Abstract
Tracheary elements (TEs) have a unique cell death program in which the rapid collapse of the vacuole triggers the beginning of nuclear degradation. Although various nucleases are known to function in nuclear DNA degradation in animal apoptosis, it is unclear what hydrolase is involved in nuclear degradation in plants. In this study, we demonstrated that an S1-type nuclease, Zinnia endonuclease 1 (ZEN1), functions directly in nuclear DNA degradation during programmed cell death (PCD) of TEs. In-gel DNase assay demonstrated the presence of a 24-kD Ca2+/Mg2+-dependent nuclease and a 40-kD Zn2+-dependent nuclease as well as ZEN1 in 60-h-cultured cells that included differentiating TEs. Such cell extracts possessed the ability to degrade the nuclear DNA isolated from Zinnia elegans cells in the presence of Zn2+, and its activity was suppressed by an anti-ZEN1 antibody, indicating that ZEN1 is a central DNase responsible for nuclear DNA degradation. The introduction of the antisense ZEN1 gene into Zinnia cells cultured for 40 h specifically suppressed the degradation of nuclear DNA in TEs undergoing PCD but did not affect vacuole collapse. Based on these results, a common mechanism between animal and plant PCD is discussed.
INTRODUCTION
In most multicellular organisms, programmed cell death (PCD) is built into the processes of normal development and growth. One key event in PCD is DNA degradation, because the degradation of the genome is considered to be a means by which the cell death program is made irreversible and facilitates the disassembly of the nucleus. Indeed, DNA degradation is a hallmark of apoptosis during PCD in animal cells (Wyllie, 1980; Jacobson et al., 1997). Apoptotic DNA degradation occurs in at least three stages (Wyllie, 1980; Oberhammer et al., 1993). Early in the process, DNA is cleaved to high molecular mass fragments (50 to 200 kb) consistent with the size of chromatin loop domains. The subsequent cleavage of DNA occurs at the internucleosomal linker region, and its products produce a 180-bp DNA ladder. Importantly, some cell lines exhibit only high molecular mass DNA cleavage (Oberhammer et al., 1993). Finally, the fragmented DNA in apoptotic cells is digested completely by an enzyme(s) such as DNase II produced by engulfing cells (McIlroy et al., 2000). To date, apoptosis-inducing factor (Susin et al., 1999), topoisomerase II (Li et al., 1999), and caspase-activated DFF/CAD-ICAD (Sakahira et al., 1999) have been implicated in the early process of DNA cleavage. On the other hand, internucleosomal cleavage is known to be associated with several endonucleases, including caspase-activated DFF/CAD-ICAD (Liu et al., 1997, 1998; Enari et al., 1998; Sakahira et al., 1998), endonuclease G (Li et al., 2001; Parrish et al., 2001), and DNase I (Oliveri et al., 2001).
In plants, the active degradation of genomic DNA has been observed in PCD that is associated with the hypersensitive response (Mittler et al., 1995, 1997; Levine et al., 1996; Ryerson and Heath, 1996; Wang et al., 1996; Tada et al., 2001), environmental stress–induced cell death (Katsuhara and Kawasaki, 1996; Katsuhara, 1997; Stein and Hansen, 1999), senescence (Orzáez and Granell, 1997a, 1997b; Yen and Yang, 1998; Xu and Hanson, 2000), the death of cereal aleurone (Wang et al., 1998; Fath et al., 2000), and tracheary element (TE) differentiation (Obara et al., 2001). In plant PCD, increased activities of several kinds of nuclease have also been reported (for review, see Sugiyama et al., 2000). They are categorized into at least four classes depending on their requirement for divalent cations: Zn2+-dependent nucleases (Brown and Ho, 1986, 1987; Thelen and Northcote, 1989; Pérez-Amador et al., 2000), Ca2+-dependent nucleases (Oleson et al., 1974, 1982; Mittler and Lam, 1995), Mg2+-dependent nucleases (Marchetti et al., 2001), and Ca2+/Mg2+-dependent nucleases (Xu and Hanson, 2000). However, there is no direct evidence implicating these PCD-related nucleases in the degradation of nuclear DNA during the process of PCD.
We isolated BEN1 and ZEN1, whose activities increase in association with endosperm degradation and TE cell death, respectively, as plant genes encoding Zn2+-dependent nucleases (Aoyagi et al., 1998). BEN1 and ZEN1 belong to the S1-type nuclease gene family. Recently, four other plant S1-type nuclease genes were reported, three of which are expressed in association with cell death processes such as senescence (DSA6 [Panavas et al., 1999] and BFN1 [Pérez-Amador et al., 2000]) and a salt stress–induced cell death process (Bnuc1 [Muramoto et al., 1999]). Because the presence of S1-type nucleases has not been confirmed in animals, S1-type nucleases may function in plant-specific cell death programs. However, it is not known how S1-type nucleases function in cell death in plants.
Terminal differentiation of TEs, which are components of the vessels and tracheids of the xylem, is a classic example of plant PCD and has been studied extensively using the Zinnia elegans cell culture system established by Fukuda and Komamine (1980). Confocal laser scanning microscopy showed that rapid degeneration of the nucleus in differentiating TEs occurred within 10 to 20 min after vacuole collapse (Obara et al., 2001). Various hydrolases, including nucleases (Thelen and Northcote, 1989; Ye and Droste, 1996; Aoyagi et al., 1998) and proteases (Minami and Fukuda, 1995; Ye and Varner, 1996; Beers and Freeman, 1997), are synthesized before the active degeneration of cellular contents and are thought to accumulate in the vacuole of TEs to sequester them from the cytoplasm. Thelen and Northcote (1989), using an in-gel assay of extracts of Zinnia cultured cells, showed the presence of at least seven active nucleases, six of which were induced specifically in TEs. Moreover, only a 43-kD nuclease from Zinnia possessed the ability to degrade both single- and double-stranded DNA in addition to RNA. However, because Thelen and Northcote (1989) focused only on nucleases requiring Zn2+, the involvement of other types of nucleases in PCD during TE differentiation is unknown.
ZEN1 was isolated as the cDNA for the 43-kD nuclease reported by Thelen and Northcote (1989) (Aoyagi et al., 1998). The ZEN1 protein possesses a putative signal peptide at the N terminus for targeting to a specific organelle or for secretion. When genomic DNA of ZEN1 was introduced into tobacco cells, active ZEN1 protein accumulated in the vacuoles (J. Ito, M. Sugiyama, and H. Fukuda, unpublished results). Because the 43-kD nuclease showed an acidic pH optimum (Thelen and Northcote, 1989), it is believed that ZEN1 functions in the vacuoles or after tonoplast breakdown.
Here, we conducted experiments to elucidate the type and role of nucleases in nuclear degradation during plant PCD using TE differentiation as a model system. We report the following observations. First, we found that, in addition to ZEN1, a novel Ca2+/Mg2+-dependent nuclease with a molecular mass of 24 kD is expressed transiently at the PCD stage. Second, in vitro experiments revealed that ZEN1, but not the 24-kD nuclease, plays a major role in nuclear degradation during PCD. Third, gene transfer experiments with an antisense ZEN1 construct indicated that ZEN1 functions in nuclear degradation in vivo.
RESULTS
DNases Responsible for TE Differentiation
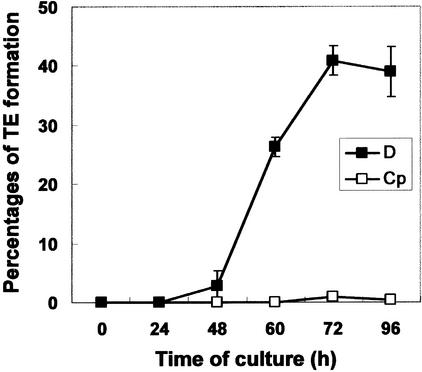
TEs characterized by secondary cell wall thickenings were formed from 48 to 72 h when isolated mesophyll cells of Zinnia were cultured in TE-inductive D medium, whereas TE differentiation was not induced in control Cp medium (see Methods) (Figure 1). To identify the DNases responsible for PCD in TE differentiation, we performed an in-gel DNase assay using various metal ions and denatured calf thymus DNA as a substrate (Figure 2). When protein extracts from cells were incubated in reaction buffer containing 1 mM EDTA and no metal ion, no DNase activity was detected (data not shown). DNase activities were observed only when metal ions were added to the reaction buffer after EDTA treatment, suggesting that DNase activities in the protein extracts are EDTA sensitive. Two Zn2+-dependent nucleases with molecular masses of 43 and 40 kD and three Ca2+/Mg2+-dependent nucleases with molecular masses of 24, 27, and 29 kD were detected in protein extracts from cells cultured in TE-inductive D medium (Figure 2). Their appearances during culture were different. A Zn2+-dependent nuclease with a molecular mass of 43 kD and a Ca2+/Mg2+-dependent nuclease with a molecular mass of 24 kD appeared transiently during TE maturation. The 43-kD Zn2+-dependent nuclease was active under both neutral and acidic conditions, but it was more active under acidic conditions. By contrast, the 24-kD Ca2+/Mg2+-dependent nuclease was active only at neutral pH. These results suggest that the 43-kD Zn2+-dependent nuclease and the 24-kD Ca2+/Mg2+-dependent nuclease, which may be activated in different subcellular compartments, play roles in DNA degradation during PCD of TEs.
Figure 1.
Time Course of TE Differentiation of Zinnia Mesophyll Cells Cultured in TE-Inductive D Medium or Control Cp Medium.
The rate of TE formation was defined as a percentage of TEs per total cells, including TEs and living cells. Each point represents the mean result from three samples (n = 500 per sample), and bars show standard deviations.
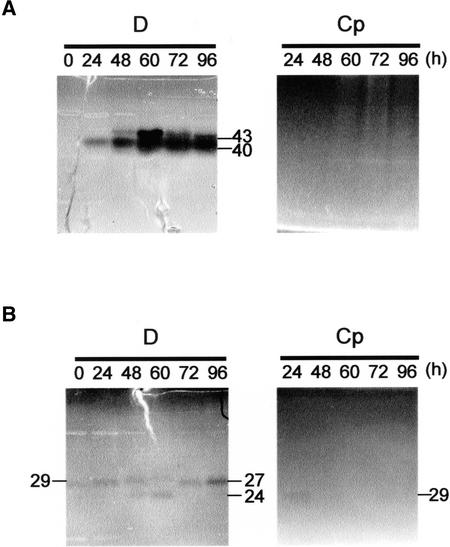
Figure 2.
Detection of DNase Activities during TE Differentiation.
Samples of protein extracts (10 μg of protein per lane) from Zinnia cells cultured for various periods in TE-inductive medium (D) or a control medium (Cp) were separated by SDS-PAGE using denatured calf thymus DNA as a substrate. After preincubation in 10 mM Tris-HCl, pH 7.5, containing 1 mM EDTA for 1 h, gels were incubated for another 12 h in different reaction buffers: 25 mM sodium acetate–acetic acid buffer, pH 5.5, containing 5 mM ZnSO4 (A) or 10 mM Tris-HCl, pH 7.5, containing 10 mM CaCl2 and 10 mM MgCl2 (B). Background substrate was stained with ethidium bromide to visualize DNase bands. The predicted molecular mass of each band is presented in kD.
Identification of Nucleases with Antibodies
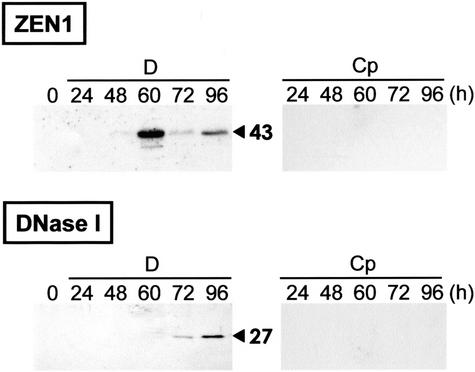
The 43-kD nuclease is known to be encoded by ZEN1 (Aoyagi et al., 1998; J. Ito, M. Sugiyama, and H. Fukuda, unpublished results). To determine whether the PCD-related increase in 43-kD nuclease activity is dependent on increased amounts of ZEN1 protein, the amount of ZEN1 during culture was examined with an anti-ZEN1 antibody (Figure 3). In Cp-cultured cells, the accumulation of ZEN1 was not detectable during the culture period. In D-cultured cells, the ZEN1 protein, recognized as a band with a molecular mass of 43 kD, began to accumulate after 48 h in culture and peaked at 60 h (Figure 3, top). The accumulation pattern of ZEN1 was similar to the change in activity of the 43-kD nuclease (Figure 2A).
Figure 3.
Immunoblot Analysis Using Antibodies against ZEN1 and DNase I.
The same samples of cell extracts (30 μg of protein per lane) used for Figure 2 were subjected to SDS-PAGE and transferred to a nitrocellulose membrane. Immunodetection was performed with anti-ZEN1 antibody (top) or anti-DNase I antibody (bottom). The predicted molecular mass of each band is presented in kD.
DNase I is one of the Ca2+/Mg2+-dependent nucleases associated with apoptosis in animals. Using an antibody against DNase I, we examined whether DNase I–type nucleases were also expressed during TE differentiation. The antibody recognized a protein with a molecular mass of 27 kD only in cells cultured in TE-inductive D medium (Figure 3, bottom). The DNase I–like protein was detected first in extracts of cells cultured for 72 h and increased during the next 24 h. This pattern of accumulation of the DNase I–like protein was consistent with the activity of the 27-kD Ca2+/Mg2+-dependent nuclease (Figures 2B and 3). These results suggest that a DNase I–like nuclease may be involved in a late step of in vitro xylem differentiation.
ZEN1 Is a Key Nuclease Degrading Nuclear DNA in PCD
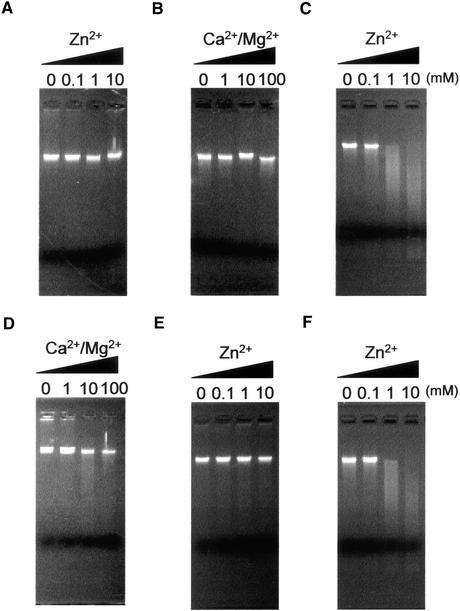
To examine nuclear DNA–degrading activity in vitro, nuclei purified from Zinnia cells after 20 h in culture were incubated with protein extracts of Zinnia cells cultured for various periods, and DNA released as a result of enzyme action was electrophoresed on an agarose gel. Protein extracts prepared from cells cultured for 24 h in TE-inductive D medium did not degrade nuclear DNA substantially (Figures 4A and 4B). However, protein extracts of cells cultured for 60 h in TE-inductive D medium exhibited high activity in degrading nuclear DNA in the presence of >1 mM Zn2+ (Figure 4C) but not in the presence of Ca2+ and/or Mg2+ (Figure 4D).
Figure 4.
In Vitro Endonuclease Activity Assay in Nuclei Isolated from Zinnia Cells 20 h after Culture.
(A) to (D) Protein extracts prepared from Zinnia cells, which had been cultured for 24 h ([A] and [B]) or 60 h ([C] and [D]) in TE-inductive D medium, were incubated with the isolated nuclei for 2 h at 27°C in two types of reaction buffer: Zn2+ reaction buffer (25 mM sodium acetate–acetic acid buffer, pH 5.5, and 1 mM EDTA with or without 0.1, 1, or 10 mM ZnSO4) ([A] and [C]) or Ca2+/Mg2+ reaction buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA with or without 1, 10, or 100 mM CaCl2 and MgCl2) ([B] and [D]). DNA was extracted and separated on a 1.5% agarose gel.
(E) and (F) To investigate the role of ZEN1 in in vitro nuclear DNA digestion, 60-h-cultured cell protein extracts were preincubated with anti-ZEN1 antibody (E) or preimmune serum (F) for 30 min at 37°C and then reacted with the isolated nuclei in Zn2+ reaction buffer.
To examine the involvement of ZEN1 in Zn2+-dependent DNA cleavage, the effect of an anti-ZEN1 antibody on nuclear DNA degradation was tested. Preincubation of the 60-h cell extract with the anti-ZEN1 antibody suppressed Zn2+-dependent nuclear DNA degradation (Figure 4E), but preimmune serum had no effect (Figure 4F).
Role of ZEN1 in Nuclear Degradation in Differentiating TEs
To examine the effect of the transient expression of ZEN1 on nuclear degradation, a β-glucuronidase (GUS) gene construct (p35SGUS) was introduced into Zinnia cells by particle bombardment. Zinnia cells were bombarded for 40 h after culture initiation and then cultured for another 26 h. As a result, ∼3 to 8% of Zinnia cells, including TEs and non-TEs, showed GUS-positive staining. The blue transformed cells were categorized into three types: differentiating TEs with the nucleus, hollow dead TEs, and non-TE cells (Figure 5). Bombardment at 40 h did not affect the progress of TE differentiation (data not shown).
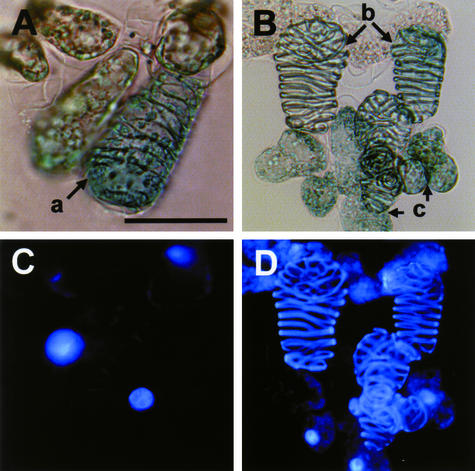
Figure 5.
Light and Fluorescence Images of Zinnia Cells Stained with X-Gluc Solution and 4′,6-Diamidino-2-Phenylindole.
p35SZEN1A and p35SGUS were introduced by particle bombardment into Zinnia cells cultured for 40 h in TE-inductive D medium. Collected cells were fixed and then incubated with X-Gluc solution (see Methods) for 12 h. 4′,6-Diamidino-2-phenylindole staining was performed for 15 min in the dark. The transformed cells, which were identified by the expression of GUS activity (blue staining), were categorized into three types by cell shape and staining pattern of 4′,6-diamidino-2-phenylindole: GUS-positive TEs with the nucleus (a); GUS-positive TEs without the nucleus (b); and GUS-positive non-TE cells with the nucleus (c). (A) and (C) show light images, and (B) and (D) show fluorescence images. Bar = 50 μm.
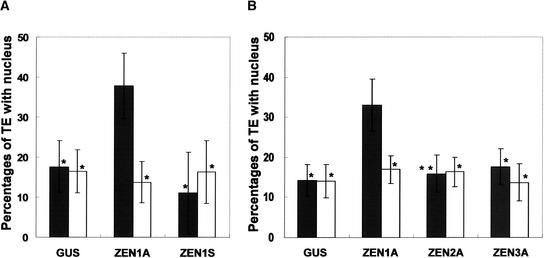
To investigate the function of ZEN1 in nuclear degradation during differentiation into TEs, sense (p35SZEN1S) and antisense (p35SZEN1A) ZEN1 gene constructs with p35SGUS were introduced into Zinnia cells that had been cultured for 40 h in TE-inductive D medium, and these treated cells were cultured for another 26 h (Figure 6A). At least 200 transformed or nontransformed TEs were counted in each bombardment, and the effects of the introduced ZEN1 constructs on nuclear degradation were evaluated by comparing nuclear degradation in GUS-expressing TEs and nontransformed TEs. At 66 h of culture (26 h after gene transfection), TEs accounted for ∼20% of the total cell population in all cultures bombarded with p35SZEN1A plus p35SGUS, p35SZEN1S plus p35SGUS, and p35SGUS. In all cultures, ∼15% of nontransformed TEs had a nucleus (Figure 6A, open columns). The introduction of p35SZEN1S plus p35SGUS or only p35SGUS did not change the number of TEs with nuclei (Figure 6A, closed columns). However, the introduction of an antisense ZEN1 construct (p35SZEN1A) suppressed nuclear degradation conspicuously, and ∼35% of TEs had prominent nuclei. This experiment was repeated at least five times, and in all the cases, similar results were obtained.
Figure 6.
Suppression of Nuclear Degradation in Zinnia TEs by the Introduction of an Antisense ZEN1 Gene.
p35SGUS (GUS) was cotransfected by bombardment of gold particles coated with p35SZEN1A (ZEN1A), p35SZEN1S (ZEN1S), p35SZEN2A (ZEN2A), or p35SZEN3A (ZEN3A) into Zinnia cells cultured for 40 h in TE-inductive D medium; then, the cells were cultured for another 26 h. The cells were stained with X-Gluc solution (see Methods) for 12 h and then for another 15 min with 4′,6-diamidino-2-phenylindole. Open columns, percentages of nontransformed TEs with the nucleus per total nontransformed TEs; closed columns, percentages of transformed TEs with the nucleus per total transformed TEs. The data represent averages and standard deviations of three replicates (n = 200 per sample). Values are significantly different between p35SZEN1A and the other constructs (single asterisks, P < 0.02 by two-sample t test; double asterisks, P < 0.05 by two-sample t test). The experiment was repeated two to five times and yielded similar results.
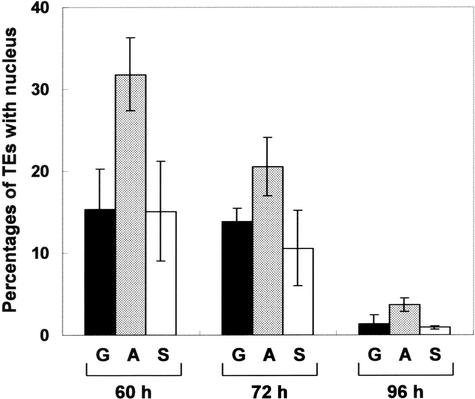
Figure 7 shows a time course of nuclear degradation in transformed TEs, supporting the observation that the antisense ZEN1 gene suppresses nuclear degradation in differentiating TEs. These data also indicated that the introduction of the antisense ZEN1 gene did not suppress nuclear degradation completely but rather delayed the loss of the nucleus. One can judge the rupture of the vacuole based on the shape of the nucleus in differentiating TEs (Obara et al., 2001). We then examined the effect of introducing the constructs described above on the rupture of the vacuole, using the nuclear shape as an index. We found that the introduction of the antisense ZEN1 gene did not delay the rupture of the vacuole compared with that in other constructs (data not shown).
Figure 7.
Time Course of Nuclear Degradation in Zinnia TEs Transformed with an Antisense ZEN1 Gene.
The experiment was performed as shown in Figure 6 except that cells were cultured for another 20, 32, and 56 h after p35SGUS (G; black columns), p35SGUS plus p35SZEN1A (A; gray columns), or p35SGUS plus p35SZEN1S (S; white columns) was introduced at 40 h of culture. Each column represents the percentage of transformed TEs with the nucleus per total transformed TEs. The data represent averages and standard deviations of three replicates (n = 200 per sample). Values are significantly different between p35SZEN1A and the other constructs in each culture period (P < 0.05 by two-sample t test). The experiment was repeated at least twice and yielded similar results.
S1-type nuclease genes form a small gene family (e.g., five genes in the Arabidopsis genome). In Zinnia, three genes (ZEN1, ZEN2, and ZEN3) are known to be expressed in TE-inductive culture (Pérez-Amador et al., 2000). Microarray analysis has suggested that ZEN2 mRNA starts accumulating earlier than ZEN1 mRNA and continues to accumulate even in the late stage of culture (T. Demura, unpublished results), although detailed expression patterns of ZEN2 and ZEN3 during xylem differentiation are still unknown. These facts suggest the possibility that ZEN2 and ZEN3 also may be involved in PCD of TEs. Therefore, to determine if the suppression of nuclear degradation by the antisense construct is specific for the ZEN1 gene, we made antisense constructs of two additional S1-type nuclease genes, ZEN2 and ZEN3. The antisense constructs of ZEN2 and ZEN3 had no effect on nuclear degradation during TE PCD, unlike the result with antisense ZEN1 (Figure 6B). This finding strongly supported our idea that ZEN1 is involved specifically in nuclear DNA degradation in vivo.
DISCUSSION
The most striking feature of PCD that is unique to TE is the rapid collapse of the large central vacuole, which leads to the final degeneration of cell contents (Fukuda, 2000; Jones, 2000). The disruption of the tonoplast is predicted to cause vacuolar enzymes to invade the cytoplasm and degrade various organelles, including the nucleus. A serial observation of nuclear DNA levels stained with a fluorescent dye in living TEs revealed that no detectable level of nuclear DNA was degraded before the vacuole collapsed, but the DNA was digested rapidly within 15 min after the vacuole collapsed (Obara et al., 2001), suggesting the presence of DNase insulated in the vacuole or activated after the vacuole collapsed. The 43-kD Zn2+-dependent S1-type nuclease, ZEN1, has been proposed as one such enzyme involved specifically in nuclear degradation, because its enzymatic properties and the pattern of accumulation of its mRNA are consistent with a role in PCD (Thelen and Northcote, 1989; Aoyagi et al., 1998). However, there has been no direct evidence to indicate that ZEN1 plays a crucial role in nuclear degradation during PCD. In this report, we have succeeded in demonstrating that ZEN1 functions directly in nuclear DNA degradation during TE PCD.
An in-gel DNase assay of cell extracts revealed the presence of several Ca2+/Mg2+-dependent nucleases and a 40-kD Zn2+-dependent nuclease as well as ZEN1 in 60-h-cultured cells that included differentiating TEs (Figure 2). Such cell extracts could degrade the DNA of isolated nuclei in the presence of Zn2+ but not in the presence of Ca2+/Mg2+ (Figures 4C and 4D). Furthermore, nuclear DNA–degrading activity was suppressed by an anti-ZEN1 antibody, which recognized only ZEN1 with a molecular mass of 43 kD (Figures 4E and 4F). By contrast, cell extracts from early-stage-cultured cells in which the ZEN1 gene was not expressed did not possess the nuclear DNA degradation activity (Figures 2, 4A, and 4B). These facts indicate clearly that ZEN1 is a central DNase responsible for nuclear DNA degradation. For this in vitro assay, we used nuclei isolated from 20-h-cultured cells, which are in the dedifferentiation stage and which have not yet been fated to cell death (Fukuda, 1997). The fact that their nuclei were degraded by the 60-h-cultured cell extracts indicated that the extracts contain a set of enzymes responsible for degrading normal nuclei.
We used a method to transiently transform cultured Zinnia cells without affecting the course of TE differentiation (Figure 5). This method allowed us to show that the introduction of antisense ZEN1 retarded nuclear degradation in differentiating TEs in vivo (Figures 6 and 7) but did not prevent vacuole collapse. To confirm that this suppression is ZEN1 specific, we also performed a similar experiment using other nuclease genes, ZEN2 and ZEN3, which also are expressed in TE-inductive culture (Pérez-Amador et al., 2000). As expected, the antisense constructs of ZEN2 and ZEN3 had no effect on nuclear degradation during TE PCD, unlike the result with antisense ZEN1 (Figure 6B). S1-type nuclease genes are known to form a small gene family. In Arabidopsis, the family is composed of only five genes, three of which (BFN1, NP-176996, and NP-567630) have all nine of the conserved amino acids necessary for the binding of zinc ions. In Zinnia, three S1-type nuclease genes that share the nine conserved amino acids (ZEN1, ZEN2, and ZEN3) have been reported. A comparison of the deduced amino acid sequences of these genes indicates that ZEN1 is similar to BFN1 and that ZEN2 and ZEN3 have high similarity to NP-176996 and NP-567630 (data not shown). Although we isolated ∼15,000 ESTs from cultured Zinnia cells differentiating into xylem cells, we found no S1-type nuclease genes other than ZEN1, ZEN2, and ZEN3. Therefore, our antisense experiments may cover most, if not all, of the ZEN-type nuclease genes that are expressed in Zinnia cells cultured in TE-inductive medium.
Taking these facts into consideration, our antisense experiments strongly suggest that that ZEN1, but not ZEN2 or ZEN3, is involved directly in nuclear degradation in vivo. However, the introduction of antisense ZEN1 did not inhibit nuclear degradation completely. This incomplete inhibition may result from the incomplete suppression of ZEN1 mRNA accumulation by the antisense gene or from the presence of DNases other than ZEN1 that are responsible for nuclear degradation during TE PCD. The latter possibility may be excluded by the fact that an anti-ZEN1 antibody repressed in vitro nuclear degradation almost completely (Figure 4). By contrast, because it is well known that an antisense construct usually causes only partial repression of the accumulation of its target mRNA, the former possibility is likely. In addition, we cannot exclude the possibility of partial gene silencing. Unfortunately, measurements of endogenous ZEN1 mRNA or of ZEN1 activity decreased by the introduction of its antisense construct in individual cells cannot be made using current techniques. Indeed, nuclear degradation in differentiating TE is a rapid event and takes only 15 min after vacuole collapse (Obara et al., 2001). Nevertheless, the retardation of nuclear degradation by the antisense ZEN1 occurred in several hours. This may be explained by the partial suppression of ZEN1 mRNA accumulation by the antisense gene.
On the other hand, the introduction of sense ZEN1 did not affect nuclear degradation significantly. Our experiments with transformed BY-2 cells indicated that overexpressed ZEN1 nuclease was accumulated in the vacuole, although in an active form, and did not affect nuclear degradation (J. Ito, M. Sugiyama, and H. Fukuda, unpublished results), suggesting that even in Zinnia TEs, overexpressed ZEN1 nuclease may not function in nuclear DNA degradation until the vacuole ruptures. In addition, the time (15 min) during which ZEN1 nuclease acts in nuclear degradation after vacuole collapse is much shorter than the time (several hours) during which the ZEN1 protein is accumulated in the vacuole (Aoyagi et al., 1998; Obara et al., 2001). Therefore, it is not surprising that the expression of the sense ZEN1 construct in developing TEs did not result in the promotion of nuclear degradation.
Is the family of S1-type nucleases generally involved in PCD in plants? The Arabidopsis genome database indicates the presence of five S1-type–like nuclease genes, all of which were confirmed to be expressed by the existence of ESTs that encode these genes. Of these genes, BFN1 is known to be induced during leaf and stem senescence (Pérez-Amador et al., 2000). The daylily DSA6, which is highly similar to BFN1, also is expressed during petal senescence (Panavas et al., 1999). In barley, two S1-type nucleases, BEN1 and Bnuc1, have activities that increase in association with endosperm degradation and salt stress–induced cell death, respectively (Brown and Ho, 1986; Muramoto et al., 1999). Bnuc1 mRNA also is expressed in senescing leaves (Muramoto et al., 1999). In Zinnia, there are two other S1-type nuclease genes, ZEN2 and ZEN3, in addition to ZEN1 (Pérez-Amador et al., 2000). ZEN1 is expressed specifically in TE PCD but not in leaf senescence or in stress-induced cell death (data not shown). By contrast, ZEN2 and ZEN3 are expressed during senescence as well as in xylem cell culture (Pérez-Amador et al., 2000). These results indicate that each member of the S1-type nuclease gene family is involved in a specific plant cell death program(s). Plant S1-type nucleases are targeted into an intracellular organelle or are secreted out of the cell. For instance, ZEN1 is targeted to the vacuole of Zinnia TEs (J. Ito, J. Nakashima, and H. Fukuda, unpublished data), and a barley nuclease (BEN1) is secreted out of the aleurone cells into the endosperm (Brown and Ho, 1986). In any case, S1-type nucleases may contribute to plant PCDs by degrading nuclear DNA, although it is unlikely that all S1-type nucleases function in plant PCD, because the activity of one S1-type nuclease is reported to be associated with DNA synthesis and repair (Grafi and Larkins, 1995).
We found a 24-kD Ca2+/Mg2+-dependent nuclease that appeared transiently at the cell-death stage of TE differentiation (Figure 2). This nuclease was most active at neutral pH but not at acidic pH, suggesting that it does not act in the vacuole or in the cytoplasm after vacuole collapse. In animal cells, once apoptosis is induced, most nucleases, including Ca2+/Mg2+-dependent nuclease, are activated and translocated to the nucleus (Zamzami and Kroemer, 1999; Li et al., 2001). Therefore, it is possible that in Zinnia, the 24-kD Ca2+/Mg2+-dependent nuclease might be targeted to the nucleus before the attack of the vacuolar-type ZEN1 DNase.
The chromosomal DNA of most animal cells forms DNA ladders during apoptosis that result from the contribution of specific kinds of nucleases, such as CAD/DFF40, endonuclease G, and the DNase I family, most of which have neutral pH optima (Shiokawa and Tanuma, 2001; Widlak and Garrard, 2001; Widlak et al., 2001). These nucleases are known to be activated or to be transported to the nucleus in association with apoptosis. By contrast, DNA ladder formation is not detected in TE PCD (Groover et al., 1997). There are no definite homologs for CAD/DFF40 and endonuclease G in the Arabidopsis genome. ZEN1 nuclease, together with cell extracts, induced a smear pattern of nuclear DNA degradation in vitro (Figure 4). Therefore, in plant TE PCD, S1-type nucleases may degrade chromosomal DNA instead of the ladder-forming nucleases seen in animal apoptosis. Interestingly, recent studies of animal apoptosis provide strong evidence for another pathway of chromosomal DNA degradation (Ferri and Kroemer, 2000, 2001; McIlroy et al., 2000; Wu et al., 2000). McIlroy et al. (2000) found that when CAD activity is suppressed, phagocytes can activate an auxiliary mode of apoptotic DNA fragmentation in which a lysosomal acid nuclease such as DNase II is involved. In Caenorhabditis elegans, a DNase II homolog from engulfing cells, NUC-1, was also found to function in chromosomal DNA degradation (Wu et al., 2000). However, DNase II and S1-type nucleases including ZEN1 show biochemical differences that include ion requirements and the mode of DNA cleavage. Because S1-type nucleases are unique to plants and DNase II–like activity is not detected during TE PCD (data not shown), it is possible that plants have evolved the use of S1-type nucleases to degrade chromosomal DNA independently of animals.
METHODS
Plant Material
The first leaves of 14-day-old seedlings of Zinnia elegans (cv Canary Bird) (Takii Shubyo, Kyoto, Japan) were used for the isolation of mesophyll cells in suspension culture according to the method of Fukuda and Komamine (1980). The culture medium contained 0.1 mg/L α-naphthylacetic acid and 0.2 mg/L benzyladenine (the tracheary element [TE]–inductive D medium) or 0.1 mg/L α-naphthylacetic acid and 0.001 mg/L benzyladenine (the noninductive Cp medium).
Protein Extraction
The cell suspension was separated into cells and medium by gentle filtration through a nylon filter (pore size of 10 μm) at various times of culture. Approximately 100 mg of cells was homogenized with 500 μL of homogenization buffer (50 mM Tris-HCl, pH 7.5, 2 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, and 50 μM leupeptin) at 4°C. The homogenate was stirred at 4°C in the presence of polyvinylpolypyrrolidone and Amberlite XAD-4 (Organo, Tokyo, Japan) and centrifuged at 10,000g for 10 min, and the supernatant was collected and stored at −80°C until use. The concentration of soluble proteins was measured by the Bradford (1977) method.
In-Gel DNase Assay
An in-gel DNase assay was performed according to Thelen and Northcote (1989), with some modification. Proteins in the sample were dissolved in SDS-PAGE sample buffer (1% [w/v] SDS, 62.5 mM Tris-HCl, pH 6.8, and 10% [w/v] glycerol) without a reducing agent, incubated at 37°C for 15 min, and subjected to SDS-PAGE on a 12% polyacrylamide gel containing 50 μg/mL fibrinogen and 30 μg/mL denatured calf thymus DNA. After electrophoresis and removal of SDS, gels were washed with washing buffer (10 mM Tris-HCl, pH 7.5, and 1 mM EDTA) and then incubated for 12 h at 45°C in reaction buffer (10 mM Tris-HCl, pH 7.5, containing 10 mM CaCl2 and 10 mM MgCl2, or 25 mM sodium acetate–acetic acid buffer, pH 5.5, containing 1 mM ZnSO4). DNase activity was visualized by staining the gel with 2 μg/mL ethidium bromide.
Immunoblot Analysis
For immunoblot analysis, proteins prepared as described above were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. The membrane was incubated with the purified anti-ZEN1 antibody or an anti-DNase I antibody (Sigma, St. Louis, MO). After further incubation of the membrane with a peroxidase-conjugated secondary antibody, signal was detected using the enhanced chemiluminescence system (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's protocol.
Isolation of Nuclei
Nuclei were prepared from Zinnia cells cultured for 20 h according to the method of Obara et al. (2001). A pellet of nuclei was resuspended in 25 mM sodium acetate–acetic acid buffer, pH 5.5, or 10 mM Tris-HCl, pH 8.0, each of which contained 40 mM KCl, 10 mM NaCl, 0.15 mM spermine, 0.15 mM spermidine, and 10% (w/v) Suc. Nuclei were prepared at a density of ∼1.5 × 107/mL. The suspension of nuclei was mixed with an equal volume of glycerol and stored at −20°C until use.
In Vitro Endonuclease Activity Assay
Isolated nuclei were incubated with 2.5 μg of protein extract for 2 h at 27°C in Zn2+ reaction buffer (25 mM sodium acetate–acetic acid buffer, pH 5.5, and 1.0 mM EDTA with or without 0.1, 1.0, or 10 mM ZnSO4) or Ca2+/Mg2+ reaction buffer (10 mM Tris-HCl, pH 8.0, and 1.0 mM EDTA with or without 1.0, 10, or 100 mM CaCl2 and MgCl2). As a negative control, nuclei were incubated for 2 h at 27°C in the same buffer without protein extracts. The reaction was stopped by adding an equal volume of 2 × lysis buffer (200 mM Tris-HCl, pH 8.0, 100 mM EDTA, 250 mM NaCl, 1% [w/v] sarkosyl, and 1 mg/mL proteinase K) and incubated further for 1 h at 55°C. Nuclear debris were spun down in a microfuge for 10 min, and the DNA in the supernatant was precipitated with 0.8 volume of isopropanol for 10 min at −20°C. The DNA precipitate was washed with 70% (w/v) ethanol, dried, and resuspended in water. The DNA was treated with 10 μg/mL RNase A at 37°C for 1 h and then analyzed by electrophoresis on a 1.5% (w/v) agarose gel containing 0.5 μg/mL ethidium bromide.
To analyze the effect of anti-ZEN1 antibody, protein extracts were preincubated with anti-ZEN1 antibody at 37°C for 1 h and added to isolated nuclei to start the reaction.
Transformation of Cultured Zinnia Cells with the ZEN1 Gene
A HindIII-EcoRI fragment of 2.2 kb containing a 35S promoter of Cauliflower mosaic virus–β-glucuronidase (GUS) cassette was removed from pBE2113 (Mitsuhara et al., 1996) and ligated into the corresponding site of pUC18, generating p35SGUS. To make antisense constructs of ZEN1, ZEN2, and ZEN3, the GUS coding region in p35SGUS was replaced with the antisense gene as follows. The ApaI sites of the 1.1-, 1.1-, and 1.4-kb ApaI-XbaI fragments containing the coding regions of ZEN1, ZEN2, and ZEN3, respectively, which were excised from the pBluescript KS+ plasmid (Stratagene, La Jolla, CA), were blunt ended by treating them with T4 DNA polymerase (Takara, Kyoto, Japan) and ligated into the XbaI–blunt-ended SacI site of p35SGUS. The resulting constructs were designated p35SZEN1A, p35SZEN2A, and p35SZEN3A, respectively. We also created a sense ZEN1 gene ligated into the XbaI-SacI site of p35SGUS, designated p35SZEN1S. The nucleotide sequences of the constructs were confirmed using an ABI 373A DNA Sequencer (Applied Biosystems, Foster City, CA).
To introduce these constructs into Zinnia cells, the particle bombardment method was adopted using a helium-driven particle accelerator (IDERA GIE-III; Tanaka, Sapporo, Japan) with all basic adjustments set according to the manufacturer's recommendations (vacuum, 665 mm Hg; distance, 10.5 cm; helium press, 4.0 kgf/cm2; and releasing time, 0.025 s). Gold particles with a diameter of 1.0 μm (Bio-Rad, Hercules, CA) were coated with objective DNAs (0.6 mg gold·μg−1 DNA·shot−1) and subjected to bombardment. A 20-mL aliquot of Zinnia cells cultured for 40 h in TE-inductive D medium was collected on a nylon filter (pore size of 10 μm) and placed on TE-inductive D medium containing 0.5% (w/v) agar until use. p35SGUS was cotransferred with p35SZEN1A, p35SZEN2A, p35SZEN3A, or p35SZEN1S into Zinnia cells with three replications. p35SGUS also was transferred into Zinnia cells as a control. The treated cells were resuspended in old medium and cultured again for the times indicated in Figures 6 and 7.
Histochemical Detection of GUS Activity and 4′,6-Diamidino-2-Phenylindole Staining
Cells were collected at various times of culture and incubated in fixation buffer (0.3% [v/v] formaldehyde, 10 mM Mes, pH 5.6, and 0.3 M mannitol) for 45 min at room temperature. After washing twice with 50 mM sodium phosphate, pH 7.0, the cells were stained in X-Gluc solution (1 mM 5-bromo-4-chloro-3-indolyl-β-glucronide [X-Gluc], 50 mM sodium phosphate, pH 7.0, and 20% [v/v] methanol) for 12 h at 37°C. The stained cells were washed with 50 mM NaPO4, pH 7.0, suspended in 3% (w/v) melting agarose, and solidified on the dish (60 × 15 mm). To observe nuclear degradation, cells were stained with 1 μg/mL 4′,6-diamidino-2-phenylindole in TAN buffer (20 mM Tris-HCl, pH 7.65, 0.5 mM EDTA, 7 mM 2-mercaptoethanol, and 0.4 mM phenylmethylsulfonyl fluoride) for 15 min in the dark after 1% (v/v) glutaraldehyde fixation. The numbers of cells and TEs with or without a nucleus, and with or without GUS staining, were measured with an epifluorescence microscope (model BX-50-FLA; Olympus, Tokyo, Japan), and at least 200 cells were examined in each sample.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are grateful to Russell L. Jones for careful reading of the manuscript. We thank Pamela J. Green for supplying the ZEN2 and ZEN3 clones. We thank Koichiro Saito and Munetaka Sugiyama for providing technical and helpful information about the antisense experiment. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sport, and Culture of Japan (Grants 14036205 and 10219201 to H.F.), from the Japan Society for the Promotion of Science (Grant 13440236 to H.F. and a Research Fellowship for Young Scientists to J.I.), and from the Ministry of Agriculture, Forest, and Fisheries (to H.F.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006411.
References
- Aoyagi, S., Sugiyama, M., and Fukuda, H. (1998). BEN1 and ZEN1 cDNAs encoding S1-type DNases that are associated with programmed cell death in plants. FEBS Lett. 429, 134–138. [DOI] [PubMed] [Google Scholar]
- Beers, E.P., and Freeman, T.B. (1997). Protease activity during tracheary element differentiation in Zinnia mesophyll cultures. Plant Physiol. 113, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1977). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brown, P.H., and Ho, T.-H.D. (1986). Barley aleurone layers secrete a nuclease in response to gibberellic acid: Purification and partial characterization of the associated ribonuclease, deoxyribonuclease, and 3′-nucleotidase activities. Plant Physiol. 82, 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P.H., and Ho, T.-H.D. (1987). Biochemical properties and hormonal regulation of barley nuclease. Eur. J. Biochem. 168, 357–364. [DOI] [PubMed] [Google Scholar]
- Enari, M., Sakahira, H., Yokoyama, H., Okawa, K., Iwamatsu, A., and Nagata, S. (1998). A caspase-activated DNase that degrades DNA during apoptosis and its inhibitor ICAD. Nature 391, 43–50. [DOI] [PubMed] [Google Scholar]
- Fath, A., Bethke, P., Lonsdale, J., Meza-Romero, R., and Jones, R. (2000). Programmed cell death in cereal aleurone. Plant Mol. Biol. 44, 255–266. [DOI] [PubMed] [Google Scholar]
- Ferri, K.F., and Kroemer, G. (2000). Control of apoptotic DNA degradation. Nat. Cell Biol. 2, 63–64. [DOI] [PubMed] [Google Scholar]
- Ferri, K.F., and Kroemer, G. (2001). Organelle-specific initiation of cell death pathways. Nat. Cell Biol. 3, 255–263. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (1997). Programmed cell death during vascular system formation. Cell Death Differ. 4, 684–688. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (2000). Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. 44, 245–253. [DOI] [PubMed] [Google Scholar]
- Fukuda, H., and Komamine, A. (1980). Establishment of an experimental system for the tracheary element differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 65, 57–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi, G., and Larkins, B.A. (1995). Activity of single-stranded DNA endonucleases in mung bean is associated with cell division. Plant Mol. Biol. 29, 703–710. [DOI] [PubMed] [Google Scholar]
- Groover, A., DeWitt, N., Heidel, A., and Jones, A. (1997). Programmed cell death of plant tracheary element differentiation in vitro. Protoplasma 196, 197–211. [Google Scholar]
- Jacobson, M.D., Weil, M., and Raff, M.C. (1997). Programmed cell death in animal development. Cell 88, 347–354. [DOI] [PubMed] [Google Scholar]
- Jones, A. (2000). Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci. 5, 225–230. [DOI] [PubMed] [Google Scholar]
- Katsuhara, M. (1997). Apoptosis-like cell death in barley roots under salt stress. Plant Cell Physiol. 38, 1087–1090. [Google Scholar]
- Katsuhara, M., and Kawasaki, T. (1996). Salt stress induced nuclear and DNA degradation in meristematic cells of barley roots. Plant Cell Physiol. 37, 169–173. [Google Scholar]
- Levine, A., Pennell, R.I., Alvarez, M.E., Palmer, R., and Lamb, C. (1996). Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr. Biol. 4, 427–437. [DOI] [PubMed] [Google Scholar]
- Li, L.Y., Luo, X., and Wang, X. (2001). Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412, 95–99. [DOI] [PubMed] [Google Scholar]
- Li, T.K., Chen, A.Y., Yu, C., Mao, Y., Wang, H., and Liu, L.F. (1999). Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev. 13, 1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Li, P., Widlak, P., Zou, H., Luo, X., Garrard, W.T., and Wang, X. (1998). The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA 95, 8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Zou, H., Slaughter, C., and Wang, X. (1997). DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89, 175–184. [DOI] [PubMed] [Google Scholar]
- Marchetti, S., Zaina, G., Chiabà, C., Pappalardo, C., and Pitotti, A. (2001). Isolation and characterization of an endonuclease synthesized by barley (Hordeum vulgare L.) uninucleate microspores. Planta 213, 199–206. [DOI] [PubMed] [Google Scholar]
- McIlroy, D., Tanaka, M., Sakahira, H., Fukuyama, H., Suzuki, M., Yamamura, K., Ohsawa, Y., Uchiyama, Y., and Nagata, S. (2000). An auxiliary mode of apoptotic DNA fragmentation provided by phagocytes. Genes Dev. 14, 549–558. [PMC free article] [PubMed] [Google Scholar]
- Minami, A., and Fukuda, H. (1995). Transient and specific expression of a cysteine endopeptidase during autolysis in differentiating tracheary elements from Zinnia mesophyll cells. Plant Cell Physiol. 36, 1599–1606. [PubMed] [Google Scholar]
- Mitsuhara, I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Mol. Physiol. 37, 49–59. [DOI] [PubMed] [Google Scholar]
- Mittler, R., and Lam, E. (1995). In situ detection of nDNA fragmentation during the differentiation of tracheary elements in higher plants. Plant Physiol. 108, 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Shulaev, V., and Lam, E. (1995). Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., Simon, L., and Lam, E. (1997). Pathogen-induced programmed cell death in tobacco. J. Cell Sci. 110, 1333–1344. [DOI] [PubMed] [Google Scholar]
- Muramoto, Y., Watanabe, A., Nakamura, T., and Takabe, T. (1999). Enhanced expression of a nuclease gene in leaves of barley plants under salt stress. Gene 234, 315–321. [DOI] [PubMed] [Google Scholar]
- Obara, K., Kuriyama, H., and Fukuda, H. (2001). Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 125, 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhammer, F., Wilson, J.W., Dive, C., Morris, I.D., Hichman, J.A., Wakeling, A.E., Walker, P.R., and Sikoska, M. (1993). Apoptotic death in epithelial cells: Cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 12, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleson, A.E., Janski, A.M., and Clark, E.T. (1974). An extracellular nuclease from suspension cultures of tobacco. Biochim. Biophys. Acta 366, 89–100. [DOI] [PubMed] [Google Scholar]
- Oleson, A.E., Janski, A.M., Fahrlander, P.D., and Wiesner, T.A. (1982). Nuclease I from suspension-cultured Nicotiana tabacum: Purification and properties of the extracellular enzyme. Arch. Biochem. Biophys. 216, 223–233. [DOI] [PubMed] [Google Scholar]
- Oliveri, M., Daga, A., Cantoni, C., Lunardi, C., Millo, R., and Puccetti, A. (2001). DNase I mediates internucleosomal DNA degradation in human cells undergoing drug-induced apoptosis. Eur. J. Immunol. 31, 743–751. [DOI] [PubMed] [Google Scholar]
- Orzáez, D., and Granell, A. (1997. a). DNA fragmentation is regulated by ethylene during carpel senescence in Pisum sativum. Plant J. 11, 137–144. [Google Scholar]
- Orzáez, D., and Granell, A. (1997. b). The plant homologue of the defender against apoptotic death gene is down-regulated during senescence of flower petals. FEBS Lett. 404, 275–278. [DOI] [PubMed] [Google Scholar]
- Panavas, T., Pikula, A., Reid, P.D., Rubinstein, B., and Walker, E.L. (1999). Identification of senescence-associated genes from daylily petals. Plant Mol. Biol. 40, 237–248. [DOI] [PubMed] [Google Scholar]
- Parrish, J., Li, L., Klotz, K., Ledwich, D., Wang, X., and Xue, D. (2001). Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412, 90–94. [DOI] [PubMed] [Google Scholar]
- Pérez-Amador, M.A., Abler, M.L., Rocher, E.J.D., Thompson, D.M., van Hoof, A., LeBrasseur, N.D., Lers, A., and Green, P.J. (2000). Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiol. 122, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryerson, D.E., and Heath, M.C. (1996). Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell 8, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira, H., Enari, M., and Nagata, S. (1998). Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391, 96–99. [DOI] [PubMed] [Google Scholar]
- Sakahira, H., Enari, M., Ohsawa, Y., Uchiyama, Y., and Nagata, S. (1999). Apoptotic nuclear morphological change without DNA fragmentation. Curr. Biol. 9, 543–546. [DOI] [PubMed] [Google Scholar]
- Shiokawa, D., and Tanuma, S. (2001). Characterization of human DNase I family endonucleases and activation of DNase γ during apoptosis. Biochemistry 40, 143–152. [DOI] [PubMed] [Google Scholar]
- Stein, J.C., and Hansen, G. (1999). Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiol. 121, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, M., Ito, J., Aoyagi, S., and Fukuda, H. (2000). Endonucleases. Plant Mol. Biol. 44, 387–397. [DOI] [PubMed] [Google Scholar]
- Susin, S.A., et al. (1999). Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397, 441–446. [DOI] [PubMed] [Google Scholar]
- Tada, Y., Hata, S., Takata, Y., Nakayashiki, H., Tosa, Y., and Mayama, S. (2001). Induction and signaling of an apoptotic response typified by DNA laddering in the defense response of oats to infection and elicitors. Mol. Plant-Microbe Interact. 14, 477–486. [DOI] [PubMed] [Google Scholar]
- Thelen, M.P., and Northcote, D.H. (1989). Identification and purification of a nuclease from Zinnia elegans L.: A potential molecular marker for xylogenesis. Planta 179, 181–195. [DOI] [PubMed] [Google Scholar]
- Wang, H., Li, J., Bostock, R.M., and Gilchrist, D.G. (1996). Apoptosis: A functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8, 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Oppedijk, B.J., Caspers, M.P.M., Lamers, G.E.M., Boot, M.J., Geerlings, D.N.G., Bakhuizen, B., Meijer, A.H., and van Duijn, B. (1998). Spatial and temporal regulation of DNA fragmentation in aleurone of germinating barley. J. Exp. Bot. 49, 1293–1309. [Google Scholar]
- Widlak, P., and Garrard, W.T. (2001). Ionic and cofactor requirements for the activity of the apoptotic endonuclease DFF40/CAD. Mol. Cell. Biochem. 218, 125–130. [DOI] [PubMed] [Google Scholar]
- Widlak, P., Li, L.Y., Wang, X., and Garrard, W.T. (2001). Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: Cooperation with exonuclease and DNase I. J. Biol. Chem. 276, 48404–48409. [DOI] [PubMed] [Google Scholar]
- Wu, Y.-C., Stanfield, G.M., and Horvitz, H.R. (2000). NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 14, 536–548. [PMC free article] [PubMed] [Google Scholar]
- Wyllie, A.H. (1980). Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284, 555–556. [DOI] [PubMed] [Google Scholar]
- Xu, Y., and Hanson, M.R. (2000). Programmed cell death during pollination-induced petal senescence in petunia. Plant Physiol. 122, 1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z.-H., and Droste, D.L. (1996). Isolation and characterization of cDNAs encoding xylogenesis-associated and wound-induced ribonucleases in Zinnia elegans. Plant Mol. Biol. 30, 697–709. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H., and Varner, J.E. (1996). Induction of cysteine and serine proteases during xylogenesis in Zinnia elegans. Plant Mol. Biol. 30, 1233–1246. [DOI] [PubMed] [Google Scholar]
- Yen, C.H., and Yang, C.H. (1998). Evidence for programmed cell death during leaf senescence in plants. Plant Cell Physiol. 39, 922–927. [Google Scholar]
- Zamzami, N., and Kroemer, G. (1999). Condensed matter in cell death. Nature 401, 127–128. [DOI] [PubMed] [Google Scholar]