Abstract
Rp1 is a complex disease resistance locus in maize that is exceptional in both allelic variability and meiotic instability. Genomic sequence analysis of three maize BACs from the Rp1 region of the B73 inbred line revealed 4 Rp1 homologs and 18 other gene-homologous sequences, of which at least 16 are truncated. Thirteen of the truncated genes are found in three clusters, suggesting that they arose from multiple illegitimate break repairs at the same sites or from complex repairs of each of these sites with multiple unlinked DNA templates. A 43-kb region that contains an Rp1 homolog, six truncated genes, and three Opie retrotransposons was found to be duplicated in this region. This duplication is relatively recent, occurring after the insertion of the three Opie elements. The breakpoints of the duplication are outside of any genes or identified repeat sequence, suggesting a duplication mechanism that did not involve unequal recombination. A physical map and partial sequencing of the Rp1 complex indicate the presence of 15 Rp1 homologs in regions of ∼250 and 300 kb in the B73 inbred line. Comparison of fully sequenced Rp1-homologous sequences in the region demonstrates a history of unequal recombination and diversifying selection within the Leu-rich repeat 2 region, resulting in chimeric gene structures.
INTRODUCTION
In plants, a major class of disease resistance genes is involved in the detection of pathogen presence, leading to the activation of a defense cascade. Most of these resistance genes encode proteins with a nucleotide binding site and a Leu-rich repeat (LRR) region. The LRR region is believed to be involved directly in the recognition of the pathogen (Ellis et al., 2000a). Resistance genes are abundant and highly diverse in plants (Michelmore and Meyers, 1998; Ellis et al., 2000a; Richter and Ronald, 2000; Dangl and Jones, 2001; Staskawicz, 2001; Staskawicz et al., 2001). Numerous mutational forces act on disease resistance genes, including point mutations, unequal recombination, gene conversion, and transposon activity, all providing the raw material on which natural selection may act.
Many disease resistance genes exhibit diversifying selection, in which nonsynonymous substitutions exceed synonymous substitutions within an LRR domain or domains. These include resistance genes belonging to the RGC2 family of lettuce, Cf-4/Cf-9 homologs of tomato, and the Xa21 family of rice (Parniske et al., 1997; Song et al., 1997; Meyers et al., 1998b). Diversifying selection appears to be particularly strong in the xxLxLxx consensus sequence of the LRR region, which encodes the part of the protein that should be exposed to solvent and involved in ligand binding (Kobe and Deisenhofer, 1995).
Many of the resistance genes in plants are arranged in clusters, including Cf-4/Cf-9, I2, and Pto of tomato, N of flax, RGC2 of lettuce, RPP5 of Arabidopsis, and Xa21 of rice (Martin et al., 1994; Parniske et al., 1997; Song et al., 1997; Meyers et al., 1998a; Simons et al., 1998; Noel et al., 1999; Dodds et al., 2001). It is probable that the resistance genes within a cluster were generated originally by unequal crossing over. Within these resistance gene clusters, meiotic instability caused by unequal recombination can generate novel alleles with new disease resistance specificities (Richter et al., 1995).
Transposable elements play an important role in the evolution of disease resistance genes (Michelmore and Meyers, 1998; Richter and Ronald, 2000), as they do for all other plant genes (Wessler et al., 1995). Two elements of the same family inserted within a resistance gene region provide an opportunity for gene duplication by unequal crossing over. Insertion of transposons can create interrupted disease resistance genes, as seen in Arabidopsis RPP5, flax N, maize Hm1, and rice Xa21 homologs (Song et al., 1997; Multani et al., 1998; Noel et al., 1999; Dodds et al., 2001). The insertion of a few amino acids in an L6 allele of flax was caused by the insertion and subsequent excision of a transposable element (Ellis et al., 1997).
Rp1 is a complex locus in maize that confers resistance to Puccinia sorghi, a fungal rust pathogen. Several Rp1 genes map within a region of ∼0.4 centimorgan (cM) near the telomere on the short arm of chromosome 10 (Saxena and Hooker, 1968). Most Rp1 genes are meiotically unstable (Bennetzen et al., 1988). Unequal recombination, including conversion, is observed frequently in the Rp1 region (Sudupak et al., 1993). Rp1-D and eight paralogs from the Rp1-D haplotype have been cloned and sequenced (Collins et al., 1999; Sun et al., 2001). One of the nine Rp1 homologs was truncated. Sequence analysis indicated that maize Rp1 genes undergo diversifying selection in an LRR region (Sun et al., 2001). Although the sequence relationships of resistance genes in the Rp1 region have been investigated, their physical organization has been estimated only crudely (Sun et al., 2001). In the absence of this structural information, it is not possible to describe accurately the chromosomal rearrangements that gave rise to current Rp1 haplotypes and that continue to generate diversity in this region.
In the present study, we sequenced and analyzed two segments that constitute 195 kb of the maize Rp1 gene region. In addition, we mapped the Rp1 complex to a series of overlapping clones that form two contigs of 300 and 450 kb. These studies reveal the genomic organization and suggest some of the molecular events that have contributed to the creation of this complex locus. Many of these modes of regional evolution have been detected at other genes, including complex resistance loci, but two detected phenomena are unprecedented in any region of any genome: nearly exact duplications of 43 kb of DNA that are separated by >300 kb, and several clusters of unrelated gene fragments inserted between the resistance genes.
RESULTS
Sequence Analysis of Maize Rp1 BACs
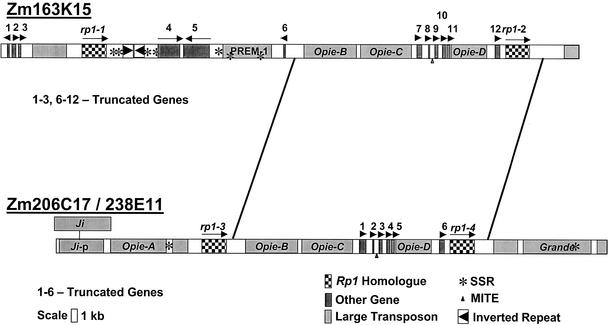
To study genomic organization at the Rp1 locus in maize, BACs Zm163K15, Zm238E11, and Zm206C17 were selected by their hybridization to a probe from the 5′ end of the Rp1-D gene (Collins et al., 1999). In the first small library that we screened, these were the only clones that were found, and preliminary restriction mapping indicated that they did not overlap. These clones were isolated from a BAC library that was constructed from B73, a standard maize inbred line. These three BACs were sequenced fully by a shotgun approach (Dubcovsky et al., 2001). The final error rate was less than one base per 10 kb, and all of the finished sequence was of high quality (PHRED value of 25 or more, see www.phrap.org/phrap.docs/phred.html). The maize DNA in BAC Zm163K15 consists of 95,078 bp and has two Rp1-homologous genes (rp1-1 and rp1-2). This BAC also contains 12 other gene-homologous sequences (of which at least 10 are truncated), 3 full-length Opie retroelements (SanMiguel et al., 1996), and 1 PREM-1 retroelement (Turcich and Mascarenhas, 1994) (Figure 1). The two Rp1 homologs are in direct orientation, ∼68 kb apart. BACs Zm206C17 and Zm238E11 were found to overlap for most of their length, yielding exactly identical sequence for 73,026 bp. The maize DNA in the Zm206C17/Zm238E11 contiguous sequence (contig) is 99,156 bp and harbors two Rp1 homologs (rp1-3 and rp1-4) plus six other gene-homologous segments (all truncated) and six retrotransposons. The two Rp1 homologs are in the same orientation, ∼38 kb apart. In addition, there are several truncated retroelements on each of these BACs. Identified retrotransposon sequences constitute ∼70% of Zm206C17/Zm238E11 and 45% of Zm163K15.
Figure 1.
Sequence Organization on Two BACs from the rp1 Complex.
The four Rp1 homologs were named rp1-1 through rp1-4. Arrows indicate genes and arrowheads indicate truncated genes, plus their size and proposed direction of transcription. Asterisks indicate simple sequence repeats (SSRs) that contain eight or more tandem repeats. Lines connecting the two BACs indicate the 43-kb duplicated region. Grande, Ji, Opie, and PREM-1 are maize retrotransposons. Miniature inverted repeat transposable elements (MITEs) are indicated by small triangles. The two large arrowheads indicate inverted repeat sequences of 2283 and 2284 bp that are 96% identical to each other (ignoring indels) and separated by 199 bp.
An ∼43-kb region (42,366 bp in Zm163K15 and 42,860 bp in Zm206C17/Zm238E11) was duplicated on these two BACs. This duplication contains one Rp1 homolog, six truncated genes, and three Opie retrotransposons. Truncated genes 7 through 12 of Zm163K15 correspond to genes 1 through 6 of Zm206C17/Zm238E11. A deletion of 488 bp corresponding to part of the truncated helicase-like transcription factor gene is observed in Zm163K15. Comparison of the duplicated region identified insertions/deletions (indels) of 3, 5, 19, and 20 bp. Most of the indels (3, 5, and 19 bp) contained perfect short direct repeats in one of the 43-kb duplicated segments. In Zm163K15, the 3′ boundary of the 43-kb duplication is located ∼2 kb downstream from the predicted stop codon of the rp1-2 gene. Ignoring indels, these 43-kb segments are >99% identical. In Zm206C17/Zm238E11, the 5′ boundary of this duplication is situated ∼1 kb downstream of the rp1-3 gene and the 3′ boundary is situated ∼2 kb downstream of the rp1-4 gene (Figure 1).
Genes and Truncated Genes
Within the Rp1 region sequenced, we identified six loci that appear to be functional genes. Four of these genes are Rp1 homologs, and they contain uninterrupted reading frames similar to the functional Rp1-D gene. Although B73 does not have any known Rp1 gene that provides resistance to any identified race of P. sorghi, it is possible that an Rp1 homolog in B73 does specify resistance to some extinct or otherwise unidentified rust pathogen race. Rp1 homologs from this study show 87 to 100% identities over an 800-bp region with maize B73 ESTs (BQ577730, AI947572, BM347712, and BI992969), indicating that some Rp1-homologous genes in this complex are expressed in this inbred line.
Unlike other complex disease resistance loci, the Rp1 region contains two apparently intact genes within the cluster that play no obvious role in disease resistance (genes 4 and 5 of Zm163K15; Figure 1). Gene 4 exhibits homology with Arabidopsis (AP002820; E value = 8e−91) and rice hypothetical proteins with no similarity to any ESTs. Gene 5 shows homology with rice unknown (AC087723; E value = 4e−19) and hypothetical proteins with 91 and 96% identity with maize ESTs (H89387 and AW424864) over a 440-bp region.
Genes were classified as truncated when the Basic Local Alignment Search Tool (BLAST) X program detected homology with only part of a protein entry in the GenBank database (Table 1). The presumed translated protein products of most of these truncated gene fragments were homologous with protein products encoded by genes in Arabidopsis. This homology was limited only to N-terminal, C-terminal, or central portions of the protein. For instance, the predicted protein product of the truncated gene 3 of Zm163K15 is homologous with amino acids 392 through 615 of the 1042 amino acids specified by a maize calcium ATPase gene (AF09687). Four of the truncated genes in Zm163K15 exhibited significant EST matches. Some of the truncated genes have well-defined introns and exons supported by hits to ESTs. For example, truncated gene 12 of Zm163K15 is homologous with exons 2, 3, and 4 plus introns 2 and 3 of the maize Suc phosphate synthase gene (M97550). Five truncated genes (genes 7 to 11) in Zm163K15 are present in a cluster of ∼5 kb.
Table 1.
Truncated and Apparently Intact Genes in BAC Zm163K15
| Gene | BLASTX Homology | Accession No. | E Value |
|---|---|---|---|
| 1a | Anthranilate phosphoribosyl transferase–like (Arabidopsis) | AB010697 | 1e−09 |
| 2a | N-Acetyl transferase hookless1 (Arabidopsis) | U50399 | 2e−23 |
| 3a | Calcium ATPase (maize) | AF09687 | 3e−46 |
| 4 | Hypothetical protein (Arabidopsis) | AP002820 | 8e−91 |
| 5 | Unknown protein (rice) | AC087723 | 4e−19 |
| 6a | Phosphoglycerate kinase (Populus nigra) | U50399 | 1e−05 |
| 7a | Helicase-like transcription factor (Arabidopsis) | BAB11535 | 3e−40 |
| 8a,b | Unknown protein (Arabidopsis) | AB007651 | 6e−07 |
| 9a | Unknown protein (Arabidopsis) | AAF63179 | 3e−29 |
| 10a | Unknown protein (Arabidopsis) | AAG52482 | 6e−18 |
| 11a | Cytosolic factor SEC14 (Arabidopsis) | BAA81767 | 2e−23 |
| 12a | Suc phosphate synthase (maize) | AAA33513 | 2e−75 |
Truncated genes.
Stop codon in the middle of the truncated gene.
Physical Order of the Rp1 Genes in Maize Inbred Line B73
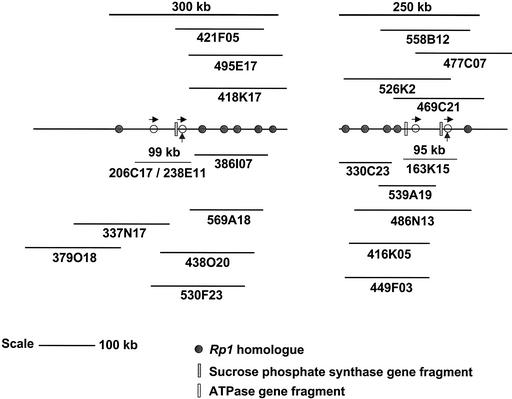
A physical map of the rp1 region was made to determine the positions of the sequenced Rp1 homologs within the genome of B73 maize. Thirty-three maize BACs that hybridized to an Rp1 probe were identified from a large insert BAC library of maize inbred line B73 (www.chori.org/bacpac). The restriction enzymes NotI, MluI, PacI, SwaI, and NcoI were used to generate a physical map based on overlapping restriction fragments. Gel blot hybridization analysis with the Rp1 5′ probe and a subclone of the truncated Suc phosphate synthase–like gene (next to rp1-2 and rp1-4) was used to confirm the physical map. Furthermore, PCR amplification using primers that amplify the truncated Suc phosphate synthase–like gene and the truncated ATPase-like gene (present near rp1-1) also was used to confirm the order of the BACs in the Rp1 gene cluster. The truncated ATPase-like gene was present in Zm163K15 but was absent in Zm206C17/Zm238E11. The truncated Suc phosphate synthase–like gene was part of the 43-kb duplicated region and therefore was present on both BACs. Interestingly, neither of these probes hybridized or amplified fragments from other regions within the rp1 complex (Figure 2). Based on the locations of these fragments, we were able to conclude that the 43-kb duplications (and the included Rp1 homologs) are part of two contigs within the B73 Rp1 gene cluster. In all, the two BAC contigs covered ∼300 and 450 kb. All of the Rp1-homologous sequences were mapped to a region of ∼250 and 300 kb within these two contigs (Figure 2). Because we do not have any definitive information concerning the relative locations of these two contigs (other than their adjacent locations on the short arm of chromosome 10 by recombinational mapping [Hulbert and Bennetzen, 1991]), we cannot say which of these two segments should be placed on the left or the right side of Figure 2, nor can we draw any conclusions about the distance between them.
Figure 2.
Physical Map of the Rp1 Region in Maize Inbred Line B73.
Each circle represents an Rp1 homolog. Open circles represent the four Rp1 homologs in BACs sequenced in the present study, and those with vertical arrows are part of the 43-kb duplicated region. The truncated Suc phosphate synthase–like and ATPase-like genes are designated by closed and open rectangles, respectively. Physical distances and BACs harboring Rp1 homologs are represented by lines above and below the rp1 complex map.
The number of Rp1 genes in the contigs was determined by hybridization of NcoI-digested BAC DNAs with the Rp1 5′ probe and low-pass sequencing of five additional BACs in the region. Our sequence analysis of the two segments of the maize genome harboring four Rp1 genes indicated the presence of unique bands hybridizing to three NcoI fragments (6.1, 4.4, and 8.3 kb). The fourth gene (rp1-4) was part of the duplicated 43-kb region, so the NcoI fragment is the same size as the NcoI fragment for rp1-2. A previous report by Sun et al. (2001) suggested that the number of NcoI hybridizing fragments corresponds to the number of Rp1 homologs. However, our data demonstrate that some Rp1 homologs can be on NcoI fragments of the same size, as seen here for rp1-2 and rp1-4. In addition, more than one hybridizing band per gene would be observed if there were an NcoI restriction site in the 5′ region of any Rp1 gene covered by the probe. As a result of the complex nature of the Rp1 locus, with multiple resistance genes in close vicinity and multiple duplications involving large genomic segments, low-pass sequencing of BACs Z486N13, Z526K02, Z337N17, Z418K17, and Z438O20 was performed to confirm the physical map. Based on sequencing and NcoI fragments harboring Rp1 homologs, we identified 15 Rp1 homologs in the Rp1 region (Figure 2).
Analysis of the Rp1 Homologs
The four Rp1 homologs sequenced in this study were compared with the Rp1-D gene that had been cloned and sequenced (Collins et al., 1999) and with each other. Rp1-D is the only sequenced Rp1 homolog that is known to provide disease resistance. The nucleotide sequence identities range from 90.8% (between rp1-1 and rp1-3) to 99.9% (between rp1-2 and rp1-4), corresponding to 351 and 1 nucleotide differences, respectively. rp1-1 was similar to rp1-3 in the N-terminal region of the gene, whereas rp1-2 and rp1-4 were most similar to Rp1-D (Figure 3) in this region. For these analyses, it is useful to evaluate several different domains in the putative RP1 proteins separately. In particular, three domains in the LRR region exhibit different repeat patterns: the LRR1 region (LRRs 1 to 11), domain C (LRRs 12 to 14), and the LRR2 region (LRRs 15 to 27) (Collins et al., 1999). In the first LRR domain, the LRR1 region, there is an apparent recombination breakpoint at which rp1-3 becomes similar to Rp1-D, rp1-2, and rp1-4 after a region in which there is a 3-bp deletion in rp1-3 and a 15-bp deletion in rp1-2 and rp1-4 compared with Rp1-D. There are several such regions downstream of the LRR1 region that give rise to a mosaic gene pattern. Nine adjacent amino acids were missing in rp1-2, rp1-3, and rp1-4 compared with Rp1-D and rp1-1 after the 13th amino acid of the fourth LRR unit. In another region downstream of LRR1, rp1-2, rp1-3, and rp1-4 had five more amino acids than Rp1-D and rp1-1 in the seventh LRR unit. In the LRR2 region, the number of amino acids was identical in all five Rp1 homologs.
Figure 3.
Mosaic Gene Structure of the Rp1 Homologs.
N-term indicates the N-terminal nucleotide binding site region (domain A), LRR1 indicates Leu-rich repeat region 1 (domain B), and LRR2 indicates Leu-rich repeat region 2 (domain D). Domain C is the region between LRR1 and LRR2. Domain E is the region between LRR2 and the end of the gene (stop codon). Recombination breakpoints are located in the LRR regions, creating chimeric gene structures. Gray and black boxes represent the highest similarity to Rp1-D and rp1-2/rp1-4, respectively. Hatched boxes represent regions that are unique to the specific Rp1 homologs.
Diversifying Selection
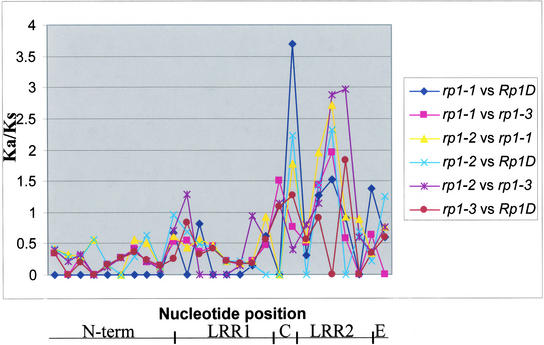
Like other plant disease resistance genes, Rp1 homologs have been shown to be under divergent or diversifying selection, especially in the LRR2 region (Sun et al., 2001). Sliding-window analysis (Proutski and Holmes, 1998) was performed with a window size of 150 bp to identify specific regions of the Rp1 homologs that are subject to diversifying selection. Our analysis shows that specific regions corresponding to the C domain and the LRR2 region are subject to diversifying selection (Figure 4). Varying degrees of selection were observed in different comparisons. For instance, comparison of rp1-1 with Rp1-D showed maximal diversifying selection in the C domain, which was found to be statistically significant (P < 0.05), even at this small window size. Purifying selection was observed throughout the nucleotide binding site and LRR1 regions for most of the pairwise comparisons. From these results, it appears that sliding window analysis is a particularly powerful tool for the identification of selected domains, with more discrimination power than a simple nonsynonymous substitution/synonymous substitution ratio for the entire LRR region.
Figure 4.
Sliding-Window Analysis Showing Diversifying and Purifying Selection across the Rp1 Homologs.
Each data point represents a window size of 150 bp. Nonsynonymous substitution/synonymous substitution (Ka/Ks) values >1, ∼1, and <1 indicate diversifying, neutral, and purifying selection, respectively. Abbreviations are as in Figure 3.
Assessment of Retrotransposon Insertion and Regional Duplication Dates
The insertion dates of full-length retrotransposons can be estimated from the divergence of their long terminal repeats (SanMiguel et al., 1998). In addition, we can use sequence divergence within the 43-kb duplicated segments to approximate when this duplication occurred. The number of substitutions per site was estimated using the Kimura two-parameter method, which corrects for multiple hits and takes into account transition and transversion substitution rates (Kimura, 1980). The synonymous substitution rate for the adh1 and adh2 genes of grasses (6.5 × 10−9 substitutions per year per site) was used to estimate the insertion and divergence dates of the retrotransposons. The Opie retroelements (Opie-B, Opie-C, and Opie-D) that are part of the duplicated 43-kb segment were estimated to have inserted within the last 1.5 million years (Table 2). The Grande element in Zm238E11 seems to be the most recent insertion, because its long terminal repeats are still identical. All of the other retrotransposons in the two BACs were inserted <1.5 million years ago. Comparison of the Opie elements within the 43-kb duplicated regions of Zm163K15 and Zm238E11 indicated that they began to diverge from each other within the last 0.2 million years (Table 2), consistent with the divergence time predicted by the one-nucleotide difference between rp1-2 and rp1-4. Based on these results, we conclude that the duplication of the 43-kb maize segment occurred within the last 0.2 million years, long after the insertion of the three retrotransposons in this duplicated region.
Table 2.
Predicted Retrotransposon Insertion and Divergence Times
| Name | Sites | T | V | K | SE | Mya | SE |
|---|---|---|---|---|---|---|---|
| Zm163K15 | |||||||
| Opie-B | 1229 | 11 | 3 | 0.011 | 0.003 | 0.85 | 0.23 |
| Opie-C | 1258 | 15 | 4 | 0.015 | 0.004 | 1.15 | 0.31 |
| Opie-D | 1225 | 5 | 4 | 0.007 | 0.002 | 0.54 | 0.15 |
| PREM-1 | 3510 | 42 | 17 | 0.017 | 0.002 | 1.31 | 0.15 |
| Zm206C17/Zm238E11 | |||||||
| Grande | 623 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ji | 1235 | 8 | 0 | 0.007 | 0.002 | 0.54 | 0.15 |
| Opie-A | 1254 | 7 | 1 | 0.006 | 0.002 | 0.46 | 0.15 |
| Opie-B | 1229 | 10 | 3 | 0.011 | 0.003 | 0.85 | 0.23 |
| Opie-C | 1239 | 11 | 2 | 0.011 | 0.003 | 0.85 | 0.23 |
| Opie-D | 1226 | 2 | 3 | 0.004 | 0.002 | 0.31 | 0.15 |
| Zm163K15/Zm238E11 | |||||||
| Opie-B | 8946 | 11 | 4 | 0.002 | 0 | 0.15 | 0 |
| Opie-C | 8959 | 9 | 6 | 0.002 | 0 | 0.15 | 0 |
| Opie-D | 5841 | 3 | 1 | 0.001 | 0 | 0.08 | 0 |
T, transition; V, transversion; K, number of substitutions per site calculated according to the Kimura two-parameter method; Mya, estimated insertion or divergence time of the retrotransposon in million years.
DISCUSSION
Rp1 is a complex disease resistance locus that shows meiotic instability as a result of unequal crossing over (Bennetzen et al., 1988; Sudupak et al., 1993; Richter et al., 1995; Collins et al., 1999). We studied the sequence organization and evolution of this region by sequencing three BACs covering ∼195 kb of the Rp1 region from maize line B73, an inbred line with no known Rp1 resistance specificity. Several interesting features were revealed that indicate the dynamic nature of this locus. The duplication of a 43-kb segment shared by the two BACs was identified. From our physical map, we conclude that these duplicated segments are separated by at least 300 kb. Although unequal recombination can create adjacent direct duplications of a very large size, from a few base pairs to hundreds of kilobases, there is no known single-step mechanism that can generate duplications of more than a few base pairs that are several kilobases apart. The boundaries of this 43-kb duplication are located outside of the Rp1 homologs. Therefore, the duplication of the 43-kb segment probably did not involve unequal recombination sited within Rp1 homologs. However, the instability of this region makes it likely that the current state of the region is the product of numerous rearrangement events. Regardless, at least two of these rearrangements must have had boundaries outside of the Rp1 homologs to generate the current B73 Rp1 region.
Comparison of the 43-kb duplicated segments revealed the presence of several indels. The presence of direct tandem repeats suggests that replication slippage, illegitimate recombination, and/or transposable element excision were responsible for these indels. The small number of differences within the 43-kb region indicates that this duplication occurred very recently, within the last 200,000 years. Recombinational studies and investigations of locus instability (Bennetzen et al., 1988; Sudupak et al., 1993; Richter et al., 1995; Sun et al., 2001) all have suggested that unequal recombination and/or other rearrangements in the Rp1 region can occur at rates of up to a few tenths of a percent per meiosis in some alleles or allelic combinations, so recent rearrangements are expected.
Several truncated gene fragments were detected next to the Rp1 homologs in the two BACs. Such truncated genes have been reported in a few disease resistance gene clusters in plants and also in duplicated segments of the human genome (Parniske et al., 1997; Meyers et al., 1998a; Noel et al., 1999; Holub, 2001; Emanuel and Shaikh, 2001). For instance, the Cf-4/Cf-9 cluster contains several copies of one truncated gene that is similar to plant lipoxygenases, whereas the Dm region of lettuce has three gene fragments that have homology with a few hundred base pairs of a cyclin gene (Parniske et al., 1997; Meyers et al., 1998a). In addition, eight truncated copies of a Ser/Thr protein kinase gene and a 5S rRNA gene were observed in the Arabidopsis RPP5 locus (Noel et al., 1999). In humans, segmental duplications involved in genomic disorders sometimes contain truncated genes (Emanuel and Shaikh, 2001). In the B73 Rp1 region, we found numerous truncated genes, often arranged in clusters. Truncated genes are relatively rare in plants, and no clustering of such genes has been reported to date. We believe that the accumulation of these elements in the Rp1 complex that we have discovered may be an outcome of the general instability of this region.
Double-stranded breaks are critical lesions in genomes that can be repaired by illegitimate or homologous recombination. In large genomes, such as that of tobacco, 40% of observed deletions in one study were accompanied by the insertion of filler sequences, whereas no large deletions or insertions were observed during double-stranded break repair in Arabidopsis (Kirik et al., 2000). Filler DNA was associated in maize with spontaneous wx and bz1 deletion alleles (Ralston et al., 1988; Wessler et al., 1990) and at the breakpoints of several spontaneous Ac deletion derivatives (Yan et al., 1999). The presence of discontinuous stretches of sequences in maize r-r alleles also was attributed to this gap-repair mechanism (Walker et al., 1995). In the illegitimate recombination process of gap repair, short stretches of heterologous DNA with very little sequence homology with the gap boundaries (often not more than 1 or 2 bp) can serve as a template to fill the gap between two broken ends. Although these fragments often consist of genomic DNA, long interspersed nuclear element DNAs and genic cDNA fragments have been inserted at the sites of DNA breaks in yeast and mammals (Moore and Haber, 1996; Teng et al., 1996). Hence, double-stranded break repair offers a mechanism for spreading sequences into new chromosomal locations. In general, though, it is rare for a filler sequence to contain an uninterrupted open reading frame over its entire length (Salomon and Puchta, 1998). In the B73 Rp1 region of maize, we do not know whether genes 4 and 5 of BAC Zm163K15 are genes that have been intrinsic to this region for many millions of years or whether they represent unusually large filler sequences.
The numerous truncated genes in the maize Rp1 region suggest that a high frequency of DNA breakage occurs in this region and that the truncated loci have been acquired as fillers in double-stranded break repair. The fact that the truncated genes are clustered also implies that the breaks must occur frequently at the same locations or that nu-merous fillers were needed to deal with the gaps that were created. The appearance of rare truncated genes at other complex resistance loci, although at lower abundances, sug-gests that double-stranded breaks may be unusually frequent in all complex resistance loci but just more so in the highly unstable Rp1 locus.
Genomic regions that exhibit high levels of recombination are usually gene rich in eukaryotes (Gill et al., 1996; Farris et al., 2000; Fu et al., 2001). The maize Rp1 complex varies tremendously in recombinational “size” from experiment to experiment, depending on the haplotypes that are paired. In a standard case, the most distal Rp1 resistance determinants are ∼0.4 cM apart (Saxena and Hooker, 1968). In our studies, the most distant Rp1 homologs are at least 550 kb apart, amounting to >1.3 Mb/cM. This finding suggests somewhat less recombination in the Rp1 area than the predicted average number for a maize genome that is ∼2400 Mb in size and has a genetic map of ∼2500 cM (Arumuganathan and Earle, 1991). However, in a previous study, markers ksu3/4 and ksu16 (bnl3.04), which are separated by ∼4 cM and which flank the Rp1 region (Hulbert and Bennetzen, 1991; Hong et al., 1993), were estimated to be ∼650 kb apart based on fluorescence in situ hybridization (Jiang et al., 1996). That study suggested a recombination rate of ∼160 kb per cM, approximate sevenfold higher than average. These results reconfirm earlier observations that the Rp1 region can be a hot spot for meiotic recombination in some homozygotes or allelic combinations but may be relatively deficient in recombination in other haplotype combinations. The low recombination rates in some combinations may be caused by dissimilar sizes and compositions of the regions between these highly divergent alleles, by inversions in some or all of the Rp1 paralogs present, or by a physically short distance between the Rp1 homologs that actually provide a scorable trait (race-specific disease resistance).
Unequal recombination can be localized within genes and is often sited in the repeats in the LRR region, as observed in mutants at the M locus in flax and in RPP5 in Arabidopsis (Anderson et al., 1997; Noel et al., 1999). However, in haplotypes of the Cf-4/Cf-9 region, unequal recombination within the complex was sited primarily at a specific location that was not within a resistance gene (Parniske et al., 1997; Parniske and Jones, 1999). The breakpoints in novel RGC2 haplotypes were between rather than within RGC2 genes (Chin et al., 2001). In the Rp1 homologs studied by Sun et al. (2001), all but two of the detected unequal recombination events were resolved within the Rp1 homologs themselves. However, each of these unequal recombination events was identified because it altered a resistance phenotype, an outcome that may be most likely to occur if recombination is sited within a resistance gene. Future studies of “unselected” recombination or rearrangement events (perhaps like the 43-kb duplication seen in this study) may provide a less biased indication of how often resistance genes are the sites of unequal recombination events.
Unequal or equal recombination within the LRR repeats can expand or contract LRR repeat numbers and make chimeric repeats, thereby potentially altering the recognition specificity of the gene (Ellis et al., 2000b). Hence, recombination at intragenic locations should provide outcomes that are most likely to be selectively advantageous. Alternatively, a specific mechanism to site recombination events within particular regions of resistance/recognition genes might evolve to promote resistance gene diversification (Bennetzen and Hulbert, 1992). In the Rp1 region, our data indicate that numerous equal or unequal recombination events have boundaries located within the LRR domains, particularly in LRR2. This finding suggests that this area is actively selected for diverse sequences.
Diversifying selection in the LRR regions allows plant disease resistance genes to keep up with rapidly evolving pathogen populations. In general, the N-terminal nucleotide binding site region shows purifying selection as expected as a result of its proposed role as an obligatory effector domain involved in signal transduction (Baker et al., 1997). The LRR regions participate in direct recognition of pathogen-derived avirulence factors, so their potential for variation is highly important. In this study, we show that sliding-window analysis can provide a particularly precise indication of the sites in resistance genes that are under diversifying or purifying selection.
Transposons are important players in the evolution of all plant genes and all aspects of genome structure (Wessler et al., 1995; Bennetzen, 2000). In some cases, resistance gene inactivation and diversification have been caused by transposable elements. For instance, an insertion in maize Hm1 was responsible for the breakdown of fungal resistance in many commercial breeding populations (Multani et al., 1998). Insertion of the transposable elements Retrofit and Truncator resulted in truncated open reading frames of rice Xa21 gene family members (Song et al., 1997). It is not known whether the retrotransposons or other transposable elements in the vicinity of the B73 rp1 cluster have any effect on Rp1 gene function or evolution.
The regions sequenced are unusual in their deficiency of miniature inverted repeat transposable elements (Bureau and Wessler, 1994). These elements are abundant throughout plant genomes, especially in genic areas (Wessler et al., 1995; Tikhonov et al., 1999; Jiang and Wessler, 2001). It is not clear why there are only two miniature inverted repeat transposable elements in this region compared with the average of three or more per gene that we observed in other regions of the maize genome (Tikhonov et al., 1999). The Zm206C17/Zm238E11 BAC contig and BAC Zm163K15 exhibit retrotransposon contents of 70 and 45%, respectively, similar to the retrotransposon contents reported in the maize adh1 region (74%) (Tikhonov et al., 1999) and for the entire maize genome (50 to 75%) (SanMiguel and Bennetzen, 1998; Meyers et al., 2001). Hence, the meiotic instability of the Rp1 region does not seem to have removed most repetitive DNAs. Future studies can use retrotransposons as tools to date the history of changes in different rp1 haplotypes and also can investigate the degree to which these abundant elements may have altered Rp1 region structure and gene function.
METHODS
BAC Selection, Restriction Map, and Sequencing
An Rp1 5′ probe, as described by Sun et al. (2001) (kindly provided by Scot Hulbert, Kansas State University, Manhattan, KS), was used to screen a maize (Zea mays) B73 BAC library (Genome Systems, St. Louis, MO). Restriction maps of BACs Zm163K15, Zm206C17, and Zm238E11 were constructed to experimentally validate sequence assembly. BACs were digested with the restriction enzymes AscI, NotI, PacI, PmeI, and SwaI. Restriction fragments were separated by field inversion gel electrophoresis on 1% agarose gels, transferred to nylon membranes, and hybridized with the Rp1 5′ probe.
Shotgun libraries for BACs Zm163K15, Zm206C17, and Zm238E11, sequencing, and analysis were as described by Dubcovsky et al. (2001). For finishing the sequences of the BACs, gaps were closed by a combination of different approaches, including the use of different sequence chemistries, use of the thermofidelase enzyme, PCR amplification of gaps, shotgun sequencing of transposon-inserted subclones that flank a gap, and direct sequencing of BAC template. When gaps were caused by repetitive regions, subclones that either started or ended in unique regions with the remaining portion in the repetitive region were assembled separately and inserted into the main assembly.
Construction of a Physical Map
A large insert B73 maize BAC library (www.chori.org/bacpac) was screened with the Rp1 5′ probe. A total of 33 positive BACs were identified, and these clones were mapped using different combinations of MluI, NcoI, NotI, and SwaI. Fragments were size-fractionated by field inversion gel electrophoresis. The DNAs in the gels were transferred to nylon membranes and hybridized with the Rp1 5′ probe and a probe that contained a truncated Suc phosphate synthase–like gene identified in the region by sequence analysis. Fragments observed on the agarose gel were compared between BACs to identify common fragments. This map also was compared with the maize genome physical map that is in preparation at the Arizona Genomics Institute (www.genome.arizona.edu/fpc/maize/).
Sequence Analysis
Annotation and sequence analysis were performed as described previously (Dubcovsky et al., 2001). FGENESH (http://www.softberry.com/berry.phtml) with the monocot training set was used for gene prediction, in addition to GENSCAN (http://genes.mit.edu/GENSCAN.html) and GeneMark.hmm (http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi). The criteria used to define a gene were as follows: (1) a match to a sequence in a protein database using BLASTX (Altschul et al., 1997); (2) a match to ESTs or cDNAs; or (3) prediction as a gene by two or more gene prediction programs. These criteria were used after excluding identified transposons.
All of the Rp1 homologs and retrotransposons were aligned using CLUSTAL X (Thompson et al., 1997). Rates of nucleotide substitution were estimated using the distance measures of Nei and Gojobori (1986) and the Jukes-Cantor correction as implemented in the MEGA2 (Molecular Evolutionary Genetic Analysis) package (Kumar et al., 2001). Synonymous substitution rates were estimated as implemented in MEGA2. Divergence times (T) were estimated for retrotransposons using k = K/2T, where k is the absolute rate of synonymous substitution per site per year for the adh1 and adh2 genes in grasses and K is the estimated number of substitutions per site between sequences using the Kimura two-parameter method (Kimura, 1980). A 2 × 2 contingency table G test was used to test for the significance of differences in synonymous and nonsynonymous substitution rates (Zhang et al., 1997).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences reported in this article are AF466931 (Zm163K15) and AF466932 (Zm206C17/Zm238E11).
Acknowledgments
This work was funded by the National Science Foundation Plant Genome Program (Grant 9975618).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006338.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 211–215. [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2000). Comparative sequence analysis of plant nuclear genomes: Microcolinearity and its many exceptions. Plant Cell 12, 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L., and Hulbert, S.H. (1992). Organization, instability and evolution of plant-disease resistance genes. Plant Mol. Biol. 20, 575–578. [PubMed] [Google Scholar]
- Bennetzen, J.L., Qin, M.M., Ingels, S., and Ellingboe, A.H. (1988). Allele-specific and mutator-associated instability at the rp1 disease-resistance locus of maize. Nature 332, 369–370. [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1994). Mobile inverted-repeat elements of the tourist family are associated with the genes of many cereal grasses. Proc. Natl. Acad. Sci. USA 91, 1411–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, D.B., Arroyo-Garcia, R., Ochoa, O.E., Kesseli, R.V., Lavelle, D.O., and Michelmore, R.W. (2001). Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157, 831–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N., Drake, J., Ayliffe, M., Sun, Q., Ellis, J., Hulbert, S., and Pryor, T. (1999). Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001). Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27, 439–453. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J., Ramakrishna, W., SanMiguel, P., Busso, C., Yan, L., Shiloff, B., and Bennetzen, J. (2001). Comparative sequence analysis of colinear barley and rice BACs. Plant Physiol. 125, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000. a). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3, 278–284. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000. b). The generation of plant disease resistance gene specificities. Trends Plant Sci. 5, 373–379. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Ayliffe, M., Anderson, P., Collins, N., Finnegan, J., Frost, D., Luck, J., and Pryor, T. (1997). Advances in the molecular genetic analysis of the flax–flax rust interaction. Annu. Rev. Phytopathol. 35, 271–291. [DOI] [PubMed] [Google Scholar]
- Emanuel, B.S., and Shaikh, T.H. (2001). Segmental duplications: An ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2, 791–800. [DOI] [PubMed] [Google Scholar]
- Farris, J.D., Haen, K., and Gill, B.S. (2000). Saturation mapping of a gene-rich recombinant hot spot region in wheat. Genetics 154, 823–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H.H., Park, W.K., Yan, X.H., Zheng, Z.W., Shen, B.Z., and Dooner, H.K. (2001). The highly recombinogenic bz locus lies in an unusually gene-rich region of the maize genome. Proc. Natl. Acad. Sci. USA 98, 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K.S., Gill, B.S., Endo, T.R., and Taylor, T. (1996). Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub, E.B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. [DOI] [PubMed] [Google Scholar]
- Hong, K.S., Richter, T.E., Bennetzen, J.L., and Hulbert, S.H. (1993). Complex duplications in maize lines. Mol. Gen. Genet. 239, 115–121. [DOI] [PubMed] [Google Scholar]
- Hulbert, S.H., and Bennetzen, J.L. (1991). Recombination at the rp1 locus of maize. Mol. Gen. Genet. 226, 377–382. [DOI] [PubMed] [Google Scholar]
- Jiang, J.M., Hulbert, S.H., Gill, B.S., and Ward, D.C. (1996). Interphase fluorescence in situ hybridization mapping: A physical mapping strategy for plant species with large complex genomes. Mol. Gen. Genet. 252, 497–502. [DOI] [PubMed] [Google Scholar]
- Jiang, N., and Wessler, S.R. (2001). Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 13, 2553–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Kirik, A., Salomon, S., and Puchta, H. (2000). Species-specific double-strand break repair and genome evolution in plants. EMBO J. 19, 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Frary, A., Wu, T.Y., Brommonschenkel, S., Chunwongse, J., Earle, E.D., and Tanksley, S.D. (1994). A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell-death. Plant Cell 6, 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Chin, D.B., Shen, K.A., Sivaramakrishnan, S., Lavelle, D.O., Zhang, Z., and Michelmore, R.W. (1998. a). The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998. b). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Moore, J.K., and Haber, J.E. (1996). Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature 383, 644–646. [DOI] [PubMed] [Google Scholar]
- Multani, D.S., Meeley, R.B., Paterson, A.H., Gray, J., Briggs, S.P., and Johal, G.S. (1998). Plant-pathogen microevolution: Molecular basis for the origin of a fungal disease in maize. Proc. Natl. Acad. Sci. USA 95, 1686–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B.H., and Jones, J.D.G. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Parniske, M., and Jones, J.D.G. (1999). Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proutski, V., and Holmes, E.C. (1998). SWAN: Sliding window analysis of nucleotide sequence variability. Bioinformatics 14, 467–468. [DOI] [PubMed] [Google Scholar]
- Ralston, E.J., English, J.J., and Dooner, H.K. (1988). Sequence of 3 bronze alleles of maize and correlation with the genetic fine-structure. Genetics 119, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, T.E., Pryor, T.J., Bennetzen, J.L., and Hulbert, S.H. (1995). New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics 141, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, T.E., and Ronald, P.C. (2000). The evolution of disease resistance genes. Plant Mol. Biol. 42, 195–204. [PubMed] [Google Scholar]
- Salomon, S., and Puchta, H. (1998). Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17, 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Ann. Bot. 82, 37–44. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- Saxena, K.M.S., and Hooker, A.L. (1968). On the structure of a gene for disease resistance in maize. Proc. Natl. Acad. Sci. USA 61, 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, G., et al. (1998). Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 10, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.-Y., Pi, L.-Y., Wang, G.-L., Gardner, J., Holsten, T., and Ronald, P.C. (1997). Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J. (2001). Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiol. 125, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz, B.J., Mudgett, M.B., Dangl, J.L., and Galan, J.E. (2001). Common and contrasting themes of plant and animal diseases. Science 292, 2285–2289. [DOI] [PubMed] [Google Scholar]
- Sudupak, M.A., Bennetzen, J.L., and Hulbert, S.H. (1993). Unequal exchange and meiotic instability of disease-resistance genes in the rp1 region of maize. Genetics 133, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q., Collins, N.C., Ayliffe, M., Smith, S.M., Drake, J., Pryor, T., and Hulbert, S.H. (2001). Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S.C., Kim, B., and Gabriel, A. (1996). Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature 383, 641–644. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., SanMiguel, P.J., Nakajima, Y., Gorenstein, N.D., Bennetzen, J.L., and Avramova, Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcich, M.P., and Mascarenhas, J.P. (1994). PREM-1, a putative maize retroelement, has LTR (long terminal repeat) sequences that are preferentially transcribed in pollen. Sex. Plant Reprod. 7, 2–11. [Google Scholar]
- Walker, E.L., Robbins, T.P., Bureau, T.E., Kermicle, J., and Dellaporta, S.L. (1995). Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize r-r complex. EMBO J. 14, 2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S., Tarpley, A., Purugganan, M., Spell, M., and Okagaki, R. (1990). Filler DNA is associated with spontaneous deletions in maize. Proc. Natl. Acad. Sci. USA 87, 8731–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler, S.R., Bureau, T.E., and White, S.E. (1995). LTR-retrotransposons and MITEs: Important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5, 814–821. [DOI] [PubMed] [Google Scholar]
- Yan, X.H., Martinez-Ferez, I.M., Kavchok, S., and Dooner, H.K. (1999). Origination of Ds elements from Ac elements in maize: Evidence for rare repair synthesis at the site of Ac excision. Genetics 152, 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Kumar, S., and Nei, M. (1997). Small-sample tests of episodic adaptive evolution: A case study of primate lysozymes. Mol. Biol. Evol. 14, 1335–1338. [DOI] [PubMed] [Google Scholar]