Abstract
DELLA proteins are nuclear repressors of plant gibberellin (GA) responses. Here, we investigate the properties of SLN1, a DELLA protein from barley that is destabilized by GA treatment. Using specific inhibitors of proteasome function, we show that proteasome-mediated protein degradation is necessary for GA-mediated destabilization of SLN1. We also show that GA responses, such as the aleurone α-amylase response and seedling leaf extension growth, require proteasome-dependent GA-mediated SLN1 destabilization. In further experiments with protein kinase and protein phosphatase inhibitors, we identify two additional signaling steps that are necessary for GA response and for GA-mediated destabilization of SLN1. Thus, GA signaling involves protein phosphorylation and dephosphorylation steps and promotes the derepression of GA responses via proteasome-dependent destabilization of DELLA repressors.
INTRODUCTION
Bioactive gibberellins (GAs) are essential regulators of plant growth and development (Hooley, 1994). For example, during the germination of cereal grains, GA is synthesized by the embryo and secreted into the aleurone. In this situation, GA regulates the synthesis and secretion of hydrolyzing enzymes (such as α-amylase) into the endosperm. The hydrolyzing enzymes then catalyze the breakdown of endosperm storage macromolecules, releasing nutrients that are used by the establishing seedling (Bethke et al., 1997; Ritchie and Gilroy, 1998; Lovegrove and Hooley, 2000).
GA is thought to elicit GA responses in the following manner. First, GA appears to be perceived on the surface of plant cells by an unidentified outward-facing plasma membrane–associated GA receptor (Hooley et al., 1991; Gilroy and Jones, 1994). The perception of GA results in rapid increases in the levels of cytosolic calcium and calmodulin (Gilroy, 1996; Schuurink et al., 1996). G-proteins, protein phosphatases, and cGMP also may play important roles during the cytoplasmic steps of the GA signal transduction chain (Kuo et al., 1996; Penson et al., 1996; Jones et al., 1998). Inside the nucleus, the DELLA proteins, a family of putative transcriptional regulators, mediate the GA signal (Dill et al., 2001; Richards et al., 2001; Itoh et al., 2002; Wen and Chang, 2002). Downstream of the DELLA proteins, GA regulates α-amylase synthesis in aleurone via a myb-like transcription factor (GAmyb) that binds to a specific region of the promoters of genes that encode α-amylase (Gubler et al., 1995). Recent work has shown that, in addition to genes that encode α-amylase, GAmyb can transactivate other GA-regulated genes (Gubler et al., 1995, 1999; Cercós et al., 1999).
Mutants of wheat, barley, and rice that are affected in GA signaling display an altered aleurone α-amylase response. For example, dominant mutations at the homoeoallelic wheat Rht-B1a and Rht-D1a loci confer dwarfism and a reduced growth response to GA (Börner et al., 1996; Peng et al., 1999). Severely dwarfing alleles, such as Rht-B1c, abolish the GA response of mutant aleurone cells (Gale and Marshall, 1975; Ho et al., 1981; Börner et al., 1996). By contrast, recessive mutations at the barley SLENDER (SLN1) and rice SLENDER RICE1 (SLR1) loci (e.g., sln1-1 and slr1-1) confer a taller-than-wild-type, slender phenotype as a result of exaggerated elongation growth (Foster, 1977; Ikeda et al., 2001; Chandler et al., 2002). The aleurone cells of these slender mutants constitutively express α-amylase in the absence of GA induction (Chandler, 1988; Lanahan and Ho, 1988; Ikeda et al., 2001; Chandler et al., 2002; Gubler et al., 2002). The accelerated growth and constitutive α-amylase expression of the slender mutants is unaffected by GA biosynthesis inhibitors (Chandler, 1988; Lanahan and Ho, 1988; Ikeda et al., 2001). Although the phenotype of barley and rice slender mutants resembles that of wild-type plants treated with GA, the endogenous bioactive GA levels in these mutants are lower than those of the wild type (Croker et al., 1990; Ikeda et al., 2001).
Wheat Rht-B1a and Rht-D1a, rice SLR1, and barley SLN1 encode proteins orthologous with Arabidopsis GAI, a member of the GRAS family of putative transcriptional regulators (Peng et al., 1997, 1999; Harberd et al., 1998; Pysh et al., 1999; Richards et al., 2000, 2001; Ikeda et al., 2001; Chandler et al., 2002; Gubler et al., 2002). The Arabidopsis genome contains four other genes that encode proteins that are closely related to GAI: RGA, RGL1, RGL2, and RGL3 (Silverstone et al., 1998; Dill and Sun, 2001; Lee et al., 2002). GAI and RGA encode proteins that act together as negative regulators of GA responses (Peng et al., 1997; Silverstone et al., 1997, 1998; Dill and Sun, 2001; King et al., 2001), and RGL1 and RGL2 also encode proteins that function in GA signaling (Lee et al., 2002; Wen and Chang, 2002). The proteins encoded by GAI, RGA, RGL1, RGL2, and RGL3, and by orthologous genes in other species, are the above-mentioned DELLA proteins, named after a motif that is highly conserved among them and that is important to their function in GA signaling (Peng et al., 1997; Wen and Chang, 2002). The DELLA proteins generally are thought to operate as repressors of GA responses, and GA is thought to induce GA responses by opposing DELLA protein action (Peng et al., 1997; Harberd et al., 1998; King et al., 2001; Richards et al., 2001).
Recent studies using DELLA proteins fused to the green fluorescent protein have shown that RGA, SLR1, and SLN1 accumulate in the nucleus of plant cells and that treatment with exogenous GA causes the disappearance of these proteins from the nucleus (Silverstone et al., 2001; Dill et al., 2001; Itoh et al., 2002; Gubler et al., 2002). These observations are compatible with previous proposals that DELLA proteins work as repressors of growth, whereas GA opposes their growth-repressing function (Peng et al., 1997; Harberd et al., 1998; Richards et al., 2001).
Here, we describe the molecular analysis of the barley SLN1 gene and the mechanism by which its product (SLN1) mediates barley GA responses. We investigated the mechanism of GA-induced SLN1 destabilization by studying the effects of a number of different inhibitory compounds on this process. In particular, we show that specific inhibitors of 26S proteasome function block both the GA-mediated destabilization of SLN1 and GA responses (the aleurone α-amylase response and seedling leaf elongation). We also demonstrate that selected protein kinase and protein phosphatase inhibitors can block the GA induction of both SLN1 destabilization and GA responses, thus implicating protein phosphorylation and dephosphorylation steps in GA signaling. In summary, our results indicate that GA stimulates GA responses by eliciting proteasome-dependent degradation of the nuclear SLN1 GA response repressor.
RESULTS
Molecular Characterization of the Barley sln1-1 Mutant Allele
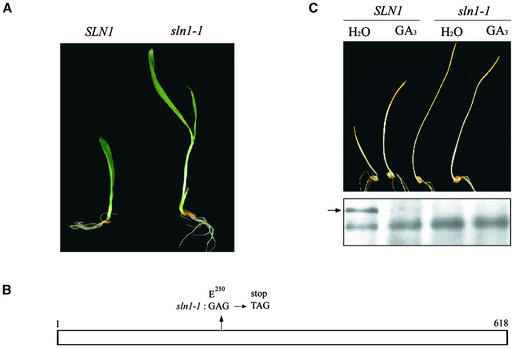
As shown in Figure 1A, recessive mutations at SLN1 (e.g., sln1-1) confer exaggerated elongation growth of barley seedlings. This phenotype persists throughout the development of the plant, resulting in adult plants that are taller than wild-type plants, with thin, pale green leaves and sterile flowers (the “slender” phenotype) (Foster, 1977; Chandler, 1988; Lanahan and Ho, 1988). In addition, the growth of sln1-1 mutants is resistant to the growth-inhibitory effects of the GA biosynthesis inhibitor paclobutrazol, suggesting that SLN1 encodes a repressor of GA responses and that loss-of-function mutations at SLN1 confer a constitutive GA response (Chandler, 1988; Lanahan and Ho, 1988).
Figure 1.
The sln1-1 Mutant Allele.
(A) Five-day-old seedlings homozygous for SLN1 or sln1-1.
(B) Scheme of SLN1/SLN1 showing the site of the mutation in sln1-1. Numbers represent amino acid positions in SLN1 (1 indicates the start Met, and 618 indicates the final Pro).
(C) Seedling phenotypes and protein gel blot analysis of SLN1 and sln1-1 plants. The seedlings were grown at 20°C for 5 days with or without 100 μM GA3. Proteins then were extracted from seedlings, and 15 μg of total protein per lane was loaded and probed with anti-GAI antibodies (see Methods). The arrow indicates SLN1; an additional, nonspecific band served as a loading control.
Because mutations at SLN1 confer altered GA responses, we reasoned that SLN1 might be a barley ortholog of the GAI/RGA/d8/Rht-B1a/Rht-D1a/SLR1 genes (genes that encode DELLA proteins from a variety of species) (Chandler et al., 2002; Gubler et al., 2002). Therefore, we amplified SLN1 using PCR primers derived from the wheat Rht-D1a sequence (see Methods). The predicted amino acid sequence of the protein encoded by the amplified SLN1 was identical to that described previously (Chandler et al., 2002; Gubler et al., 2002; X. Fu and D.E. Richards, unpublished data). We also amplified SLN1 from the sln1-1 mutant (see Methods) (Figure 1A). DNA sequencing showed that the sln1-1 mutation is a single nucleotide substitution (GAG to TAG) that converts the codon encoding Glu-250 to a stop codon (Figure 1B).
Immunoblot analysis showed that sln1-1 plants lack detectable SLN1 protein. As shown in Figure 1C, the shoots of SLN1 seedlings germinated and grown in the presence of exogenous GA3 were longer than those germinated in water, whereas there was no effect of GA on sln1-1 seedlings. Total proteins extracted from these seedlings were electrophoretically fractionated and analyzed using anti-GAI antibodies. These experiments identified an ∼65-kD immunoreactive protein that was detectable in water-treated SLN1 seedlings but not in water-treated sln1-1 seedlings, indicating that the protein identified is SLN1 (the predicted molecular mass of SLN1 is 65.2 kD) (Figure 1C). As shown previously, SLN1 was not detectable in GA-treated SLN1 seedlings, showing that SLN1 disappears in response to exogenous GA (Chandler et al., 2002; Gubler et al., 2002) (Figure 1C).
GA-Induced Disappearance of SLN1 Is Unaffected by the Protease Inhibitors Pefabloc SC, Aprotinin, and Phenylmethylsulfonyl Fluoride
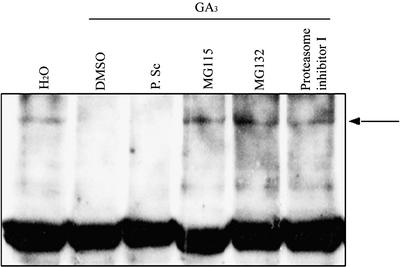
We sought to determine a mechanism for the GA-induced disappearance of SLN1, testing initially the effects of the cell-permeable protease inhibitors Pefabloc SC, aprotinin, and phenylmethylsulfonyl fluoride (for details of inhibitors, see Methods). As described previously, SLN1 was detected in extracts from SLN1 seedlings treated with water but not in extracts treated with GA (Figure 2). SLN1 also was not detected in SLN1 seedlings treated for 2 h with GA and Pefabloc SC, a general inhibitor of Ser proteases (Figure 2). Similar results were obtained with GA and Pefabloc SC treatments of 30 min and 24 h (data not shown). Thus, Pefabloc SC had no detectable effect on GA-induced SLN1 disappearance (Figure 2). Similarly, treatments with other protease inhibitors, such as aprotinin and phenylmethylsulfonyl fluoride, failed to block the GA-induced disappearance of SLN1 (data not shown).
Figure 2.
The Proteasome Pathway Mediates GA-Induced SLN1 Protein Degradation.
Extracts were prepared from 5-day-old SLN1 seedlings, which had been treated for 2 h with GA3 and 1% DMSO with or without protease or proteasome inhibitors (Pefabloc SC [P. Sc], MG115, MG132, or proteasome inhibitor I). Extracts from water-treated SLN1 seedlings were used as a positive control. Total protein (15 μg/lane) was loaded and probed with anti-GAI antibodies. The arrow indicates SLN1. The strong bottom band in all lanes represents the nonspecific background protein described for Figure 1, and the weak lower bands may indicate SLN1 degradation products.
GA-Induced Disappearance of SLN1 Is Affected by Proteasome Inhibitors
In contrast with the results reported above, the GA-induced disappearance of SLN1 was affected by the addition of five different cell-permeable proteasome-specific inhibitors: MG115, MG132, proteasome inhibitor I, proteasome inhibitor II, and lactacystin (for details of inhibitors, see Methods). As shown in Figure 2, extracts from SLN1 seedlings treated with both GA and MG115, MG132, or proteasome inhibitor I contained detectable levels of SLN1. Similar results were obtained with proteasome inhibitor II and lactacystin and from samples treated with GA and proteasome inhibitors for 30 min or 24 h (data not shown). These results show that proteasome inhibitors can prevent the GA-induced disappearance of SLN1, suggesting that this disappearance might be attributable to proteasome-mediated degradation.
GA-Regulated Seedling Leaf Extension Growth Is Dependent on Proteasome-Mediated SLN1 Degradation
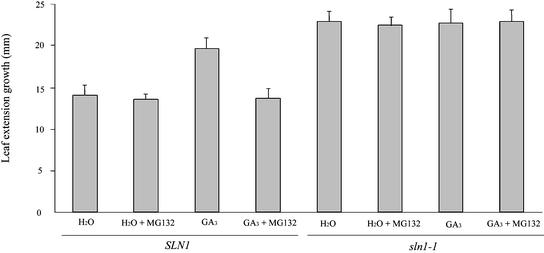
Because proteasome inhibitors block the GA-dependent destabilization of SLN1 in barley seedlings, we tried to determine whether GA-promoted seedling leaf extension growth was affected by MG132. Three-day-old SLN1 and sln1-1 seedlings were grown in the presence of GA and/or MG132, and first leaf lengths were measured before and after treatment (Figure 3). The leaves of SLN1 seedlings grown in the presence of GA grew longer than those of water-treated controls, whereas MG132 blocked the response of SLN1 seedlings to GA. The growth of sln1-1 seedlings was little affected by either GA or MG132, indicating that the inhibitory effect of MG132 on the growth of GA-treated SLN1 seedlings was not caused by general metabolic poisoning (Figure 3). Together, these results indicate that GA promotes the extension growth of barley seedling leaves via the proteasome-dependent destabilization of SLN1.
Figure 3.
GA-Promoted Leaf Extension Growth Requires Proteasome-Dependent GA-Mediated SLN1 Destabilization.
Seedlings were germinated and grown for 3 days on water. The length of the first leaf (from leaf tip to seed) of each seedling was measured, and treatment (combinations of water, GA, and MG132 as shown) was begun. Twelve hours after the initiation of treatment, the length of the first leaf was measured again. The results shown are mean differences (n = 30; error bars represent standard errors) between the first and second measurements.
α-Amylase Induction in Aleurone Cells Is Dependent on Proteasome-Mediated SLN1 Degradation
GA-mediated destruction of SLN1 in aleurone cells is associated with the GA induction of α-amylase (Gubler et al., 2002). Therefore, we examined the effects of a range of inhibitors on the α-amylase responses of de-embryonated SLN1 and sln1-1 half-grains (see Methods). As described previously, sln1-1 half-grains produce comparable amounts of α-amylase activity in the presence or absence of GA (Chandler, 1988; Lanahan and Ho, 1988) (Table 1). None of the inhibitors tested (Pefabloc SC, MG115, MG132, and proteasome inhibitor I) affected the production of α-amylase by sln1-1 half-grains in the presence or absence of GA (Table 1). By contrast, the proteasome inhibitors MG115 and MG132 largely blocked the GA induction of α-amylase activity from SLN1 half-grains (Table 1). The fact that MG115 and MG132 blocked the α-amylase response in SLN1 half-grains but did not inhibit the α-amylase production of sln1-1 half-grains shows that the observed effects of these inhibitors on GA responses is not attributable to nonspecific effects or the poisoning of cellular metabolism. Rather, MG115 and MG132 block the α-amylase response of SLN1 half-grains by inhibiting proteasome activity, and proteasome-dependent degradation of SLN1 is necessary for the induction of α-amylase activity.
Table 1.
Effects of the Different Inhibitors on α-Amylase Production in Barley Aleurone Layers
| α-Amylase Produced (milliunits/g)
|
||||
|---|---|---|---|---|
|
SLN1
|
sln1-1
|
|||
| Inhibitors | Water | GA3 | Water | GA3 |
| Control | 0.08 ± 0.01 | 9.60 ± 0.03 | 6.24 ± 0.07 | 7.21 ± 0.05 |
| MG115 | 0.07 ± 0.02 | 0.29 ± 0.04 | 5.87 ± 0.06 | 6.37 ± 0.07 |
| MG132 | 0.07 ± 0.03 | 0.34 ± 0.09 | 5.99 ± 0.08 | 6.29 ± 0.10 |
| Pefabloc SC | 0.09 ± 0.02 | 6.48 ± 0.11 | 5.47 ± 0.12 | 6.91 ± 0.08 |
| SV | 0.08 ± 0.03 | 0.45 ± 0.03 | 5.93 ± 0.11 | 6.72 ± 0.09 |
| AG555 | 0.54 ± 0.09 | 0.81 ± 0.03 | 7.18 ± 0.05 | 7.89 ± 0.09 |
| PP2 | 0.09 ± 0.02 | 8.36 ± 0.02 | 6.69 ± 0.10 | 7.03 ± 0.08 |
| Staurosporine | 0.10 ± 0.03 | 9.24 ± 0.09 | 6.03 ± 0.09 | 7.11 ± 0.07 |
Each value shown is the mean ± se from 12 half-grains.
Protein Kinase and Phosphatase Inhibitors Block GA-Induced SLN1 Protein Degradation and α-Amylase Production
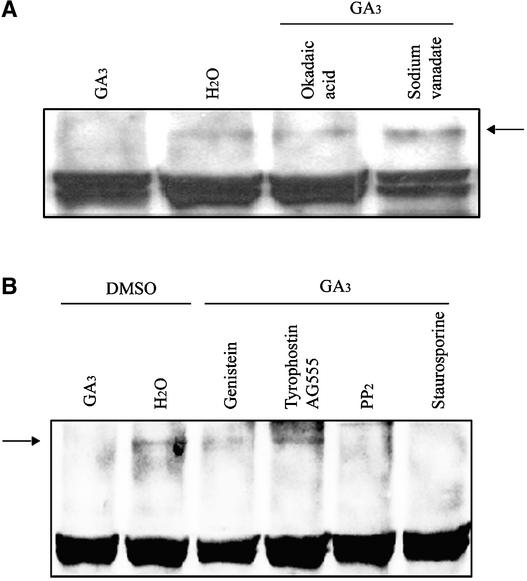
To identify additional steps in the GA signal transduction pathway, we tested the effects of various protein phosphorylation and dephosphorylation inhibitors on GA-induced SLN1 degradation. Previous experiments have shown that the Ser/Thr protein phosphatase inhibitor okadaic acid (OA) is effective at blocking the GA-induced production of α-amy-lase by wheat aleurone cells (Kuo et al., 1996). We examined the effects of OA and sodium vanadate (SV), a widely used general inhibitor of protein phosphatases (for details of these inhibitors, see Methods), on the GA-induced degradation of SLN1. SLN1 was detected in extracts from SLN1 seedlings treated with water or with GA and either OA or SV for 2 h (Figure 4A) or 24 h (data not shown) and was not detected in GA-only controls. Thus, treatment with OA or SV blocked the GA-induced degradation of SLN1. We also showed that both OA and SV blocked the GA-mediated induction of α-amylase activity in SLN1 half-grains but did not block the constitutive production of α-amylase by sln1-1 half-grains (Table 1). These results show that OA and SV affect GA responses by perturbing the signaling chain associated with SLN1 (or they affect SLN1 itself), making SLN1 resistant to GA-mediated destabilization.
Figure 4.
Effect of Protein Phosphatase and Protein Kinase Inhibitors on the GA-Induced Degradation of SLN1.
(A) Effect of protein phosphatase inhibitors on GA-induced SLN1 degradation. Extracts were prepared from 5-day-old SLN1 seedlings treated for 2 h with GA3 with or without OA or SV. Water-treated SLN1 seedlings were used as a positive control. The arrow indicates the SLN1 protein. Additional nonspecific bands served as loading controls.
(B) Effect of protein kinase inhibitors on GA-induced SLN1 degradation. Extracts were prepared from 5-day-old SLN1 seedlings treated for 2 h with GA3 and 1% DMSO with or without Tyrophostin B46 (AG555), PP2, genistein, or staurosporine. Extracts also were prepared from control SLN1 seedlings treated with water and 1% DMSO. Total protein (15 μg/lane) was loaded and probed with anti-GAI antibodies. The arrow indicates the SLN1 protein. The bottom band in all lanes represents a nonspecific background protein as described for Figure 1.
Staurosporine is a broad-range inhibitor of Ser/Thr protein kinases, whereas protein phosphatase 2 (PP2) is a selective protein kinase inhibitor (see Methods). Treatment with either staurosporine or PP2 failed to block the GA-induced degradation of SLN1 or the production of α-amylase activity (Figure 4B, Table1). By contrast, treatment with two protein Tyr kinase inhibitors, genistein (a broad-range protein kinase inhibitor) and Tyrophostin B46 (AG555; for details, see Methods), blocked the GA-induced degradation of SLN1 (Figure 4B). Figure 4 shows extracts from seedlings treated for 2 h; similar results were obtained for each inhibitor after treatments for 30 min and 24 h (data not shown). Genistein and Tyrophostin B46 also blocked GA-induced α-amylase production in SLN1 half-grains but did not block constitutive α-amylase production in sln1-1 half-grains (Table 1). Together, the results described here suggest that protein kinases and protein phosphatases mediate GA-induced degradation of SLN1, thus eliciting GA responses.
DISCUSSION
Here, we show that several well-defined, cell-permeable inhibitors of proteasome function can block the GA-induced disappearance of SLN1. Thus, proteasome function is necessary for the destabilization of SLN1 in response to the GA signal. Furthermore, we show that two GA responses, leaf extension growth and the aleurone α-amylase response, are blocked by proteasome inhibitors. Our results suggest that GA induces GA responses in barley via proteasome-dependent degradation of SLN1.
To identify additional steps in the GA signaling pathway, we tested the effects of a range of protease, kinase, and phosphatase inhibitors on GA responses and on the GA-mediated destabilization of SLN1. None of these inhibitors (and none of the proteasome inhibitors described above) inhibited the production of α-amylase activity by sln1-1 aleurones. In addition, the growth of sln1-1 seedling leaves was unaffected by the proteasome inhibitor MG132. These are important observations because they enable us to discount a criticism that often is leveled at studies using inhibitors—that is, that the effects of inhibitors are nonspecific and may be the result of general poisoning of cellular metabolism. This cannot be the case in our experiments, because if it had been, the inhibitors would have blocked the constitutive GA responses exhibited by the sln1-1 mutant. Any affect of the inhibitors used in our experiments on the GA responses of SLN1 plants must be attributable to a specific effect of that inhibitor on the activity of SLN1, either via a direct effect on SLN1 itself or via an effect on steps in the GA signaling pathway leading to the destabilization of SLN1.
We identified two additional steps in GA signaling. First, we showed that protein phosphorylation inhibitors can block GA responses and the GA-mediated destabilization of SLN1, implying a protein phosphorylation step in GA signaling. The inhibitors that were effective at blocking GA responses and SLN1 destabilization are Tyr kinase inhibitors, making it possible that phosphotyrosine is involved in GA signaling. It has been suggested previously that the DELLA proteins are structurally, and perhaps functionally, related to the STAT proteins (which signal via Tyr phosphorylation) (Peng et al., 1999; Richards et al., 2000). In addition, GRAS proteins contain a C-terminal sequence that is related to a consensus Tyr phosphorylation site (Bolle et al., 2000). Second, we showed that protein phosphatase inhibitors can block GA responses and the GA-mediated destabilization of SLN1, implying a protein dephosphorylation step in GA signaling. This step was identified previously because of the effect of OA on GA-induced α-amylase production by wheat aleurone cells (Kuo et al., 1996). Here, we have extended this finding by showing that the protein dephosphorylation step is necessary for the GA-mediated destabilization of SLN1.
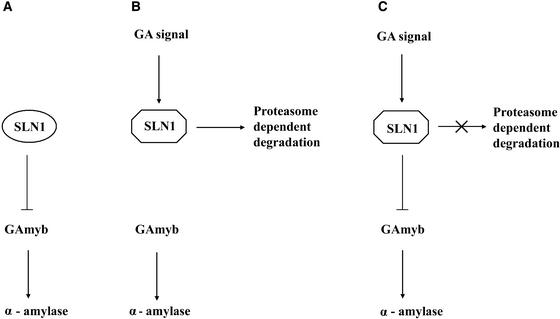
We propose the following model to explain the phenomena reported in this article (Figure 5). First, GA interacts with an unknown plasma membrane–associated specific receptor (Hooley et al., 1991; Gilroy and Jones, 1994). This interaction stimulates a signal transduction cascade that may involve the phosphorylation or dephosphorylation of proteins on Ser, Thr, or Tyr (Kuo et al., 1996). Eventually, the signal reaches the nuclear SLN1 protein. SLN1 acts as a repressor of GA responses, inhibiting for example the transcription of the gene encoding the GAmyb activator of the α-amylase response (Gómez-Cadenas et al., 2001). The GA signal alters SLN1, resulting in proteasome-dependent SLN1 destabilization and the release of GA responses from SLN1-mediated restraint.
Figure 5.
Scheme of SLN1 Function in GA Signaling.
Bioactive GA interacts with the membrane-associated GA receptor (not shown), thus activating signal transduction via second messengers, perhaps mediated by protein phosphorylation or dephosphorylation (GA signal). SLN1 is in the nucleus, in an active form that represses GA responses (e.g., SLN1 represses the accumulation of transcripts that encode GAmyb, as shown in [A]). The arrival of the GA signal causes the modification of active SLN1 into a form that is destroyed via a proteasome-dependent mechanism (B), resulting in the activation of GAmyb transcription and α-amylase production. Inhibition of the proteasome pathway (C) prevents the destruction of SLN1, which therefore persists and continues to repress GAmyb transcription and α-amylase production. Because protein kinase and protein phosphatase inhibitors also block the destruction of SLN1, it is possible that protein phosphorylation or dephosphorylation is required for the modification of SLN1 or before the destruction of the modified form can occur.
It has become apparent that targeted proteasome-mediated protein degradation is crucial to many signaling pathways in plants. For example, the accumulation of AUX/IAA proteins is key to auxin signaling (Rouse et al., 1998). AUX/IAA accumulation is regulated via auxin-mediated targeting of AUX/IAAs for destruction via the proteasome (Gray et al., 2001). Other signaling pathways are regulated at the level of targeted protein destruction, either via the proteasome or the related COP9 signalosome complex, including jasmonic acid signaling (Xie et al., 1998), photomorphogenesis (Osterlund et al., 2000), floral development pathways (Samach et al., 1999), and disease resistance pathways (Austin et al., 2002; Azevedo et al., 2002). Here, we have shown that GA signaling in barley operates via proteasome-dependent, GA-elicited destabilization of SLN1. The fact that targeted protein destruction is key to so many different plant signaling pathways may explain why they often operate negatively, via derepression.
METHODS
Plant Material and Growth Conditions
The experiments described here used the barley (Hordeum vulgare) cv Herta as the source of the wild-type SLN1 allele. The sln1-1 mutant allele was induced by diethyl sulfate treatment of Herta (Foster, 1977), and the segregating material used was the selfed progeny of a line backcrossed multiple times onto the Herta genetic background.
Seeds were surface-sterilized by washing first with 70% ethanol for 2 min, then with sodium hypochlorite for 30 min, and finally with sterile distilled water. Sterilized seeds then were grown at 20°C (16-h photoperiod) on moistened filter paper. In tests of the seedling growth response to gibberellin (GA), seeds were germinated in 100 μM GA3 (Sigma). In further tests of GA-promoted leaf extension growth, 3-day-old seedlings were incubated with water or 100 μM GA3 in the presence or absence of 100 μM MG132. In tests of GA-induced SLN1 protein degradation, 5-day-old seedlings were treated with 100 μM GA3 (Sigma) or with water in the presence or absence of different pharmacological agents in the presence of 1% DMSO.
Cloning and Sequence Analysis of SLN1 Alleles
Total RNA was isolated from SLN1 seedlings using Trizol reagent (Gibco BRL). Genomic DNA was isolated from single SLN1 or sln1-1 seedlings according to the method of Edwards et al. (1991). Based on sequence conservation of the DELLA subfamily of GRAS regulator genes, sequence encoding the N-terminal end of SLN1 was obtained by 5′ rapid amplification of cDNA ends (Gibco BRL) using primers A1 (5′-TCGAGCTGCTCCAGCTTCCTG-3′), A2 (5′-ACGGTG-TCCGTGGCGAGGTG-3′), and A3 (5′-CGTTGAGCTCGGACAGCA-TG-3′). Sequence encoding the C-terminal end of SLN1 was obtained by 3′ rapid amplification of cDNA ends (Gibco BRL) using primers B1 (5′-TCGAGAAGGTCCTGGGCACG-3′), B2 (5′-TGA-CCGTGGTCGAGCAGGAG-3′), and B3 (5′-CTGCACTACTACTCC-ACCATG-3′). Primers A1, A2, A3, B1, B2, and B3 were derived from conserved sequences in Rht-B1a and Rht-D1a (Peng et al., 1999). PCR products from SLN1 and sln1-1 were amplified using the Expand Long Template PCR System (Roche Molecular Biochemicals, East Sussex, UK) using primers BA1 (5′-GATGGGGATCCGAGATGA-AGCGCGAGTACCAGGACGGC-3′) and BA2 (5′-CTTCGAATTCCC-TATCACGGCGCGGCGAGGCGCCATGC-3′). The PCR products were digested with BamHI and EcoRI and then cloned into the pGEM-T vector (Promega). Sequencing was performed using the ABI Prism Big-Dye Terminator Cycle Sequencing Ready Reaction Kit (PE-Applied Biosystems, Foster City, CA).
Production of Anti-GAI Polyclonal Antibodies
A full-length GAI cDNA was cloned into the BamHI and PstEI sites of pQE-30 vector (Qiagen, Crawley, UK) to fuse a His tag at the N terminus of the cloned sequence. The His-tagged GAI recombinant protein was overexpressed in the Escherichia coli host strain BL21(DE3) (Novagen, Madison, WI). Cells were grown at 30°C to an OD600 of 1.0, induced by 0.8 mM isopropylthio-β-galactoside for 90 min, and then harvested. Cells were resuspended in a buffer containing 50 mM Tris-HCl, pH 8.0, 0.2 mM EDTA, 100 mg/mL lysozyme, and 0.1% Triton X-100 and sonicated on ice. After centrifugation at 10,000 rpm for 20 min at 4°C, inclusion bodies were resuspended in the binding buffer solution containing 6 M urea, 5 mM imidazole, 50 mM NaCl, and 20 mM Tris-HCl, pH 8.0. The GAI recombinant protein was affinity-purified with nickel–nitrilotriacetic acid agarose beads (Qiagen). The purified GAI recombinant protein solution then was dialyzed against PBS solution overnight at 4°C before being used to raise antibodies in a rabbit.
The antisera obtained, although prepared against E. coli–expressed Arabidopsis GAI, were capable of detecting the SLN1 protein. A band that approximated the size expected for SLN1 was detected in extracts from SLN1 plants but not in extracts from sln1-1 plants.
Protein Gel Blot Analysis
Barley seedlings were treated with or without 100 μM GA3 in the presence or absence of inhibitors. Whole seedlings were harvested and frozen in liquid nitrogen, after which total plant proteins were extracted using Trizol reagent (Gibco BRL) and dissolved into buffer E (125 mM Tris-HCl, pH 8.8, 1% SDS, 10% glycerol, and 50 mM Na2S2O5). The resulting mixture was centrifuged (in a microcentrifuge) for 10 min at 4°C. The supernatant was transferred to new tubes, and protein concentrations were determined by the Bradford assay (Sigma). Total proteins (15 μg) from each extract were fractionated by 10% SDS-PAGE and analyzed on immunoblots using a 500-fold dilution of the anti-GAI polyclonal antiserum and a 10,000-fold dilution of peroxidase-conjugated goat anti-rabbit IgG (Amersham Pharmacia Biotech). The blots then were incubated with ECL protein gel blotting detection reagents (Amersham Pharmacia Biotech), and the signals were detected by chemiluminescence. For each blotted gel, a duplicate gel was run and then stained with Coomassie Brilliant Blue R250 to act as a loading control.
Inhibitor Studies
Three broad-range Ser protease inhibitors were used: Pefabloc SC (4-[2-aminoethyl]-benzensulfonyl-flouride hydrochloride), phenylmethylsulfonyl fluoride, and aprotinin (all three from Roche Molecular Biochemicals).
MG115 is a potent, reversible proteasome inhibitor that specifically inhibits the chymotrypsin-like activity of the proteasome (Peptides International, Louisville, KY). MG132 (also from Peptides International) is a tripeptide aldehyde, a potent, reversible proteasome inhibitor (Callis and Vierstra, 2000). Proteasome inhibitor I is an inhibitor of the multicatalytic proteinase complex (20S proteasome) (A.G. Scientific, San Diego, CA). Proteasome inhibitor II is a potent proteasome inhibitor that inhibits the chymotrypsin-like activity of the multicatalytic proteinase complex (20S proteasome) (A.G. Scientific). Lactacystin is an irreversible proteasome inhibitor that specifically inhibits the 26S proteasome and blocks proteasome activity by targeting the catalytic β-subunit (A.G. Scientific).
Okadaic acid (Calbiochem) is an inhibitor of the Ser/Thr protein phosphatases PP1 and PP2B (Bialojan and Takai, 1988). Vanadium ions (purchased as sodium orthovanadate; Sigma) are widely used as general inhibitors of protein phosphatases (Lau et al., 1989). Staurosporine (Calbiochem) is a broad-range inhibitor of Ser/Thr protein kinases and a potent inhibitor of Tyr kinases (Tamaoki, 1991). Genistein (Calbiochem) is a specific inhibitor of Tyr-specific protein kinases (Akiyama et al., 1987). Tyrophostin B46 (AG555; Calbiochem) and PP2 (Calbiochem) also are Tyr kinase inhibitors (Gazit et al., 1991; Hanke et al., 1996).
For the inhibitor analyses, 5-day-old seedlings were transferred to 100 μM GA3 (Sigma) for 30 min, 2 h, or 24 h in the presence of 1% DMSO (control) or 1% DMSO with inhibitor at the following concentrations: MG115 (100 μM), MG132 (100 μM), Pefabloc SC (0.5 mg/mL), okadaic acid (1 μM), sodium vanadate (3 mM), AG555 (10 μg/mL), staurosporine (50 μM), genistein (50 μg/mL), PP2 (10 μg/mL), and proteasome inhibitor I (100 μM). After treatment, the seedlings were harvested and extracted for immunoblot analysis as described above. Data shown are representative of the results of three independent experiments.
α-Amylase Assays
Barley seeds were surface-sterilized by washing with 70% ethanol for 2 min, with sodium hypochlorite for 30 min, and then with sterile distilled water. De-embryonated SLN1 half-grains were used for the measurement of α-amylase activity. To identify seeds homozygous for sln1-1, the progeny seeds of self-pollinated SLN1/sln1-1 heterozygotes were cut in half. The half-seeds containing the embryo were transferred to Murashige and Skoog (1962) medium and germinated in a cold room (4°C) to facilitate the identification of sln1-1 mutant homozygotes (the stems and leaves of the barley sln1-1 mutant grow faster than those of wild-type controls, and this effect is particularly pronounced at lower temperatures [Harrison et al., 1998]). De-embryonated half-grains corresponding to sln1-1 seedlings then were used for the measurement of α-amylase activity. The de-embryonated half-grains were transferred aseptically to 5 mL of aqueous buffer (Fu et al., 2001) and incubated in 5 μM GA3 or without GA3 in the presence or absence of each pharmacological agent for 18 h (inhibitors were at the same concentration as described above). α-Amylase activity in the incubation medium was measured as described previously (Fu et al., 2001).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank David Laurie for helpful discussion, advice on the growth of barley, and provision of plant material, and Patrick Achard and Kathryn King for discussion of the manuscript. This work was supported by a Core Strategic Grant from the Biotechnology and Biological Science Research Council to the John Innes Centre and by Biotechnology and Biological Science Research Council Grant 208/P15108 to N.P.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006197.
References
- Akiyama, T., Ishida, J., Nakagawa, S., Ogawara, H., Watanabe, S.-i., Itoh, N., Shibuya, M., and Fukami, Y. (1987). Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 262, 5592–5595. [PubMed] [Google Scholar]
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D.G., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defences. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bethke, P.C., Schuurink, R., and Jones, R.L. (1997). Hormonal signalling in cereal aleurone. J. Exp. Bot. 48, 1337–1356. [Google Scholar]
- Bialojan, C., and Takai, A. (1988). Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Biochem. J. 256, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle, C., Koncz, C., and Chua, N.-H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Börner, A., Plaschke, J., Korzun, V., and Worland, A.J. (1996). The relationships between the dwarfing genes of wheat and rye. Euphytica 89, 69–75. [Google Scholar]
- Callis, J., and Vierstra, R.D. (2000). Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- Cercós, M., Gómez-Cadenas, A., and Ho, T.-H.D. (1999). Hormonal regulation of a cysteine proteinase gene, EBP1, in barley aleurone layers: Cis and trans-acting elements involved in the coordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19, 107–118. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M. (1988). Hormonal regulation of gene expression in the “slender” mutant of barley (Hordeum vulgare L.). Planta 175, 115–120. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya: Molecular and physiological characterization. Plant Physiol. 129, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker, S.J., Hedden, P., Lenton, J.R., and Stoddart, J.L. (1990). Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol. 94, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Jung, H.-S., and Sun, T.-p. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T.-p. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, L., Johnston, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 98, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, C.A. (1977). Slender: An accelerated extension growth mutant of barley. Barley Genet. Newsl. 7, 24–27. [Google Scholar]
- Fu, X., Sudhakar, D., Peng, J., Richards, D.E., Christou, P., and Harberd, N.P. (2001). Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 13, 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Marshall, G.A. (1975). The nature and genetic control of gibberellin insensitivity in dwarf wheat grain. Heredity 35, 55–65. [Google Scholar]
- Gazit, A., Osherov, N., Posner, I., Yaish, P., Poradosu, E., Gilon, C., and Levitzki, A. (1991). Tyrophostins. 2. Heterocyclic and alpha- substituted benzylidenemalononitrile tyrophostins as potent inhibitors of EGF receptor and ErbB2/neu tyrosine kinases. J. Med. Chem. 34, 1896–1907. [DOI] [PubMed] [Google Scholar]
- Gilroy, S. (1996). Signal transduction in barley aleurone protoplasts is calcium dependent and independent. Plant Cell 8, 2193–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy, S., and Jones, R.L. (1994). Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol. 104, 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas, A., Zentella, R., Walker-Simmonds, M.K., and Ho, T.-H.D. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13, 667–679. [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P.M., White, R.G., Llewellyn, D.J., and Jacobsen, J.V. (2002). Gibberellin signaling in barley aleurone cells: Control of SLN1 and GAMYB expression. Plant Physiol. 129, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17, 1–9. [DOI] [PubMed] [Google Scholar]
- Hanke, J.H., Gardner, J.P., Dow, R.L., Changelian, P.S., Brissette, W.H., Weringer, E.J., Pollok, B.A., and Connelly, P.A. (1996). Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. J. Biol. Chem. 271, 695–701. [DOI] [PubMed] [Google Scholar]
- Harberd, N.P., King, K.E., Carol, P., Cowling, R.J., Peng, J., and Richards, D.E. (1998). Gibberellin: Inhibitor of an inhibitor of … ? Bioessays 20, 1001–1008. [DOI] [PubMed] [Google Scholar]
- Harrison, J., Nicot, C., and Ougham, H. (1998). The effect of low temperature on patterns of cell division in developing second leaves of wild-type and slender mutant barley (Hordeum vulgare L.). Plant Cell Environ. 21, 79–86. [Google Scholar]
- Ho, T.-H.D., Nolan, R.C., and Shute, D.E. (1981). Characterisation of a gibberellin-insensitive dwarf wheat, D6899. Plant Physiol. 67, 1026–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley, R. (1994). Gibberellins: Perception, transduction and responses. Plant Mol. Biol. 26, 1529–1555. [DOI] [PubMed] [Google Scholar]
- Hooley, R., Beale, M.H., and Smith, S.J. (1991). Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta 183, 274–280. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, H.D., Smith, S.J., Desikan, R., Plakidou-Dymock, S., Lovegrove, A., and Hooley, R. (1998). Heterotrimeric G proteins are implicated in gibberellin induction of α-amylase gene expression in wild oat aleurone. Plant Cell 10, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.E., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, A., Cappelluti, S., Cervantes-Cervantes, M., Rodriguez, M., and Bush, D.S. (1996). Okadaic acid, a protein phosphatase inhibitor, blocks calcium changes, gene expression, and cell death induced by gibberellin in wheat aleurone cells. Plant Cell 8, 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan, M.B., and Ho, T.-H.D. (1988). Slender barley: A constitutive gibberellin-response mutant. Planta 175, 107–114. [DOI] [PubMed] [Google Scholar]
- Lau, K.H., Farley, J.R., and Baylink, D.J. (1989). Phosphotyrosyl protein phosphatases. Biochem. J. 257, 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove, A., and Hooley, R. (2000). Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 5, 102–110. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Osterlund, M., Hardtke, C., Wei, N., and Deng, X.-W. (2000). Targetted destabilization of HY5 during light regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Penson, S.P., Schuurink, R.C., Fath, A., Gubler, F., Jacobsen, J.V., and Jones, R.L. (1996). cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 8, 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signalling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 67–88. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., Peng, J., and Harberd, N.P. (2000). Plant GRAS and metazoan STATs: One family? Bioessays 22, 573–577. [DOI] [PubMed] [Google Scholar]
- Ritchie, S., and Gilroy, S. (1998). Gibberellins: Regulating genes and germination. New Phytol. 140, 363–383. [DOI] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Rissueeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Schuurink, P.C., Chan, P.V., and Jones, R.L. (1996). Modulation of calmodulin mRNA and protein levels in barley aleurone. Plant Physiol. 111, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.-S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.-p. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13, 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y.A., Martinez, E.C., and Sun, T.-p. (1997). The RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki, T. (1991). Use and specificity of staurosporine, UCN-01, and calphostin C as protein kinase inhibitors. Methods Enzymol. 201, 340–347. [DOI] [PubMed] [Google Scholar]
- Wen, C.-K., and Chang, C. (2002). Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., NietoRostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]