were separated on an 8% SDS–polyacrylamide gel until a 25-kD prestained size maker was run off the gel and subjected to immunoblot analysis.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Tomiko Chikada for excellent technical assistance. This work was funded in part by Grants-in-Aid for Scientific Research on Priority Areas A (Grant 12037208) and B (Grant 12138204) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology and by a Research for the Future Grant (JSPS-ooL1603) of the Japan Society for the Promotion of Sciences.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.005272.
References
- Busk, K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Chiu, W.-I., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Ciceri, P., Gianazza, E., Lazzari, G., Lippoli, G., Genga, A., Hoscheck, G., Schmidt, R.J., and Viotti, A. (1997). Phosphorylation of Opaque2 changes diurnally and impacts its DNA binding activity. Plant Cell 9, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droge-Laser, W., Kaiser, A., Lindsay, W.P., Halkier, B.A., Loake, G.J., Doerner, P., Dixon, R.A., and Lamb, C. (1997). Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 16, 726–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA insensitive loci are similar to the abi3 mutations. Plant J. 5, 765–771. [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Parcy, F., Bertauche, N., Gosti, F., Leung, J., Morris, P.C., Bouvier-Durand, M., and Vartanian, N. (1994). Current advances in abscisic acid action and signaling. Plant Mol. Biol. 26, 1557–1577. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan, M.J., Marcotte, W.R., and Quatrano, R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250, 267–270. [DOI] [PubMed] [Google Scholar]
- Ishida, N., Kitagawa, M., Hatakeyama, S., and Nakayama, K. (2000). Phosphorylation at serine 10, a major phosphorylation site of p27(Kip1), increases its protein stability. J. Biol. Chem. 275, 25146–25154. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19, 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harter, K., Kircher, S., Frohnmeyer, H., Krenz, M., Nagy, F., and Schaefer, E. (1994). Light-regulated modification and nuclear translocation of cytosolic G-box binding factors in parsley. Plant Cell 6, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, T., Terada, T., and Hamasuna, S. (1994). Sequence and functional analyses of the rice gene homologous to the maize Vp1. Plant Mol. Biol. 24, 805–810. [DOI] [PubMed] [Google Scholar]
- Hattori, T., Terada, T., and Hamasuna, S. (1995). Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: Analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J. 7, 913–925. [DOI] [PubMed] [Google Scholar]
- Hattori, T., Vasil, V., Rosenkrans, L., Hannah, L.C., McCarty, D.R., and Vasil, I.K. (1992). The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev. 6, 609–618. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 16, 271–282. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999. a). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Kowyama, Y., and Hattori, T. (1999. b). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96, 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps, M.P., and Sefton, B.M. (1989). Acid and base hydrolysis of phosphoproteins bound to Immobilon facilitates the analysis of phosphoamino acids in gel-fractionated proteins. Anal. Biochem. 176, 22–27. [DOI] [PubMed] [Google Scholar]
- Kang, J.-y., Choi, H.-i., Im, M.-y., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol, B., Malec, P., and Chamovitz, D.A. (1999). Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell 11, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y., Chung, H.J., and Thomas, T.L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Wellmer, F., Nick, P., Rügner, A., Schäfer, E., and Harter, K. (1999). Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J. Cell Biol. 144, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak, L.J., Schindler, U., and Cashmore, A.R. (1992). DNA binding activity of the Arabidopsis G-box binding factor GBF1 is stimulated by phosphorylation by casein kinase II from broccoli. Plant Cell 4, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka, J., and Shimamoto, K. (1991). Transformation and regeneration of rice protoplasts. In Plant Tissue Culture Manual, B1, K. Lindsey, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–16.
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 246, 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Wang, X.-Q., Watson, M.B., and Assmann, S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287, 300–303. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., and Chua, N.H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte, W.R., Jr., Russel, S.H., and Quatrano, R.S. (1989). Abscisic acid-responsive sequences from the Em gene of wheat. Plant Cell 1, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, D.R. (1995). Genetic control and integration of maturation and germination pathways in seed development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 71–93. [Google Scholar]
- McCarty, D.R., Hattori, T., Carson, C.B., Vasil, V., and Vasil, I.K. (1991). The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66, 895–905. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J. 25, 295–303. [DOI] [PubMed] [Google Scholar]
- Meyer, K., Leube, M.P., and Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 246, 1452–1455. [DOI] [PubMed] [Google Scholar]
- Mundy, J., and Chua, N.H. (1988). Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 7, 2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy, J., Yamaguchi-Shinozaki, K., and Chua, N.-H. (1990). Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. USA 87, 1406–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, H., Ohmiya, K., and Hattori, T. (1996). A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J. 9, 217–227. [DOI] [PubMed] [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Nantel, A., and Quatrano, R.S. (1996). Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J. Biol. Chem. 271, 31296–31305. [DOI] [PubMed] [Google Scholar]
- Oeda, K., Salinas, J., and Chua, N.H. (1991). A tobacco bZip transcription activator (TAF-1) binds to G-box-like motif conserved in plant genes. EMBO J. 10, 1793–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, S., Hattori, T., Morikami, A., and Nakamura, K. (1991). High-level expression of a sweet potato sporamin gene promoter: β-Glucuronidase (GUS) fusion gene in the stems of transgenic tobacco plants is conferred by multiple cell type-specific regulatory elements. Mol. Gen. Genet. 225, 369–378. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 in light development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Ritchie, S., and Gilroy, S. (1998). Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 95, 2697–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, J.P., and Chua, N.H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13, 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1998). Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA 95, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., and Ho, T.-H.D. (1995). Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and novel cis-acting element. Plant Cell 7, 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., Zhang, P., and Ho, T.-H.D. (1996). Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver, K., Olsen, F.L., Roger, J., and Mundy, J. (1991). Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88, 7266–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., Cocciolone, S., and McCarty, D.R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28, 409–418. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., and McCarty, D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil, V., Marcotte, W.R., Jr., Rosenkrans, L., Cocciolone, S.M., Vasil, I.K., Quatrano, R.S., and McCarty, D.R. (1995). Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7, 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Chua, K., Seghezzi, W., Lees, E., Gozani, O., and Reed, R. (1998). Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 12, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer, F., Kircher, S., Rugner, A., Frohnmeyer, H., Schafer, E., and Harter, K. (1999). Phosphorylation of the parsley bZIP transcription factor CPRF2 is regulated by light. J. Biol. Chem. 274, 29476–29482. [DOI] [PubMed] [Google Scholar]
- Wellmer, F., Schafer, E., and Harter, K. (2001). The DNA binding properties of the parsley bZIP transcription factor CPRF4a are regulated by light. J. Biol. Chem. 276, 6274–6279. [DOI] [PubMed] [Google Scholar]
- Wu, Y., Kuzma, J., Marechal, E., Graeff, R., Lee, H.C., Foster, R., and Chua, N.H. (1997). Abscisic acid signaling through cyclic ADP-ribose in plants. Science 278, 2126–2130. [DOI] [PubMed] [Google Scholar]